Trypsin Genes Are Regulated through the miRNA Bantam and Associated with Drug Sensitivity in the Sea Louse Caligus rogercresseyi
Abstract
1. Introduction
2. Results
2.1. Identification of Trypsin Genes and Bantam miRNAs in the C. rogercresseyi Genome
2.2. Expression Analyses of Bantam and Trypsin Genes
2.3. Expression Value Correlations
2.4. Expression Validation through qPCR
2.5. Validation of Trypsin Binding Site for Bantam through Luciferase Assay
3. Discussion
4. Materials and Methods
4.1. Identification of Trypsin Genes in Sea Louse Genome
4.2. Identification of Bantam miRNAs in Sea Louse Genome
4.3. Expression Analyses of Trypsin and Bantam Genes
4.4. Gene Expression Validation by qPCR
4.5. Trypsin–Bantam Interaction Analyses through Co-Expression and Target Prediction Analyses
4.6. Target Prediction Validation through Luciferase Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gallardo-Escárate, C.; Arriagada, G.; Carrera, C.; Gonçalves, A.T.; Nuñez-Acuña, G.; Valenzuela-Miranda, D.; Valenzuela-Muñoz, V. The race between host and sea lice in the Chilean salmon farming: A genomic approach. Rev. Aquac. 2019, 11, 325–339. [Google Scholar] [CrossRef]
- Pike, A. Sea lice—Major pathogens of farmed Atlantic salmon. Parasitol. Today 1989, 5, 291–297. [Google Scholar] [CrossRef]
- Costello, M.J. How sea lice from salmon farms may cause wild salmonid declines in Europe and North America and be a threat to fishes elsewhere. Proc. R. Soc. B: Boil. Sci. 2009, 276, 3385–3394. [Google Scholar] [CrossRef]
- Costello, M.J. The global economic cost of sea lice to the salmonid farming industry. J. Fish Dis. 2009, 32, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Dresdner, J.; Chávez, C.; Quiroga, M.; Jiménez, D.; Artacho, P.; Tello, A. Impact of Caligus treatments on unit costs of heterogeneous salmon farms in Chile. Aquac. Econ. Manag. 2019, 23, 1–27. [Google Scholar] [CrossRef]
- Roth, M.; Richards, R.H.; Sommerville, C. Current practices in the chemotherapeutic control of sea lice infestations in aquaculture: A review. J. Fish Dis. 1993, 16, 1–26. [Google Scholar] [CrossRef]
- Liu, Y.; Bjelland, H. Estimating costs of sea lice control strategy in Norway. Prev. Vet.-Med. 2014, 117, 469–477. [Google Scholar] [CrossRef]
- Aaen, S.M.; Helgesen, K.O.; Bakke, M.J.; Kaur, K.; Horsberg, T.E. Drug resistance in sea lice: A threat to salmonid aquaculture. Trends Parasitol. 2015, 31, 72–81. [Google Scholar] [CrossRef]
- Marín, S.; Ibarra, R.; Medina, M.; Jansen, P. Sensitivity of Caligus rogercresseyi (Boxshall and Bravo 2000) to pyrethroids and azamethiphos measured using bioassay tests—A large scale spatial study. Prev. Vet.-Med. 2015, 122, 33–41. [Google Scholar] [CrossRef]
- Arriagada, G.; Figueroa, J.; Marín, S.L.; Arriagada, A.M.; Lara, M.; Gallardo-Escárate, C. First report of the reduction in treatment efficacy of the organophosphate azamethiphos against the sea lice Caligus rogercresseyi (Boxshall & Bravo, 2000). Aquac. Res. 2019, 51, 436–439. [Google Scholar] [CrossRef]
- Núñez-Acuña, G.; Sáez-Vera, C.; Valenzuela-Muñoz, V.; Valenzuela-Miranda, D.; Arriagada, G.; Gallardo-Escárate, C. Tackling the Molecular Drug Sensitivity in the Sea Louse Caligus rogercresseyi Based on mRNA and lncRNA Interactions. Genes 2020, 11, 857. [Google Scholar] [CrossRef]
- Bass, C.; Field, L. Gene amplification and insecticide resistance. Pest Manag. Sci. 2011, 67, 886–890. [Google Scholar] [CrossRef]
- Colmegna, B.; Morosi, L.; D’Incalci, M. Molecular and pharmacological mechanisms of drug resistance: An evolving paradigm. In Mechanisms of Drug Resistance in Cancer Therapy; Springer: Berlin/Heidelberg, Germany, 2017; pp. 1–12. [Google Scholar]
- Faucon, F.; Gaude, T.; Dusfour, I.; Navratil, V.; Corbel, V.; Juntarajumnong, W.; Girod, R.; Poupardin, R.; Boyer, F.; Reynaud, S.; et al. In the hunt for genomic markers of metabolic resistance to pyrethroids in the mosquito Aedes aegypti: An integrated next-generation sequencing approach. PLoS Negl. Trop. Dis. 2017, 11, e0005526. [Google Scholar] [CrossRef] [PubMed]
- Kaur, K.; Jansen, P.A.; Aspehaug, V.T.; Horsberg, T.E. Phe362Tyr in AChE: A Major Factor Responsible for Azamethiphos Resistance in Lepeophtheirus salmonis in Norway. PLoS ONE 2016, 11, e0149264. [Google Scholar] [CrossRef] [PubMed]
- Fjørtoft, H.B.; Nilsen, F.; Besnier, F.; Stene, A.; Tveten, A.-K.; Bjørn, P.A.; Aspehaug, V.T.; Glover, K.A. Losing the ‘arms race’: Multiresistant salmon lice are dispersed throughout the North Atlantic Ocean. R. Soc. Open Sci. 2021, 8. [Google Scholar] [CrossRef]
- Valenzuela-Muñoz, V.; Chavez-Mardones, J.; Gallardo-Escárate, C. RNA-seq analysis evidences multiple gene responses in Caligus rogercresseyi exposed to the anti-salmon lice drug azamethiphos. Aquaculture 2015, 446, 156–166. [Google Scholar] [CrossRef]
- Valenzuela-Muñoz, V.; Sturm, A.; Gallardo-Escárate, C. Transcriptomic insights on the ABC transporter gene family in the salmon louse Caligus rogercresseyi. Parasites Vectors 2015, 8, 209. [Google Scholar] [CrossRef]
- Valenzuela-Miranda, D.; Gallardo-Escárate, C. Caligus rogercresseyi serine proteases: Transcriptomic analysis in response to delousing drugs treatments. Aquaculture 2016, 465, 65–77. [Google Scholar] [CrossRef]
- Fast, M.D.; Johnson, S.C.; Eddy, T.D.; Pinto, D.; Ross, N.W. Lepeophtheirus salmonis secretory/excretory products and their effects on Atlantic salmon immune gene regulation. Parasite Immunol. 2007, 29, 179–189. [Google Scholar] [CrossRef]
- Lehiy, C.J.; Drolet, B.S. The salivary secretome of the biting midge, Culicoides sonorensis. PeerJ 2014, 2, e426. [Google Scholar] [CrossRef]
- Haenisch, S.; Cascorbi, I. miRNAs as mediators of drug resistance. Epigenomics 2012, 4, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Etebari, K.; Afrad, M.H.; Tang, B.; Silva, R.; Furlong, M.J.; Asgari, S. Involvement of microRNA miR-2b-3p in regulation of metabolic resistance to insecticides inPlutella xylostella. Insect Mol. Biol. 2018, 27, 478–491. [Google Scholar] [CrossRef]
- Hong, S.; Guo, Q.; Wang, W.; Hu, S.; Fang, F.; Lv, Y.; Yu, J.; Zou, F.; Lei, Z.; Ma, K.; et al. Identification of differentially expressed microRNAs in Culex pipiens and their potential roles in pyrethroid resistance. Insect Biochem. Mol. Biol. 2014, 55, 39–50. [Google Scholar] [CrossRef]
- Lei, Z.; Lv, Y.; Wang, W.; Guo, Q.; Zou, F.; Hu, S.; Fang, F.; Tian, M.; Liu, B.; Liu, X.; et al. MiR-278-3p regulates pyrethroid resistance in Culex pipiens pallens. Parasitol. Res. 2014, 114, 699–706. [Google Scholar] [CrossRef]
- Gallardo-Escárate, C.; Valenzuela-Muñoz, V.; Boltaña, S.; Nuñez-Acuña, G.; Valenzuela-Miranda, D.; Gonçalves, A.; Détrée, C.; Tarifeño-Saldivia, E.; Farlora, R.; Roberts, S.; et al. The Caligus rogercresseyi miRNome: Discovery and transcriptome profiling during the sea lice ontogeny. Agri Gene 2017, 4, 8–22. [Google Scholar] [CrossRef]
- Núñez-Acuña, G.; Valenzuela-Muñoz, V.; Valenzuela-Miranda, D.; Gallardo-Escárate, C. Comprehensive Transcriptome Analyses in Sea Louse Reveal Novel Delousing Drug Responses Through MicroRNA regulation. Mar. Biotechnol. 2021, 23, 710–723. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela-Miranda, D.; Etebari, K.; Asgari, S.; Gallardo-Escárate, C. Long noncoding RNAs: Unexplored players in the drug response of the sea louse Caligus rogercresseyi. Agri Gene 2017, 4, 1–7. [Google Scholar] [CrossRef][Green Version]
- Lomate, P.R.; Mahajan, N.S.; Kale, S.M.; Gupta, V.S.; Giri, A.P. Identification and expression profiling of Helicoverpa armigera microRNAs and their possible role in the regulation of digestive protease genes. Insect Biochem. Mol. Biol. 2014, 54, 129–137. [Google Scholar] [CrossRef]
- Agusti-Ridaura, C.; Dondrup, M.; Horsberg, T.E.; Leong, J.; Koop, B.F.; Bravo, S.; Mendoza, J.; Kaur, K. Caligus rogercresseyi acetylcholinesterase types and variants: A potential marker for organophosphate resistance. Parasites Vectors 2018, 11, 570. [Google Scholar] [CrossRef]
- Gallardo-Escárate, C.; Valenzuela-Muñoz, V.; Nuñez-Acuña, G.; Valenzuela-Miranda, D.; Gonçalves, A.T.; Escobar-Sepulveda, H.; Liachko, I.; Nelson, B.; Roberts, S.; Warren, W. Chromosome-scale genome assembly of the sea louse Caligus rogercresseyi by SMRT sequencing and Hi-C analysis. Sci. Data 2021, 8, 60. [Google Scholar] [CrossRef] [PubMed]
- Poley, J.D.; Braden, L.M.; Messmer, A.M.; Igboeli, O.O.; Whyte, S.K.; Macdonald, A.; Rodriguez, J.; Gameiro, M.; Rufener, L.; Bouvier, J.; et al. High level efficacy of lufenuron against sea lice (Lepeophtheirus salmonis) linked to rapid impact on moulting processes. Int. J. Parasitol. Drugs Drug Resist. 2018, 8, 174–188. [Google Scholar] [CrossRef]
- Schröder, A.; Klein, K.; Winter, S.; Schwab, M.; Bonin, M.; Zell, A.; Zanger, U.M. Genomics of ADME gene expression: Mapping expression quantitative trait loci relevant for absorption, distribution, metabolism and excretion of drugs in human liver. Pharm. J. 2011, 13, 12–20. [Google Scholar] [CrossRef]
- Dorrah, M.; Elmelegi, M.; Bassal, T. Trypsin-and chymotrypsin esterase-like activity in the midgut regions of the larval Parasarcophaga hertipes (Sarcophagidae, Cyclorrhapha, Diptera). Efflatoun’s J. Entomol. 2008, 8, 1–8. [Google Scholar]
- Xiong, C.; Fang, F.; Chen, L.; Yang, Q.; He, J.; Zhou, D.; Shen, B.; Ma, L.; Sun, Y.; Zhang, D.; et al. Trypsin-Catalyzed Deltamethrin Degradation. PLoS ONE 2014, 9, e89517. [Google Scholar] [CrossRef] [PubMed]
- Sáez-Vera, C.; Núñez-Acuña, G.; Gallardo-Escárate, C. Sensitivity assessment to azamethiphos by time-to-response bioassay and biomarkers in the sea louse Caligus rogercresseyi. Aquaculture 2021, 546, 737340. [Google Scholar] [CrossRef]
- Rungruangsak-Torrissen, K.; Moss, R.; Andresen, L.H.; Berg, A.; Waagbo, R. Different expressions of trypsin and chymotrypsin in relation to growth in Atlantic salmon (Salmo salar L.). Fish Physiol. Biochem. 2006, 32, 7–23. [Google Scholar] [CrossRef]
- Hipfner, D.R.; Weigmann, K.; Cohen, S.M. The bantam Gene Regulates Drosophila Growth. Genetics 2002, 161, 1527–1537. [Google Scholar] [CrossRef]
- Banerjee, A.; Roy, J.K. Dicer-1 regulates proliferative potential of Drosophila larval neural stem cells through bantam miRNA based down-regulation of the G1/S inhibitor Dacapo. Dev. Biol. 2017, 423, 57–65. [Google Scholar] [CrossRef]
- Huang, H.; Li, J.; Hu, L.; Ge, L.; Ji, H.; Zhao, Y.; Zhang, L. Bantam is essential for Drosophila intestinal stem cell proliferation in response to Hippo signaling. Dev. Biol. 2014, 385, 211–219. [Google Scholar] [CrossRef]
- Brennecke, J.; Hipfner, D.R.; Stark, A.; Russell, R.B.; Cohen, S.M. Bantam Encodes a Developmentally Regulated microRNA that Controls Cell Proliferation and Regulates the Proapoptotic Gene hid in Drosophila. Cell 2003, 113, 25–36. [Google Scholar] [CrossRef]
- Nolo, R.; Morrison, C.M.; Tao, C.; Zhang, X.; Halder, G. The bantam MicroRNA Is a Target of the Hippo Tumor-Suppressor Pathway. Curr. Biol. 2006, 16, 1895–1904. [Google Scholar] [CrossRef] [PubMed]
- Herranz, H.; Hong, X.; Cohen, S.M. Mutual Repression by Bantam miRNA and Capicua Links the EGFR/MAPK and Hippo Pathways in Growth Control. Curr. Biol. 2012, 22, 651–657. [Google Scholar] [CrossRef]
- Núñez-Acuña, G.; Gallardo-Escárate, C. Characterization of the salmon louse Lepeophtheirus salmonis miRNome: Sex-biased differences related to the coding and non-coding RNA interplay. Mar. Genom. 2019, 45, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Jayachandran, B.; Hussain, M.; Asgari, S. An insect trypsin-like serine protease as a target of microRNA: Utilization of microRNA mimics and inhibitors by oral feeding. Insect Biochem. Mol. Biol. 2013, 43, 398–406. [Google Scholar] [CrossRef]
- Katsuda, Y. Progress and Future of Pyrethroids. Top. Curr. Chem. 2011, 314, 1–30. [Google Scholar] [CrossRef]
- Jin, Y.; Chen, Z.; Liu, X.; Zhou, X. Evaluating the microRNA targeting sites by luciferase reporter gene assay. In MicroRNA Protocols; Springer: Berlin/Heidelberg, Germany, 2013; pp. 117–127. [Google Scholar]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Wei, T.; Simko, V.; Levy, M.; Xie, Y.; Jin, Y.; Zemla, J. Package ‘corrplot’. Statistician 2017, 56, 316–324. [Google Scholar]
- Miranda, K.C.; Huynh, T.; Tay, Y.; Ang, Y.-S.; Tam, W.L.; Thomson, A.M.; Lim, B.; Rigoutsos, I. A Pattern-Based Method for the Identification of MicroRNA Binding Sites and Their Corresponding Heteroduplexes. Cell 2006, 126, 1203–1217. [Google Scholar] [CrossRef]
- Rehmsmeier, M.; Steffen, P.; Höchsmann, M.; Giegerich, R. Fast and effective prediction of microRNA/target duplexes. RNA 2004, 10, 1507–1517. [Google Scholar] [CrossRef] [PubMed]
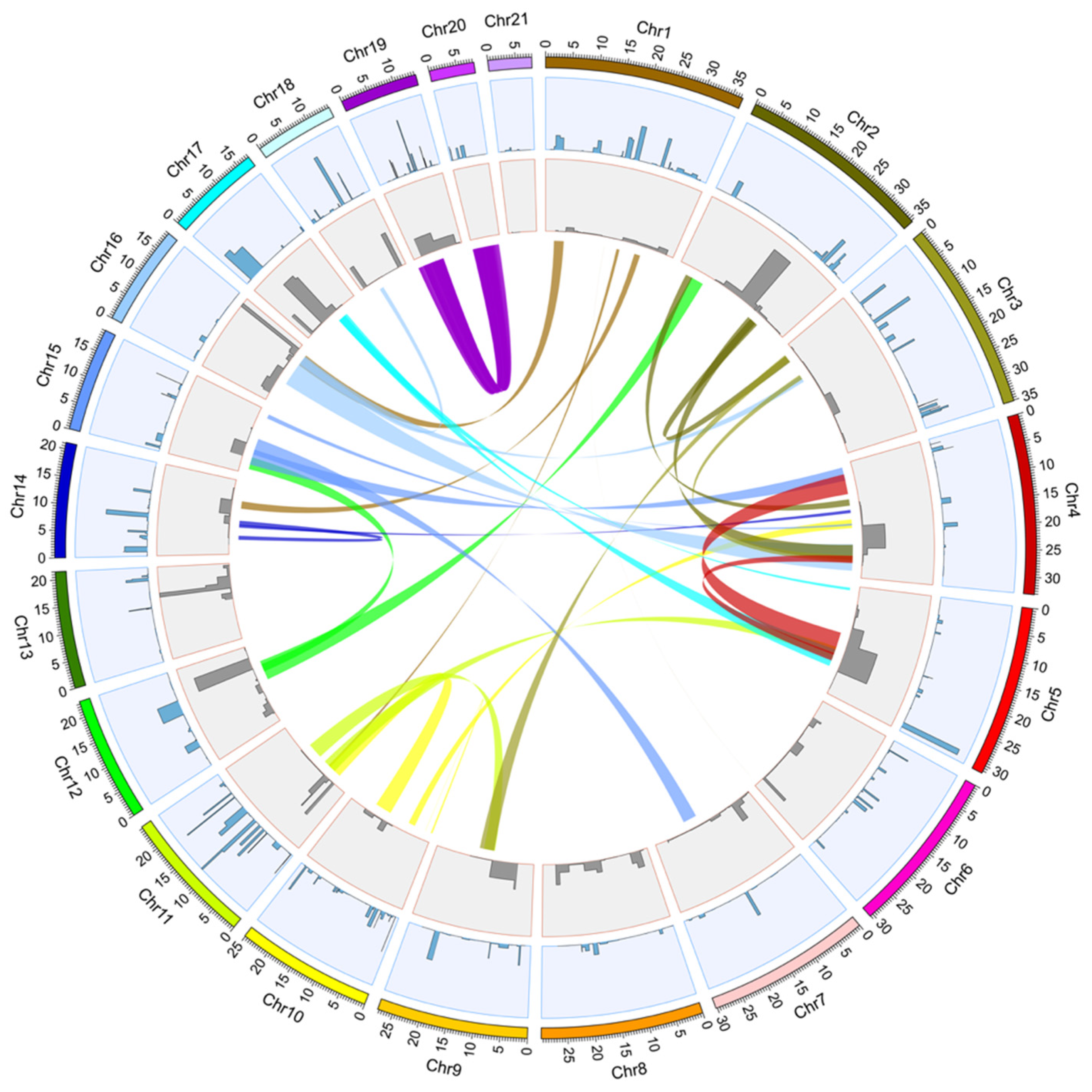
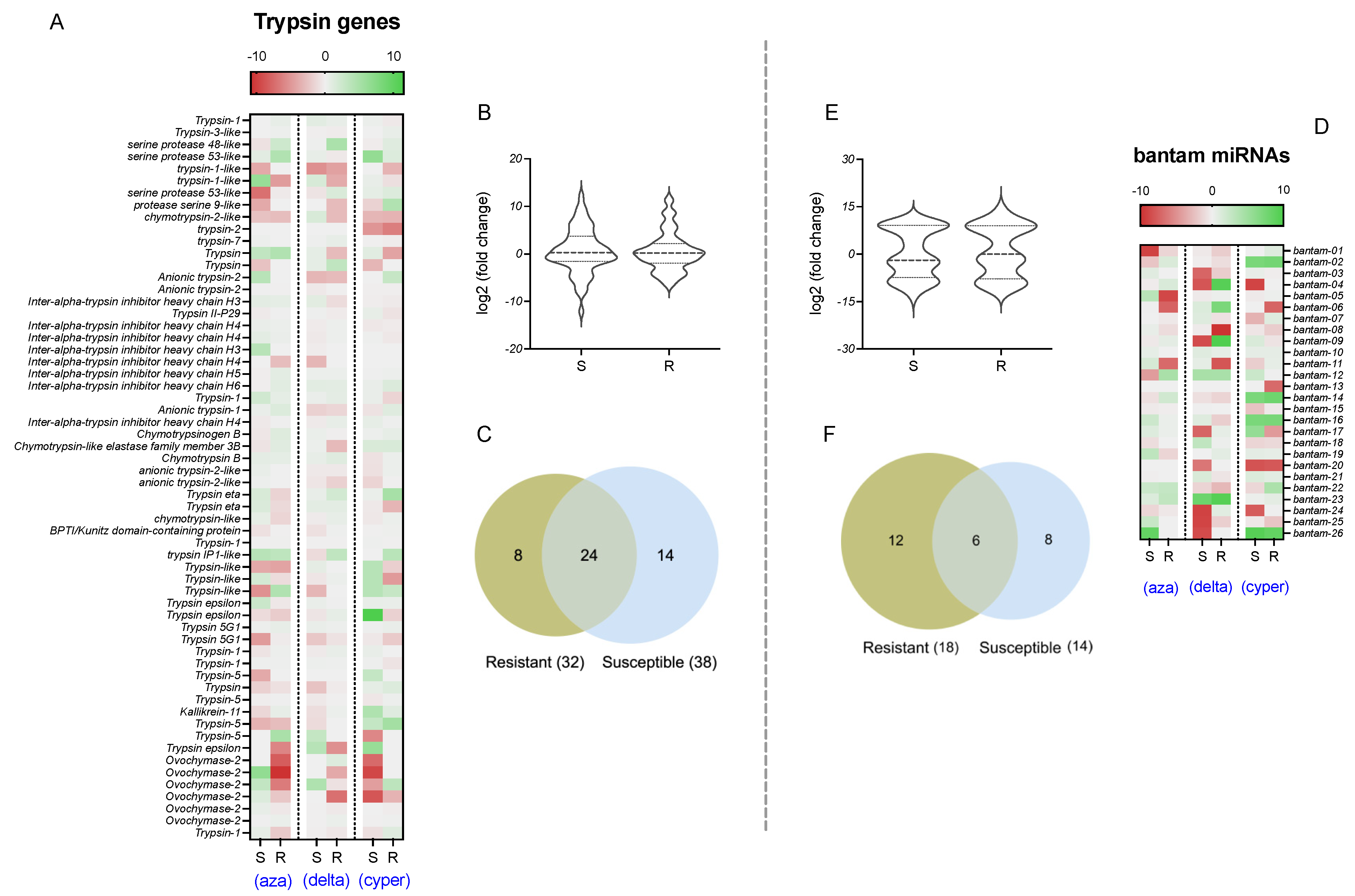
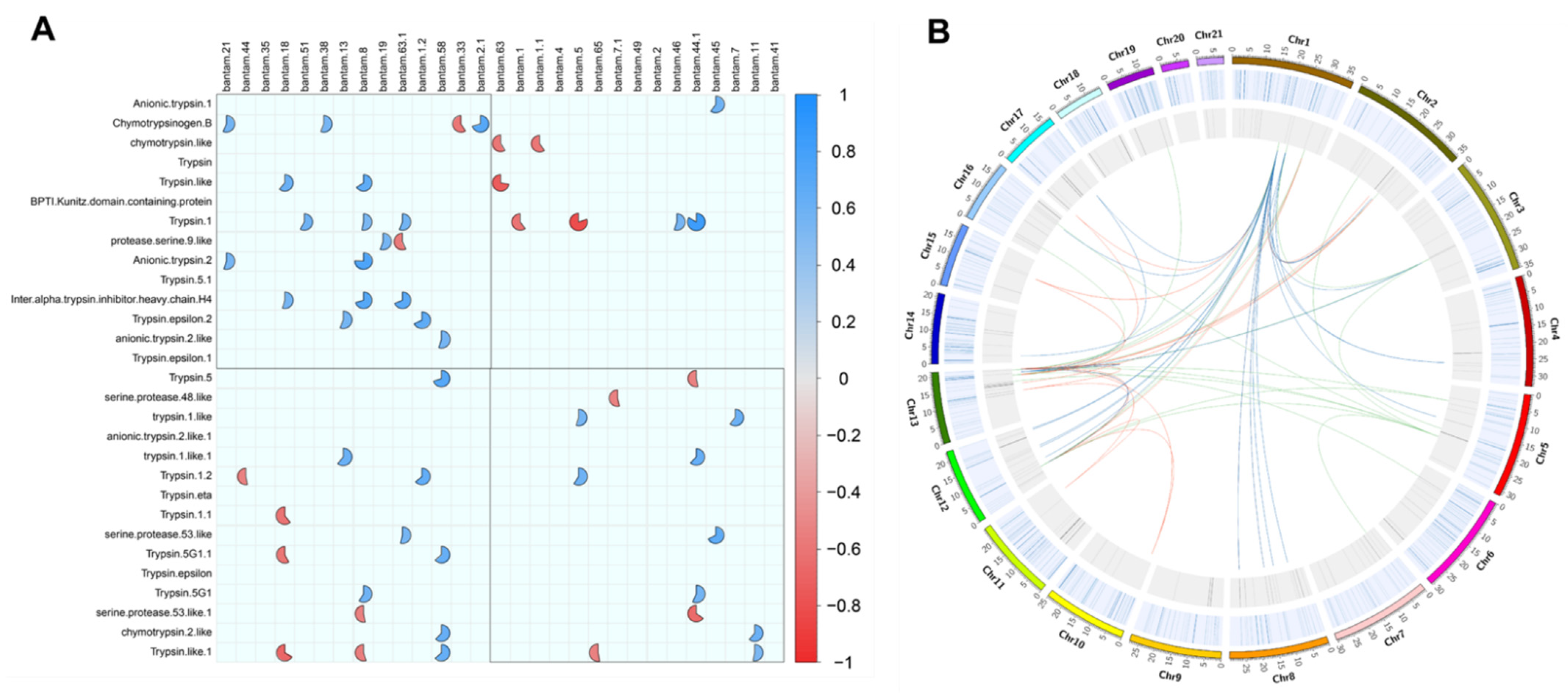


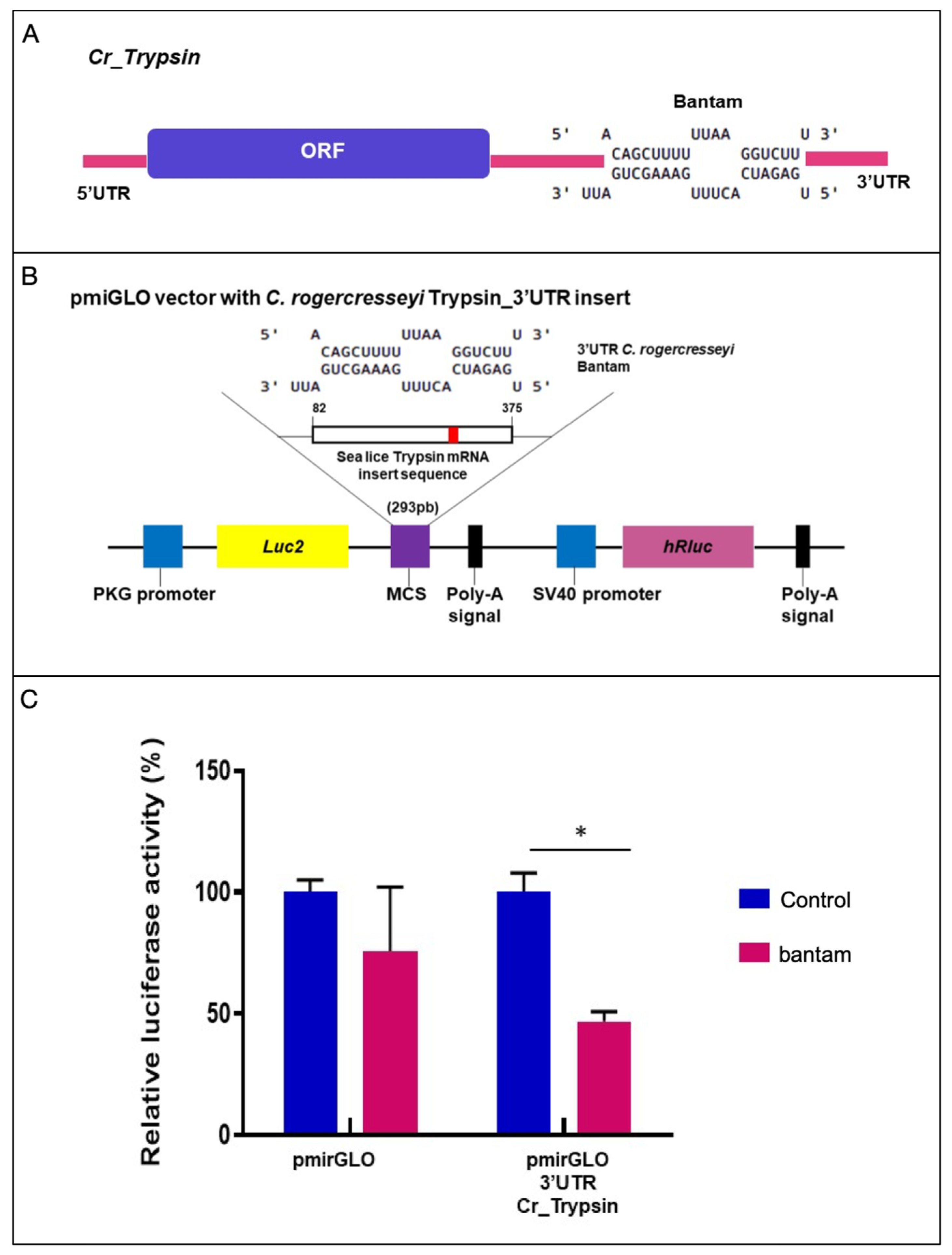
| Gene Name | Chromosome | Length (bp) | Strand | Transcripts | Transcript Length |
|---|---|---|---|---|---|
| Trypsin-1 | Chr2 | 5343 | − | 1 | 871 |
| Trypsin-3-like | Chr5 | 5134 | − | 1 | 1330 |
| Serine protease 48-like | Chr5 | 4377 | + | 7 | 508 |
| Serine protease 53-like | Chr5 | 4377 | + | 7 | 508 |
| Trypsin-1-like | Chr5 | 4377 | + | 7 | 508 |
| Trypsin-1-like | Chr5 | 4377 | + | 7 | 508 |
| Serine protease 53-like | Chr5 | 4377 | + | 7 | 508 |
| Protease serine 9-like | Chr5 | 4377 | + | 7 | 508 |
| Chymotrypsin-2-like | Chr5 | 4377 | + | 7 | 508 |
| Trypsin-2 | Chr6 | 7501 | + | 2 | 862 |
| Trypsin-7 | Chr6 | 7501 | + | 2 | 862 |
| Trypsin | Chr6 | 1885 | − | 2 | 559 |
| Trypsin | Chr6 | 1885 | − | 2 | 559 |
| Anionic trypsin-2 | Chr10 | 365 | ? | 1 | 365 |
| Anionic trypsin-2 | Chr10 | 1397 | + | 1 | 1268 |
| Inter-alpha-trypsin H3 | Chr15 | 8195 | + | 2 | 2453 |
| Trypsin II-P29 | Chr18 | 5255 | + | 1 | 1290 |
| Inter-alpha-trypsin H4 | Chr19 | 4865 | + | 2 | 4522 |
| Inter-alpha-trypsin H4 | Chr19 | 4865 | + | 2 | 4522 |
| Inter-alpha-trypsin H3 | Chr19 | 329 | − | 4 | 240 |
| Inter-alpha-trypsin H4 | Chr19 | 329 | − | 4 | 240 |
| Inter-alpha-trypsin H5 | Chr19 | 329 | − | 4 | 240 |
| Inter-alpha-trypsin H6 | Chr19 | 329 | − | 4 | 240 |
| Trypsin-1 | Chr1 | 1208 | + | 1 | 1018 |
| Anionic trypsin-1 | Chr1 | 831 | − | 1 | 703 |
| Inter-alpha-trypsin H4 | Chr3 | 3430 | ? | 1 | 3430 |
| Chymotrypsinogen B | Chr5 | 334 | ? | 1 | 334 |
| Chymotrypsin-like Elastase | Chr5 | 315 | ? | 1 | 315 |
| Chymotrypsin B | Chr5 | 448 | + | 1 | 386 |
| Anionic trypsin-2-like | Chr5 | 614 | + | 1 | 545 |
| Anionic trypsin-2-like | Chr5 | 496 | − | 1 | 427 |
| Trypsin eta | Chr5 | 807 | ? | 2 | 593 |
| Trypsin eta | Chr5 | 807 | ? | 2 | 593 |
| Chymotrypsin-like | Chr10 | 7596 | − | 1 | 985 |
| BPTI/Kunitz protein | Chr10 | 445 | + | 1 | 385 |
| Trypsin-1 | Chr12 | 646 | ? | 1 | 646 |
| trypsin IP1-like | Chr12 | 668 | ? | 1 | 668 |
| Trypsin-like | Chr13 | 6442 | + | 1 | 840 |
| Trypsin-like | Chr13 | 2764 | − | 2 | 345 |
| Trypsin-like | Chr13 | 2764 | − | 2 | 345 |
| Trypsin epsilon | Chr13 | 540 | ? | 2 | 540 |
| Trypsin epsilon | Chr13 | 540 | ? | 2 | 540 |
| Trypsin 5G1 | Chr13 | 518 | + | 2 | 434 |
| Trypsin 5G1 | Chr13 | 518 | + | 2 | 434 |
| Trypsin-1 | Chr13 | 44817 | − | 2 | 833 |
| Trypsin-1 | Chr13 | 44817 | − | 2 | 833 |
| Trypsin-5 | Chr13 | 440 | ? | 1 | 440 |
| Trypsin | Chr13 | 1143 | ? | 1 | 1143 |
| Trypsin-5 | Chr13 | 521 | ? | 2 | 521 |
| Kallikrein-11 | Chr13 | 521 | ? | 2 | 521 |
| Trypsin-5 | Chr13 | 729 | ? | 1 | 729 |
| Trypsin-5 | Chr13 | 691 | ? | 1 | 591 |
| Trypsin epsilon | Chr13 | 560 | ? | 1 | 560 |
| Ovochymase-2-like | Chr15 | 9412 | − | 6 | 2904 |
| Ovochymase-2-like | Chr15 | 9412 | − | 6 | 2904 |
| Ovochymase-2-like | Chr15 | 9412 | − | 6 | 2904 |
| Ovochymase-2-like | Chr15 | 9412 | − | 6 | 2904 |
| Ovochymase-2-like | Chr15 | 9412 | − | 6 | 2904 |
| Ovochymase-2-like | Chr15 | 9412 | − | 6 | 2904 |
| Trypsin-1 | Chr18 | 1467 | + | 1 | 427 |
| Gene | Direction | Sequence (5′-3′) | R (5′) | Q (3′) |
|---|---|---|---|---|
| B-tubulin | Forward | GGCTTCCAGCAACTACAACCG | ||
| Reverse | TTTCGTAAATAGGGTACCCGT | |||
| Probe | ACATGGCACTTGGGCCATGCG | HEX | BHQ1 | |
| Trypsin 2 | Forward | CCAAGGTGACGGACCTTTGA | ||
| Reverse | AACGCTGTGACGAGGTTCAT | |||
| Probe | CCTAAAGGTTAGCCCCGAGG | FAM | BHQ1 | |
| Trypsin 5 | Forward | TCCATGGTCCCAAGGCAAGTT | ||
| Reverse | CATTCGTTTGACCAAGGACCT | |||
| Probe | CGCCATCAATCCTCCGCCGCA | TAMRA | BHQ2 | |
| Bantam | Undirected | TCAGCGTTCACAATGATCTCA | ||
| miR-2796 | Undirected | GTGTCCCAATTATAATGTCCA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Núñez-Acuña, G.; Valenzuela-Muñoz, V.; Carrera-Naipil, C.; Sáez-Vera, C.; Benavente, B.P.; Valenzuela-Miranda, D.; Gallardo-Escárate, C. Trypsin Genes Are Regulated through the miRNA Bantam and Associated with Drug Sensitivity in the Sea Louse Caligus rogercresseyi. Non-Coding RNA 2021, 7, 76. https://doi.org/10.3390/ncrna7040076
Núñez-Acuña G, Valenzuela-Muñoz V, Carrera-Naipil C, Sáez-Vera C, Benavente BP, Valenzuela-Miranda D, Gallardo-Escárate C. Trypsin Genes Are Regulated through the miRNA Bantam and Associated with Drug Sensitivity in the Sea Louse Caligus rogercresseyi. Non-Coding RNA. 2021; 7(4):76. https://doi.org/10.3390/ncrna7040076
Chicago/Turabian StyleNúñez-Acuña, Gustavo, Valentina Valenzuela-Muñoz, Crisleri Carrera-Naipil, Constanza Sáez-Vera, Bárbara P. Benavente, Diego Valenzuela-Miranda, and Cristian Gallardo-Escárate. 2021. "Trypsin Genes Are Regulated through the miRNA Bantam and Associated with Drug Sensitivity in the Sea Louse Caligus rogercresseyi" Non-Coding RNA 7, no. 4: 76. https://doi.org/10.3390/ncrna7040076
APA StyleNúñez-Acuña, G., Valenzuela-Muñoz, V., Carrera-Naipil, C., Sáez-Vera, C., Benavente, B. P., Valenzuela-Miranda, D., & Gallardo-Escárate, C. (2021). Trypsin Genes Are Regulated through the miRNA Bantam and Associated with Drug Sensitivity in the Sea Louse Caligus rogercresseyi. Non-Coding RNA, 7(4), 76. https://doi.org/10.3390/ncrna7040076







