Regulation of Neuroendocrine-like Differentiation in Prostate Cancer by Non-Coding RNAs
Abstract
1. Introduction
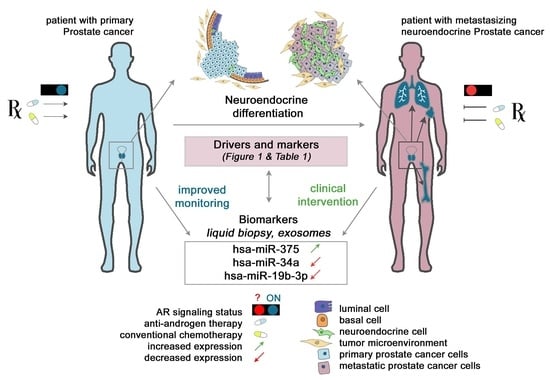
Neuroendocrine Prostate Cancer
2. Regulatory Circuits Driving Neuroendocrine Differentiation in Prostate Cancer
2.1. Signalling and Genetic Hallmarks of mCRPC Samples
2.2. ‘Lost and Found’ Protein Keys Unlocking Neuroendocrine Trans-Differentiation of Prostate Cancer
2.3. Rewiring, Remodelling, and Reshaping—A Vicious Program Turned on
3. miRNAs as Multifaceted Crossroads Driving Neuroendocrine Prostate Cancer Development
3.1. miRNA Biogenesis and Mechanisms of Action
3.2. miRNAs in the Regulation of NED and Prostate Cancer Progression
3.3. miRNAs Associated with Neuroendocrine Prostate Cancer
3.3.1. hsa-miR-194
3.3.2. hsa-miR-375
3.3.3. hsa-miR-301
3.3.4. hsa-miR-106a~363 Cluster
3.3.5. hsa-miR-106b, miR-93, and miR-25 Cluster
3.3.6. hsa-let-7
3.4. miRNAs Associated with Neuroendocrine-like Changes in Prostate Cancer Models
3.4.1. hsa-miR-17/92 Cluster
3.4.2. hsa-miR-663
3.4.3. hsa-miR-32
3.4.4. hsa-miR-204 and hsa-miR-34a
3.4.5. hsa-miR-221
3.5. Additional miRNAs Implicated in the Modulation of Key Positive and Negative Regulators of NEPC
3.5.1. miRNAs Implicated in the Regulation of AR
3.5.2. miRNAs Implicated in the Regulation of AKT and MYCN
3.5.3. miRNAs Implicated in the Regulation of MYCN
3.6. LncRNAs Implicated in NEPC
4. Clinical Significance of Non-Coding RNAs as Biomarkers and Therapeutic Targets in NEPC
5. Conclusions and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ADT | androgen-deprivation therapy |
| Ago-HITS-CLIP | high-throughput sequencing of RNA isolated by cross-linking immunoprecipitation of Argonaute protein |
| AI | androgen independence |
| AKT1 AR | AKT Serine/threonine Kinase 1 androgen receptor |
| ARPIs ASCL1 AURKA | AR pathway inhibitors Achaete-Scute Family BHLH Transcription Factor 1 Aurora kinase A |
| BCR | biochemical recurrence |
| BPH BRN2 BRN4 CgA (CHGA) | benign prostatic hyperplasia POU Class 3 Homeobox 2 POU Class 3 Homeobox 4 chromogranin A |
| CRPC | castration-resistant prostate cancer |
| CTC | circulating tumour cell |
| DCX E2F1 EHF EMT | Doublecortin E2F Transcription Factor 1 ETS Homologous Factor epithelial-to-mesenchymal transition |
| EVs | extracellular vesicles |
| EZH2 FOXA1 FOXA2 HOTAIR HRPCa | Enhancer Of Zeste 2 Polycomb Repressive Complex 2 Subunit Forkhead box A1 Forkhead box A2 HOX transcript antisense RNA hormone refractory prostate cancer |
| LINC00261 lncRNA | Long Intergenic Non-Protein Coding RNA 261 long non-coding RNA |
| lncRNA-PCAT6 m-CRPC | long non-coding RNA prostate cancer-associated transcript 6 metastatic castration-resistant prostate cancer |
| MALAT1 miRNA | Metastasis Associated Lung Adenocarcinoma Transcript 1 microRNA |
| MUC1-C N-MYC (MYCN) NE cells | Mucin 1 C-terminal part Neuroblastoma MYC oncogene neuroendocrine cells |
| NE-like cancer cells | neuroendocrine-like cancer cells |
| NED | neuroendocrine differentiation |
| NEPC | neuroendocrine prostate cancer |
| NSE (ENO2) | neuron-specific enolase |
| ONECUT2 PARP1 PCa | One cut homeobox 2 Poly (ADP-ribose) polymerase 1 prostate cancer |
| PEG10 PSA | Paternally Expressed 10 prostate specific antigen |
| PTEN RB1 REST SCC | Phosphatase and tensin homolog RB Transcriptional Corepressor 1 RE1 Silencing Transcription Factor small cell carcinoma |
| SOX2 SRRM4 STAT3 Syp | Sex-determining region Y2 Serine/Arginine Repetitive Matrix 4 Signal Transducer And Activator Of Transcription 3 synaptophysin |
| t-NEPC TF Trop-2 | treatment-emergent neuroendocrine prostate cancer transcription factor Tumor Associated Calcium Signal Transducer 2 |
References
- Patel, G.K.; Chugh, N.; Tripathi, M. Neuroendocrine Differentiation of Prostate Cancer—An Intriguing Example of Tumor Evolution at Play. Cancers 2019, 11, 1405. [Google Scholar] [CrossRef] [PubMed]
- Vanacore, D.; Boccellino, M.; Rossetti, S.; Cavaliere, C.; D’Aniello, C.; Di Franco, R.; Romano, F.J.; Montanari, M.; La Mantia, E.; Piscitelli, R.; et al. Micrornas in prostate cancer: An overview. Oncotarget 2017, 8, 50240–50251. [Google Scholar] [CrossRef] [PubMed]
- Ding, M.; Lin, B.; Li, T.; Liu, Y.; Li, Y.; Zhou, X.; Miao, M.; Gu, J.; Pan, H.; Yang, F.; et al. A dual yet opposite growth-regulating function of miR-204 and its target XRN1 in prostate adenocarcinoma cells and neuroendocrine-like prostate cancer cells. Oncotarget 2015, 6, 7686–7700. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Gao, F.; Yu, X.; Zhang, Q.; Sun, Z.; He, Y.; Guo, W. LINC00261: A burgeoning long noncoding RNA related to cancer. Cancer Cell Int. 2021, 21, 274. [Google Scholar] [CrossRef]
- Mather, R.L.; Parolia, A.; Carson, S.E.; Venalainen, E.; Roig-Carles, D.; Jaber, M.; Chu, S.C.; Alborelli, I.; Wu, R.; Lin, D.; et al. The evolutionarily conserved long non-coding RNA LINC00261 drives neuroendocrine prostate cancer proliferation and metastasis via distinct nuclear and cytoplasmic mechanisms. Mol. Oncol. 2021, 15, 1921–1941. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Parker, C.; Castro, E.; Fizazi, K.; Heidenreich, A.; Ost, P.; Procopio, G.; Tombal, B.; Gillessen, S. Prostate cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2020, 31, 1119–1134. [Google Scholar] [CrossRef]
- Watson, P.A.; Arora, V.K.; Sawyers, C.L. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat. Rev. Cancer 2015, 15, 701–711. [Google Scholar] [CrossRef]
- Nevedomskaya, E.; Baumgart, S.J.; Haendler, B. Recent Advances in Prostate Cancer Treatment and Drug Discovery. Int. J. Mol. Sci. 2018, 19, 1359. [Google Scholar] [CrossRef]
- Davies, A.H.; Beltran, H.; Zoubeidi, A. Cellular plasticity and the neuroendocrine phenotype in prostate cancer. Nat. Rev. Urol. 2018, 15, 271–286. [Google Scholar] [CrossRef]
- Clermont, P.-L.; Ci, X.; Pandha, H.; Wang, Y.; Crea, F. Treatment-emergent neuroendocrine prostate cancer: Molecularly driven clinical guidelines. Int. J. Endocr. Oncol. 2019, 6, IJE20. [Google Scholar] [CrossRef]
- Taylor, R.A.; Toivanen, R.; Risbridger, G.P. Stem cells in prostate cancer: Treating the root of the problem. Endocr.-Relat. Cancer 2010, 17, R273–R285. [Google Scholar] [CrossRef]
- Szczyrba, J.; Niesen, A.; Wagner, M.; Wandernoth, P.M.; Aumuller, G.; Wennemuth, G. Neuroendocrine Cells of the Prostate Derive from the Neural Crest. J. Biol. Chem. 2017, 292, 2021–2031. [Google Scholar] [CrossRef]
- Abrahamsson, P.A.; di Sant’Agnese, P.A. Neuroendocrine cells in the human prostate gland. J. Androl. 1993, 14, 307–309. [Google Scholar]
- Huang, J.; Yao, J.L.; di Sant’Agnese, P.A.; Yang, Q.; Bourne, P.A.; Na, Y. Immunohistochemical characterization of neuroendocrine cells in prostate cancer. Prostate 2006, 66, 1399–1406. [Google Scholar] [CrossRef]
- Abrahamsson, P.A. Neuroendocrine cells in tumour growth of the prostate. Endocr.-Relat. Cancer 1999, 6, 503–519. [Google Scholar] [CrossRef]
- Huang, Y.H.; Zhang, Y.Q.; Huang, J.T. Neuroendocrine cells of prostate cancer: Biologic functions and molecular mechanisms. Asian J. Androl. 2019, 21, 291–295. [Google Scholar] [CrossRef]
- Cindolo, L.; Cantile, M.; Vacherot, F.; Terry, S.; de la Taille, A. Neuroendocrine differentiation in prostate cancer: From lab to bedside. Urol. Int. 2007, 79, 287–296. [Google Scholar] [CrossRef]
- Li, Z.; Chen, C.J.; Wang, J.K.; Hsia, E.; Li, W.; Squires, J.; Sun, Y.; Huang, J. Neuroendocrine differentiation of prostate cancer. Asian J. Androl. 2013, 15, 328–332. [Google Scholar] [CrossRef]
- Ozawa, H.; Takata, K. The granin family—Its role in sorting and secretory granule formation. Cell Struct. Funct. 1995, 20, 415–420. [Google Scholar] [CrossRef]
- Bartolomucci, A.; Possenti, R.; Mahata, S.K.; Fischer-Colbrie, R.; Loh, Y.P.; Salton, S.R. The extended granin family: Structure, function, and biomedical implications. Endocr. Rev. 2011, 32, 755–797. [Google Scholar] [CrossRef] [PubMed]
- Abrahamsson, P.A.; Falkmer, S.; Falt, K.; Grimelius, L. The course of neuroendocrine differentiation in prostatic carcinomas. An immunohistochemical study testing chromogranin A as an “endocrine marker”. Pathol. Res. Pr. 1989, 185, 373–380. [Google Scholar] [CrossRef]
- Berruti, A.; Mosca, A.; Tucci, M.; Terrone, C.; Torta, M.; Tarabuzzi, R.; Russo, L.; Cracco, C.; Bollito, E.; Scarpa, R.M.; et al. Independent prognostic role of circulating chromogranin A in prostate cancer patients with hormone-refractory disease. Endocr.-Relat. Cancer 2005, 12, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Ather, M.H.; Abbas, F.; Faruqui, N.; Israr, M.; Pervez, S. Correlation of three immunohistochemically detected markers of neuroendocrine differentiation with clinical predictors of disease progression in prostate cancer. BMC Urol. 2008, 8, 21. [Google Scholar] [CrossRef] [PubMed]
- Hong, P.; Guo, R.Q.; Song, G.; Yang, K.W.; Zhang, L.; Li, X.S.; Zhang, K.; Zhou, L.Q. Prognostic role of chromogranin A in castration-resistant prostate cancer: A meta-analysis. Asian J. Androl. 2018, 20, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Pruneri, G.; Galli, S.; Rossi, R.S.; Roncalli, M.; Coggi, G.; Ferrari, A.; Simonato, A.; Siccardi, A.G.; Carboni, N.; Buffa, R. Chromogranin A and B and secretogranin II in prostatic adenocarcinomas: Neuroendocrine expression in patients untreated and treated with androgen deprivation therapy. Prostate 1998, 34, 113–120. [Google Scholar] [CrossRef]
- Yuan, T.C.; Veeramani, S.; Lin, F.F.; Kondrikou, D.; Zelivianski, S.; Igawa, T.; Karan, D.; Batra, S.K.; Lin, M.F. Androgen deprivation induces human prostate epithelial neuroendocrine differentiation of androgen-sensitive LNCaP cells. Endocr.-Relat. Cancer 2006, 13, 151–167. [Google Scholar] [CrossRef]
- Xu, C.M.; Luo, Y.L.; Li, S.; Li, Z.X.; Jiang, L.; Zhang, G.X.; Owusu, L.; Chen, H.L. Multifunctional neuron-specific enolase: Its role in lung diseases. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef]
- Muoio, B.; Pascale, M.; Roggero, E. The role of serum neuron-specific enolase in patients with prostate cancer: A systematic review of the recent literature. Int. J. Biol. Markers 2018, 33, 10–21. [Google Scholar] [CrossRef]
- Kamiya, N.; Akakura, K.; Suzuki, H.; Isshiki, S.; Komiya, A.; Ueda, T.; Ito, H. Pretreatment serum level of neuron specific enolase (NSE) as a prognostic factor in metastatic prostate cancer patients treated with endocrine therapy. Eur. Urol. 2003, 44, 309–314, discussion 314. [Google Scholar] [CrossRef]
- Szarvas, T.; Csizmarik, A.; Fazekas, T.; Huttl, A.; Nyirady, P.; Hadaschik, B.; Grunwald, V.; Pullen, L.; Juranyi, Z.; Kocsis, Z.; et al. Comprehensive analysis of serum chromogranin A and neuron-specific enolase levels in localized and castration-resistant prostate cancer. BJU Int. 2021, 127, 44–55. [Google Scholar] [CrossRef]
- El Far, O.; Betz, H. Synaptophysins: Vesicular cation channels? J. Physiol. 2002, 539, 332. [Google Scholar] [CrossRef]
- Sainio, M.; Visakorpi, T.; Tolonen, T.; Ilvesaro, J.; Bova, G.S. Expression of neuroendocrine differentiation markers in lethal metastatic castration-resistant prostate cancer. Pathol. Res. Pr. 2018, 214, 848–856. [Google Scholar] [CrossRef]
- Pal, S.K.; He, M.; Chen, L.; Yang, L.; Pillai, R.; Twardowski, P.; Hsu, J.; Kortylewski, M.; Jones, J.O. Synaptophysin expression on circulating tumor cells in patients with castration resistant prostate cancer undergoing treatment with abiraterone acetate or enzalutamide. Urol. Oncol. 2018, 36, 162.e161–162.e166. [Google Scholar] [CrossRef]
- Van Acker, H.H.; Capsomidis, A.; Smits, E.L.; Van Tendeloo, V.F. CD56 in the Immune System: More Than a Marker for Cytotoxicity? Front. Immunol. 2017, 8, 892. [Google Scholar] [CrossRef]
- Mlika, M.; Zendah, I.; Braham, E.; El Mezni, F. CD56 antibody: Old-fashioned or still trendy in endocrine lung tumors. J. Immunoass. Immunochem. 2015, 36, 414–419. [Google Scholar] [CrossRef]
- Lee, J.K.; Bangayan, N.J.; Chai, T.; Smith, B.A.; Pariva, T.E.; Yun, S.; Vashisht, A.; Zhang, Q.; Park, J.W.; Corey, E.; et al. Systemic surfaceome profiling identifies target antigens for immune-based therapy in subtypes of advanced prostate cancer. Proc. Natl. Acad. Sci. USA 2018, 115, E4473–E4482. [Google Scholar] [CrossRef]
- Bertoldi, M. Mammalian Dopa decarboxylase: Structure, catalytic activity and inhibition. Arch. Biochem. Biophys 2014, 546, 1–7. [Google Scholar] [CrossRef]
- Wafa, L.A.; Cheng, H.; Rao, M.A.; Nelson, C.C.; Cox, M.; Hirst, M.; Sadowski, I.; Rennie, P.S. Isolation and identification of L-dopa decarboxylase as a protein that binds to and enhances transcriptional activity of the androgen receptor using the repressed transactivator yeast two-hybrid system. Biochem. J. 2003, 375, 373–383. [Google Scholar] [CrossRef]
- Wafa, L.A.; Palmer, J.; Fazli, L.; Hurtado-Coll, A.; Bell, R.H.; Nelson, C.C.; Gleave, M.E.; Cox, M.E.; Rennie, P.S. Comprehensive expression analysis of L-dopa decarboxylase and established neuroendocrine markers in neoadjuvant hormone-treated versus varying Gleason grade prostate tumors. Hum. Pathol. 2007, 38, 161–170. [Google Scholar] [CrossRef]
- Margiotti, K.; Wafa, L.A.; Cheng, H.; Novelli, G.; Nelson, C.C.; Rennie, P.S. Androgen-regulated genes differentially modulated by the androgen receptor coactivator L-dopa decarboxylase in human prostate cancer cells. Mol. Cancer 2007, 6, 38. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mariani, M.; Karki, R.; Spennato, M.; Pandya, D.; He, S.; Andreoli, M.; Fiedler, P.; Ferlini, C. Class III beta-tubulin in normal and cancer tissues. Gene 2015, 563, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Terry, S.; Ploussard, G.; Allory, Y.; Nicolaiew, N.; Boissiere-Michot, F.; Maille, P.; Kheuang, L.; Coppolani, E.; Ali, A.; Bibeau, F.; et al. Increased expression of class III beta-tubulin in castration-resistant human prostate cancer. Br. J. Cancer 2009, 101, 951–956. [Google Scholar] [CrossRef] [PubMed]
- Ploussard, G.; Terry, S.; Maille, P.; Allory, Y.; Sirab, N.; Kheuang, L.; Soyeux, P.; Nicolaiew, N.; Coppolani, E.; Paule, B.; et al. Class III beta-tubulin expression predicts prostate tumor aggressiveness and patient response to docetaxel-based chemotherapy. Cancer Res. 2010, 70, 9253–9264. [Google Scholar] [CrossRef]
- Thomas, R.P.; Hellmich, M.R.; Townsend, C.M., Jr.; Evers, B.M. Role of gastrointestinal hormones in the proliferation of normal and neoplastic tissues. Endocr. Rev. 2003, 24, 571–599. [Google Scholar] [CrossRef]
- Ischia, J.; Patel, O.; Bolton, D.; Shulkes, A.; Baldwin, G.S. Expression and function of gastrin-releasing peptide (GRP) in normal and cancerous urological tissues. BJU Int. 2014, 113 (Suppl. S2), 40–47. [Google Scholar] [CrossRef]
- Qiao, J.; Grabowska, M.M.; Forestier-Roman, I.S.; Mirosevich, J.; Case, T.C.; Chung, D.H.; Cates, J.M.; Matusik, R.J.; Manning, H.C.; Jin, R. Activation of GRP/GRP-R signaling contributes to castration-resistant prostate cancer progression. Oncotarget 2016, 7, 61955–61969. [Google Scholar] [CrossRef]
- Solorzano, S.R.; Imaz-Rosshandler, I.; Camacho-Arroyo, I.; Garcia-Tobilla, P.; Morales-Montor, G.; Salazar, P.; Arena-Ortiz, M.L.; Rodriguez-Dorantes, M. GABA promotes gastrin-releasing peptide secretion in NE/NE-like cells: Contribution to prostate cancer progression. Sci. Rep. 2018, 8, 10272. [Google Scholar] [CrossRef]
- Li, X.; Cai, H.; Wu, X.; Li, L.; Wu, H.; Tian, R. New Frontiers in Molecular Imaging Using Peptide-Based Radiopharmaceuticals for Prostate Cancer. Front. Chem. 2020, 8, 583309. [Google Scholar] [CrossRef]
- Russell, F.A.; King, R.; Smillie, S.J.; Kodji, X.; Brain, S.D. Calcitonin gene-related peptide: Physiology and pathophysiology. Physiol. Rev. 2014, 94, 1099–1142. [Google Scholar] [CrossRef]
- Warrington, J.I.; Richards, G.O.; Wang, N. The Role of the Calcitonin Peptide Family in Prostate Cancer and Bone Metastasis. Curr. Mol. Biol. Rep. 2017, 3, 197–203. [Google Scholar] [CrossRef]
- Suzuki, K.; Kobayashi, Y.; Morita, T. Significance of serum calcitonin gene-related peptide levels in prostate cancer patients receiving hormonal therapy. Urol. Int. 2009, 82, 291–295. [Google Scholar] [CrossRef]
- Nagakawa, O.; Ogasawara, M.; Fujii, H.; Murakami, K.; Murata, J.; Fuse, H.; Saiki, I. Effect of prostatic neuropeptides on invasion and migration of PC-3 prostate cancer cells. Cancer Lett. 1998, 133, 27–33. [Google Scholar] [CrossRef]
- Martinez, A.; Zudaire, E.; Portal-Nunez, S.; Guedez, L.; Libutti, S.K.; Stetler-Stevenson, W.G.; Cuttitta, F. Proadrenomedullin NH2-terminal 20 peptide is a potent angiogenic factor, and its inhibition results in reduction of tumor growth. Cancer Res. 2004, 64, 6489–6494. [Google Scholar] [CrossRef]
- Jimenez, N.; Calvo, A.; Martinez, A.; Rosell, D.; Cuttitta, F.; Montuenga, L.M. Expression of adrenomedullin and proadrenomedullin N-terminal 20 peptide in human and rat prostate. J. Histochem. Cytochem. 1999, 47, 1167–1178. [Google Scholar] [CrossRef]
- Calvo, A.; Abasolo, I.; Jimenez, N.; Wang, Z.; Montuenga, L. Adrenomedullin and proadrenomedullin N-terminal 20 peptide in the normal prostate and in prostate carcinoma. Microsc. Res. Tech. 2002, 57, 98–104. [Google Scholar] [CrossRef]
- Larrayoz, I.M.; Martinez-Herrero, S.; Garcia-Sanmartin, J.; Ochoa-Callejero, L.; Martinez, A. Adrenomedullin and tumour microenvironment. J. Transl. Med. 2014, 12, 339. [Google Scholar] [CrossRef]
- Rocchi, P.; Boudouresque, F.; Zamora, A.J.; Muracciole, X.; Lechevallier, E.; Martin, P.M.; Ouafik, L. Expression of adrenomedullin and peptide amidation activity in human prostate cancer and in human prostate cancer cell lines. Cancer Res. 2001, 61, 1196–1206. [Google Scholar]
- Berenguer, C.; Boudouresque, F.; Dussert, C.; Daniel, L.; Muracciole, X.; Grino, M.; Rossi, D.; Mabrouk, K.; Figarella-Branger, D.; Martin, P.M.; et al. Adrenomedullin, an autocrine/paracrine factor induced by androgen withdrawal, stimulates ‘neuroendocrine phenotype’ in LNCaP prostate tumor cells. Oncogene 2008, 27, 506–518. [Google Scholar] [CrossRef]
- Maj, M.; Wagner, L.; Tretter, V. 20 Years of Secretagogin: Exocytosis and Beyond. Front. Mol. Neurosci. 2019, 12, 29. [Google Scholar] [CrossRef]
- Adolf, K.; Wagner, L.; Bergh, A.; Stattin, P.; Ottosen, P.; Borre, M.; Birkenkamp-Demtroder, K.; Orntoft, T.F.; Torring, N. Secretagogin is a new neuroendocrine marker in the human prostate. Prostate 2007, 67, 472–484. [Google Scholar] [CrossRef] [PubMed]
- Naafs, M. Parathyroid Hormone Related Peptide (PTHrP): A Mini-Review. Endocrinol. Metab. Int. J. 2017, 5, 1–9. [Google Scholar] [CrossRef]
- Asadi, F.; Farraj, M.; Sharifi, R.; Malakouti, S.; Antar, S.; Kukreja, S. Enhanced expression of parathyroid hormone-related protein in prostate cancer as compared with benign prostatic hyperplasia. Hum. Pathol. 1996, 27, 1319–1323. [Google Scholar] [CrossRef]
- DaSilva, J.; Gioeli, D.; Weber, M.J.; Parsons, S.J. The neuroendocrine-derived peptide parathyroid hormone-related protein promotes prostate cancer cell growth by stabilizing the androgen receptor. Cancer Res. 2009, 69, 7402–7411. [Google Scholar] [CrossRef]
- Cui, Y.; Sun, Y.; Hu, S.; Luo, J.; Li, L.; Li, X.; Yeh, S.; Jin, J.; Chang, C. Neuroendocrine prostate cancer (NEPCa) increased the neighboring PCa chemoresistance via altering the PTHrP/p38/Hsp27/androgen receptor (AR)/p21 signals. Oncogene 2016, 35, 6065–6076. [Google Scholar] [CrossRef]
- Park, S.I.; Lee, C.; Sadler, W.D.; Koh, A.J.; Jones, J.; Seo, J.W.; Soki, F.N.; Cho, S.W.; Daignault, S.D.; McCauley, L.K. Parathyroid hormone-related protein drives a CD11b+Gr1+ cell-mediated positive feedback loop to support prostate cancer growth. Cancer Res. 2013, 73, 6574–6583. [Google Scholar] [CrossRef]
- Ongkeko, W.M.; Burton, D.; Kiang, A.; Abhold, E.; Kuo, S.Z.; Rahimy, E.; Yang, M.; Hoffman, R.M.; Wang-Rodriguez, J.; Deftos, L.J. Parathyroid hormone related-protein promotes epithelial-to-mesenchymal transition in prostate cancer. PLoS ONE 2014, 9, e85803. [Google Scholar] [CrossRef]
- Tyler-McMahon, B.M.; Boules, M.; Richelson, E. Neurotensin: Peptide for the next millennium. Regul. Pept. 2000, 93, 125–136. [Google Scholar] [CrossRef]
- Sehgal, I.; Powers, S.; Huntley, B.; Powis, G.; Pittelkow, M.; Maihle, N.J. Neurotensin is an autocrine trophic factor stimulated by androgen withdrawal in human prostate cancer. Proc. Natl. Acad. Sci. USA 1994, 91, 4673–4677. [Google Scholar] [CrossRef]
- Vias, M.; Burtt, G.; Culig, Z.; Veerakumarasivam, A.; Neal, D.E.; Mills, I.G. A role for neurotensin in bicalutamide resistant prostate cancer cells. Prostate 2007, 67, 190–202. [Google Scholar] [CrossRef]
- Zhu, S.; Tian, H.; Niu, X.; Wang, J.; Li, X.; Jiang, N.; Wen, S.; Chen, X.; Ren, S.; Xu, C.; et al. Neurotensin and its receptors mediate neuroendocrine transdifferentiation in prostate cancer. Oncogene 2019, 38, 4875–4884. [Google Scholar] [CrossRef]
- He, T.; Wang, M.; Wang, H.; Tan, H.; Tang, Y.; Smith, E.; Wu, Z.; Liao, W.; Hu, S.; Li, Z. Evaluation of neurotensin receptor 1 as potential biomarker for prostate cancer theranostic use. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 2199–2207. [Google Scholar] [CrossRef]
- Morgat, C.; Chastel, A.; Molinie, V.; Schollhammer, R.; Macgrogan, G.; Velasco, V.; Malavaud, B.; Fernandez, P.; Hindie, E. Neurotensin Receptor-1 Expression in Human Prostate Cancer: A Pilot Study on Primary Tumors and Lymph Node Metastases. Int. J. Mol. Sci. 2019, 20, 1721. [Google Scholar] [CrossRef]
- Apte, R.S.; Chen, D.S.; Ferrara, N. VEGF in S.Signaling and Disease: Beyond Discovery and Development. Cell 2019, 176, 1248–1264. [Google Scholar] [CrossRef]
- Harper, M.E.; Glynne-Jones, E.; Goddard, L.; Thurston, V.J.; Griffiths, K. Vascular endothelial growth factor (VEGF) expression in prostatic tumours and its relationship to neuroendocrine cells. Br. J. Cancer 1996, 74, 910–916. [Google Scholar] [CrossRef]
- Grobholz, R.; Bohrer, M.H.; Siegsmund, M.; Jünemann, K.-P.; Bleyl, U.; Woenckhaus, M. Correlation between neovascularisation and neuroendocrine differentiation in prostatic carcinoma. Pathol.-Res. Pract. 2000, 196, 277–284. [Google Scholar] [CrossRef]
- Shariat, S.F.; Anwuri, V.A.; Lamb, D.J.; Shah, N.V.; Wheeler, T.M.; Slawin, K.M. Association of Preoperative Plasma Levels of Vascular Endothelial Growth Factor and Soluble Vascular Cell Adhesion Molecule-1 With Lymph Node Status and Biochemical Progression After Radical Prostatectomy. J. Clin. Oncol. 2004, 22, 1655–1663. [Google Scholar] [CrossRef]
- Polge, A.; Gaspard, C.; Mottet, N.; Guitton, C.; Boyer, J.C.; Choquet, A.; Combettes, S.; Bancel, E.; Costa, P.; Bali, J.P. Neurohormonal stimulation of histamine release from neuroendocrine cells of the human adenomatous prostate. Prostate 1998, 34, 1–9. [Google Scholar] [CrossRef]
- Chen, J.; Hu, X.Y. Inhibition of histamine receptor H3R suppresses prostate cancer growth, invasion and increases apoptosis via the AR pathway. Oncol. Lett. 2018, 16, 4921–4928. [Google Scholar] [CrossRef]
- Dizeyi, N.; Hedlund, P.; Bjartell, A.; Tinzl, M.; Austild-Tasken, K.; Abrahamsson, P.A. Serotonin activates MAP kinase and PI3K/Akt signaling pathways in prostate cancer cell lines. Urol. Oncol. 2011, 29, 436–445. [Google Scholar] [CrossRef]
- Shinka, T.; Onodera, D.; Tanaka, T.; Shoji, N.; Miyazaki, T.; Moriuchi, T.; Fukumoto, T. Serotonin synthesis and metabolism-related molecules in a human prostate cancer cell line. Oncol. Lett. 2011, 2, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Oikawa, S.; Inuzuka, C.; Kuroki, M.; Matsuoka, Y.; Kosaki, G.; Nakazato, H. Cell adhesion activity of non-specific cross-reacting antigen (NCA) and carcinoembryonic antigen (CEA) expressed on CHO cell surface: Homophilic and heterophilic adhesion. Biochem. Biophys Res. Commun. 1989, 164, 39–45. [Google Scholar] [CrossRef]
- Taheri, M.; Saragovi, U.; Fuks, A.; Makkerh, J.; Mort, J.; Stanners, C.P. Self recognition in the Ig superfamily. Identification of precise subdomains in carcinoembryonic antigen required for intercellular adhesion. J. Biol. Chem. 2000, 275, 26935–26943. [Google Scholar] [CrossRef]
- Thompson, J.A.; Grunert, F.; Zimmermann, W. Carcinoembryonic antigen gene family: Molecular biology and clinical perspectives. J. Clin. Lab. Anal. 1991, 5, 344–366. [Google Scholar] [CrossRef] [PubMed]
- DeLucia, D.C.; Cardillo, T.M.; Ang, L.; Labrecque, M.P.; Zhang, A.; Hopkins, J.E.; De Sarkar, N.; Coleman, I.; da Costa, R.M.G.; Corey, E.; et al. Regulation of CEACAM5 and Therapeutic Efficacy of an Anti-CEACAM5-SN38 Antibody-drug Conjugate in Neuroendocrine Prostate Cancer. Clin. Cancer Res. 2021, 27, 759–774. [Google Scholar] [CrossRef] [PubMed]
- Aloe, L.; Rocco, M.L.; Bianchi, P.; Manni, L. Nerve growth factor: From the early discoveries to the potential clinical use. J. Transl. Med. 2012, 10, 239. [Google Scholar] [CrossRef] [PubMed]
- Di Donato, M.; Cernera, G.; Migliaccio, A.; Castoria, G. Nerve Growth Factor Induces Proliferation and Aggressiveness In Prostate Cancer Cells. Cancers 2019, 11, 784. [Google Scholar] [CrossRef] [PubMed]
- Di Donato, M.; Cernera, G.; Auricchio, F.; Migliaccio, A.; Castoria, G. Cross-talk between androgen receptor and nerve growth factor receptor in prostate cancer cells: Implications for a new therapeutic approach. Cell Death Discov. 2018, 4, 5. [Google Scholar] [CrossRef]
- Chen, W.Y.; Wen, Y.C.; Lin, S.R.; Yeh, H.L.; Jiang, K.C.; Chen, W.H.; Lin, Y.S.; Zhang, Q.; Liew, P.L.; Hsiao, M.; et al. Nerve growth factor interacts with CHRM4 and promotes neuroendocrine differentiation of prostate cancer and castration resistance. Commun. Biol. 2021, 4, 22. [Google Scholar] [CrossRef]
- Tilan, J.; Kitlinska, J. Neuropeptide Y (NPY) in tumor growth and progression: Lessons learned from pediatric oncology. Neuropeptides 2016, 55, 55–66. [Google Scholar] [CrossRef]
- Alshalalfa, M.; Nguyen, P.L.; Beltran, H.; Chen, W.S.; Davicioni, E.; Zhao, S.G.; Rebbeck, T.R.; Schaeffer, E.M.; Lotan, T.L.; Feng, F.Y.; et al. Transcriptomic and Clinical Characterization of Neuropeptide Y Expression in Localized and Metastatic Prostate Cancer: Identification of Novel Prostate Cancer Subtype with Clinical Implications. Eur. Urol. Oncol. 2019, 2, 405–412. [Google Scholar] [CrossRef]
- Ding, Y.; Lee, M.; Gao, Y.; Bu, P.; Coarfa, C.; Miles, B.; Sreekumar, A.; Creighton, C.J.; Ayala, G. Neuropeptide Y nerve paracrine regulation of prostate cancer oncogenesis and therapy resistance. Prostate 2021, 81, 58–71. [Google Scholar] [CrossRef]
- Nadal, R.; Schweizer, M.; Kryvenko, O.N.; Epstein, J.I.; Eisenberger, M.A. Small cell carcinoma of the prostate. Nat. Rev. Urol. 2014, 11, 213–219. [Google Scholar] [CrossRef]
- Wang, W.; Epstein, J.I. Small cell carcinoma of the prostate. A morphologic and immunohistochemical study of 95 cases. Am. J. Surg. Pathol. 2008, 32, 65–71. [Google Scholar] [CrossRef]
- Beltran, H.; Tagawa, S.T.; Park, K.; MacDonald, T.; Milowsky, M.I.; Mosquera, J.M.; Rubin, M.A.; Nanus, D.M. Challenges in recognizing treatment-related neuroendocrine prostate cancer. J. Clin. Oncol. 2012, 30, e386–e389. [Google Scholar] [CrossRef]
- Beltran, H.; Hruszkewycz, A.; Scher, H.I.; Hildesheim, J.; Isaacs, J.; Yu, E.Y.; Kelly, K.; Lin, D.; Dicker, A.; Arnold, J.; et al. The Role of Lineage Plasticity in Prostate Cancer Therapy Resistance. Clin. Cancer Res. 2019, 25, 6916–6924. [Google Scholar] [CrossRef]
- Zou, M.; Toivanen, R.; Mitrofanova, A.; Floch, N.; Hayati, S.; Sun, Y.; Le Magnen, C.; Chester, D.; Mostaghel, E.A.; Califano, A.; et al. Transdifferentiation as a Mechanism of Treatment Resistance in a Mouse Model of Castration-Resistant Prostate Cancer. Cancer Discov. 2017, 7, 736–749. [Google Scholar] [CrossRef]
- Dong, B.; Miao, J.; Wang, Y.; Luo, W.; Ji, Z.; Lai, H.; Zhang, M.; Cheng, X.; Wang, J.; Fang, Y.; et al. Single-cell analysis supports a luminal-neuroendocrine transdifferentiation in human prostate cancer. Commun. Biol. 2020, 3, 778. [Google Scholar] [CrossRef]
- Nouri, M.; Caradec, J.; Lubik, A.A.; Li, N.; Hollier, B.G.; Takhar, M.; Altimirano-Dimas, M.; Chen, M.; Roshan-Moniri, M.; Butler, M.; et al. Therapy-induced developmental reprogramming of prostate cancer cells and acquired therapy resistance. Oncotarget 2017, 8, 18949–18967. [Google Scholar] [CrossRef]
- Gupta, K.; Gupta, S. Neuroendocrine differentiation in prostate cancer: Key epigenetic players. Transl. Cancer Res. 2017, 6, S104–S108. [Google Scholar] [CrossRef]
- Davies, A.; Zoubeidi, A.; Selth, L.A. The epigenetic and transcriptional landscape of neuroendocrine prostate cancer. Endocr.-Relat. Cancer 2020, 27, R35–R50. [Google Scholar] [CrossRef] [PubMed]
- Ge, R.; Wang, Z.; Montironi, R.; Jiang, Z.; Cheng, M.; Santoni, M.; Huang, K.; Massari, F.; Lu, X.; Cimadamore, A.; et al. Epigenetic modulations and lineage plasticity in advanced prostate cancer. Ann. Oncol. 2020, 31, 470–479. [Google Scholar] [CrossRef] [PubMed]
- Park, J.W.; Lee, J.K.; Sheu, K.M.; Wang, L.; Balanis, N.G.; Nguyen, K.; Smith, B.A.; Cheng, C.; Tsai, B.L.; Cheng, D.; et al. Reprogramming normal human epithelial tissues to a common, lethal neuroendocrine cancer lineage. Science 2018, 362, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Beltran, H. Clinical and Biological Features of Neuroendocrine Prostate Cancer. Curr. Oncol. Rep. 2021, 23, 15. [Google Scholar] [CrossRef]
- Ayala, G.E.; Dai, H.; Powell, M.; Li, R.; Ding, Y.; Wheeler, T.M.; Shine, D.; Kadmon, D.; Thompson, T.; Miles, B.J.; et al. Cancer-related axonogenesis and neurogenesis in prostate cancer. Clin. Cancer Res. 2008, 14, 7593–7603. [Google Scholar] [CrossRef]
- Mauffrey, P.; Tchitchek, N.; Barroca, V.; Bemelmans, A.P.; Firlej, V.; Allory, Y.; Romeo, P.H.; Magnon, C. Progenitors from the central nervous system drive neurogenesis in cancer. Nature 2019, 569, 672–678. [Google Scholar] [CrossRef]
- Sejda, A.; Sigorski, D.; Gulczyński, J.; Wesołowski, W.; Kitlińska, J.; Iżycka-Świeszewska, E. Complexity of Neural Component of Tumor Microenvironment in Prostate Cancer. Pathobiology 2020, 87 (Suppl. S2), 87–99. [Google Scholar] [CrossRef]
- Robinson, D.; Van Allen, E.M.; Wu, Y.M.; Schultz, N.; Lonigro, R.J.; Mosquera, J.M.; Montgomery, B.; Taplin, M.E.; Pritchard, C.C.; Attard, G.; et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015, 161, 1215–1228. [Google Scholar] [CrossRef]
- Wu, C.T.; Altuwaijri, S.; Ricke, W.A.; Huang, S.P.; Yeh, S.; Zhang, C.; Niu, Y.; Tsai, M.Y.; Chang, C. Increased prostate cell proliferation and loss of cell differentiation in mice lacking prostate epithelial androgen receptor. Proc. Natl. Acad. Sci. USA 2007, 104, 12679–12684. [Google Scholar] [CrossRef]
- Schroeder, A.; Herrmann, A.; Cherryholmes, G.; Kowolik, C.; Buettner, R.; Pal, S.; Yu, H.; Muller-Newen, G.; Jove, R. Loss of androgen receptor expression promotes a stem-like cell phenotype in prostate cancer through STAT3 signaling. Cancer Res. 2014, 74, 1227–1237. [Google Scholar] [CrossRef]
- Zhang, X.; Coleman, I.M.; Brown, L.G.; True, L.D.; Kollath, L.; Lucas, J.M.; Lam, H.M.; Dumpit, R.; Corey, E.; Chery, L.; et al. SRRM4 Expression and the Loss of REST Activity May Promote the Emergence of the Neuroendocrine Phenotype in Castration-Resistant Prostate Cancer. Clin. Cancer Res. 2015, 21, 4698–4708. [Google Scholar] [CrossRef]
- Jernberg, E.; Bergh, A.; Wikstrom, P. Clinical relevance of androgen receptor alterations in prostate cancer. Endocr. Connect. 2017, 6, R146–R161. [Google Scholar] [CrossRef]
- Fujita, K.; Nonomura, N. Role of Androgen Receptor in Prostate Cancer: A Review. World J. Mens. Health 2019, 37, 288–295. [Google Scholar] [CrossRef]
- Formaggio, N.; Rubin, M.A.; Theurillat, J.P. Loss and revival of androgen receptor signaling in advanced prostate cancer. Oncogene 2021, 40, 1205–1216. [Google Scholar] [CrossRef]
- Jamaspishvili, T.; Berman, D.M.; Ross, A.E.; Scher, H.I.; De Marzo, A.M.; Squire, J.A.; Lotan, T.L. Clinical implications of PTEN loss in prostate cancer. Nat. Rev. Urol. 2018, 15, 222–234. [Google Scholar] [CrossRef]
- Soundararajan, R.; Aparicio, A.M.; Logothetis, C.J.; Mani, S.A.; Maity, S.N. Function of Tumor Suppressors in Resistance to Antiandrogen Therapy and Luminal Epithelial Plasticity of Aggressive Variant Neuroendocrine Prostate Cancers. Front. Oncol. 2018, 8, 69. [Google Scholar] [CrossRef]
- Kallio, H.M.L.; Hieta, R.; Latonen, L.; Brofeldt, A.; Annala, M.; Kivinummi, K.; Tammela, T.L.; Nykter, M.; Isaacs, W.B.; Lilja, H.G.; et al. Constitutively active androgen receptor splice variants AR-V3, AR-V7 and AR-V9 are co-expressed in castration-resistant prostate cancer metastases. Br. J. Cancer 2018, 119, 347–356. [Google Scholar] [CrossRef]
- Dehm, S.M.; Tindall, D.J. Ligand-independent androgen receptor activity is activation function-2-independent and resistant to antiandrogens in androgen refractory prostate cancer cells. J. Biol. Chem. 2006, 281, 27882–27893. [Google Scholar] [CrossRef]
- Zhang, Y.; Coillie, S.V.; Fang, J.Y.; Xu, J. Gain of function of mutant p53: R282W on the peak? Oncogenesis 2016, 5, e196. [Google Scholar] [CrossRef]
- Ma, S.; McGuire, M.H.; Mangala, L.S.; Lee, S.; Stur, E.; Hu, W.; Bayraktar, E.; Villar-Prados, A.; Ivan, C.; Wu, S.Y.; et al. Gain-of-function p53 protein transferred via small extracellular vesicles promotes conversion of fibroblasts to a cancer-associated phenotype. Cell Rep. 2021, 34, 108726. [Google Scholar] [CrossRef]
- Beltran, H.; Rickman, D.S.; Park, K.; Chae, S.S.; Sboner, A.; MacDonald, T.Y.; Wang, Y.; Sheikh, K.L.; Terry, S.; Tagawa, S.T.; et al. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov. 2011, 1, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Dardenne, E.; Beltran, H.; Benelli, M.; Gayvert, K.; Berger, A.; Puca, L.; Cyrta, J.; Sboner, A.; Noorzad, Z.; MacDonald, T.; et al. N-Myc Induces an EZH2-Mediated Transcriptional Program Driving Neuroendocrine Prostate Cancer. Cancer Cell 2016, 30, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Huang, J. Phosphatidylinositol 3-kinase-AKT-mammalian target of rapamycin pathway is essential for neuroendocrine differentiation of prostate cancer. J. Biol. Chem. 2007, 282, 3571–3583. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Sun, A.; Youn, H.; Hong, Y.; Terranova, P.F.; Thrasher, J.B.; Xu, P.; Spencer, D. Conditional Akt activation promotes androgen-independent progression of prostate cancer. Carcinogenesis 2007, 28, 572–583. [Google Scholar] [CrossRef]
- Cangemi, R.; Mensah, A.; Albertini, V.; Jain, A.; Mello-Grand, M.; Chiorino, G.; Catapano, C.V.; Carbone, G.M. Reduced expression and tumor suppressor function of the ETS transcription factor ESE-3 in prostate cancer. Oncogene 2008, 27, 2877–2885. [Google Scholar] [CrossRef]
- Albino, D.; Longoni, N.; Curti, L.; Mello-Grand, M.; Pinton, S.; Civenni, G.; Thalmann, G.; D’Ambrosio, G.; Sarti, M.; Sessa, F.; et al. ESE3/EHF controls epithelial cell differentiation and its loss leads to prostate tumors with mesenchymal and stem-like features. Cancer Res. 2012, 72, 2889–2900. [Google Scholar] [CrossRef]
- Long, Z.; Deng, L.; Li, C.; He, Q.; He, Y.; Hu, X.; Cai, Y.; Gan, Y. Loss of EHF facilitates the development of treatment-induced neuroendocrine prostate cancer. Cell Death Dis. 2021, 12, 46. [Google Scholar] [CrossRef]
- Lovnicki, J.; Gan, Y.; Feng, T.; Li, Y.; Xie, N.; Ho, C.H.; Lee, A.R.; Chen, X.; Nappi, L.; Han, B.; et al. LIN28B promotes the development of neuroendocrine prostate cancer. J. Clin. Investig. 2020, 130, 5338–5348. [Google Scholar] [CrossRef]
- Albino, D.; Civenni, G.; Dallavalle, C.; Roos, M.; Jahns, H.; Curti, L.; Rossi, S.; Pinton, S.; D’Ambrosio, G.; Sessa, F.; et al. Activation of the Lin28/let-7 Axis by Loss of ESE3/EHF Promotes a Tumorigenic and Stem-like Phenotype in Prostate Cancer. Cancer Res. 2016, 76, 3629–3643. [Google Scholar] [CrossRef]
- Raj, B.; O’Hanlon, D.; Vessey, J.P.; Pan, Q.; Ray, D.; Buckley, N.J.; Miller, F.D.; Blencowe, B.J. Cross-regulation between an alternative splicing activator and a transcription repressor controls neurogenesis. Mol. Cell 2011, 43, 843–850. [Google Scholar] [CrossRef]
- Chang, Y.T.; Lin, T.P.; Campbell, M.; Pan, C.C.; Lee, S.H.; Lee, H.C.; Yang, M.H.; Kung, H.J.; Chang, P.C. REST is a crucial regulator for acquiring EMT-like and stemness phenotypes in hormone-refractory prostate cancer. Sci. Rep. 2017, 7, 42795. [Google Scholar] [CrossRef]
- Flores-Morales, A.; Bergmann, T.B.; Lavallee, C.; Batth, T.S.; Lin, D.; Lerdrup, M.; Friis, S.; Bartels, A.; Kristensen, G.; Krzyzanowska, A.; et al. Proteogenomic Characterization of Patient-Derived Xenografts Highlights the Role of REST in Neuroendocrine Differentiation of Castration-Resistant Prostate Cancer. Clin. Cancer Res. 2019, 25, 595–608. [Google Scholar] [CrossRef]
- Yang, Y.A.; Yu, J. Current perspectives on FOXA1 regulation of androgen receptor signaling and prostate cancer. Genes Dis. 2015, 2, 144–151. [Google Scholar] [CrossRef]
- Yu, X.; Gupta, A.; Wang, Y.; Suzuki, K.; Mirosevich, J.; Orgebin-Crist, M.C.; Matusik, R.J. Foxa1 and Foxa2 interact with the androgen receptor to regulate prostate and epididymal genes differentially. Ann. N. Y. Acad. Sci. 2005, 1061, 77–93. [Google Scholar] [CrossRef]
- Pomerantz, M.M.; Li, F.; Takeda, D.Y.; Lenci, R.; Chonkar, A.; Chabot, M.; Cejas, P.; Vazquez, F.; Cook, J.; Shivdasani, R.A.; et al. The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat. Genet. 2015, 47, 1346–1351. [Google Scholar] [CrossRef]
- Jin, H.J.; Zhao, J.C.; Wu, L.; Kim, J.; Yu, J. Cooperativity and equilibrium with FOXA1 define the androgen receptor transcriptional program. Nat. Commun. 2014, 5, 3972. [Google Scholar] [CrossRef]
- Kim, J.; Jin, H.; Zhao, J.C.; Yang, Y.A.; Li, Y.; Yang, X.; Dong, X.; Yu, J. FOXA1 inhibits prostate cancer neuroendocrine differentiation. Oncogene 2017, 36, 4072–4080. [Google Scholar] [CrossRef]
- Amador-Arjona, A.; Cimadamore, F.; Huang, C.T.; Wright, R.; Lewis, S.; Gage, F.H.; Terskikh, A.V. SOX2 primes the epigenetic landscape in neural precursors enabling proper gene activation during hippocampal neurogenesis. Proc. Natl. Acad. Sci. USA 2015, 112, E1936–E1945. [Google Scholar] [CrossRef]
- Vasconcelos, F.F.; Castro, D.S. Transcriptional control of vertebrate neurogenesis by the proneural factor Ascl1. Front. Cell Neurosci. 2014, 8, 412. [Google Scholar] [CrossRef]
- Couillard-Despres, S.; Winner, B.; Schaubeck, S.; Aigner, R.; Vroemen, M.; Weidner, N.; Bogdahn, U.; Winkler, J.; Kuhn, H.G.; Aigner, L. Doublecortin expression levels in adult brain reflect neurogenesis. Eur. J. Neurosci. 2005, 21, 1–14. [Google Scholar] [CrossRef]
- Arenas, E. Foxa2: The rise and fall of dopamine neurons. Cell Stem. Cell 2008, 2, 110–112. [Google Scholar] [CrossRef] [PubMed]
- Hashizume, K.; Yamanaka, M.; Ueda, S. POU3F2 participates in cognitive function and adult hippocampal neurogenesis via mammalian-characteristic amino acid repeats. Genes Brain Behav. 2018, 17, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Cheng, X.; Zhang, L.; Wang, L.; Mao, Y.; Tian, G.; Xu, W.; Wu, Y.; Ma, Z.; Qin, J.; et al. Exploration of the Brn4-regulated genes enhancing adult hippocampal neurogenesis by RNA sequencing. J. Neurosci. Res. 2017, 95, 2071–2079. [Google Scholar] [CrossRef] [PubMed]
- Knoepfler, P.S.; Cheng, P.F.; Eisenman, R.N. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev. 2002, 16, 2699–2712. [Google Scholar] [CrossRef]
- van der Raadt, J.; van Gestel, S.H.C.; Nadif Kasri, N.; Albers, C.A. ONECUT transcription factors induce neuronal characteristics and remodel chromatin accessibility. Nucleic Acids Res. 2019, 47, 5587–5602. [Google Scholar] [CrossRef]
- Mu, P.; Zhang, Z.; Benelli, M.; Karthaus, W.R.; Hoover, E.; Chen, C.C.; Wongvipat, J.; Ku, S.Y.; Gao, D.; Cao, Z.; et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science 2017, 355, 84–88. [Google Scholar] [CrossRef]
- Rapa, I.; Ceppi, P.; Bollito, E.; Rosas, R.; Cappia, S.; Bacillo, E.; Porpiglia, F.; Berruti, A.; Papotti, M.; Volante, M. Human ASH1 expression in prostate cancer with neuroendocrine differentiation. Mod. Pathol. 2008, 21, 700–707. [Google Scholar] [CrossRef]
- Rapa, I.; Volante, M.; Migliore, C.; Farsetti, A.; Berruti, A.; Vittorio Scagliotti, G.; Giordano, S.; Papotti, M. Human ASH-1 promotes neuroendocrine differentiation in androgen deprivation conditions and interferes with androgen responsiveness in prostate cancer cells. Prostate 2013, 73, 1241–1249. [Google Scholar] [CrossRef]
- Fraser, J.A.; Sutton, J.E.; Tazayoni, S.; Bruce, I.; Poole, A.V. hASH1 nuclear localization persists in neuroendocrine transdifferentiated prostate cancer cells, even upon reintroduction of androgen. Sci. Rep. 2019, 9, 19076. [Google Scholar] [CrossRef]
- Tabrizi, S.; Alshalalfa, M.; Mahal, B.A.; Davicioni, E.; Liu, Y.; Mouw, K.W.; Feng, F.; Nguyen, P.L.; Muralidhar, V. Doublecortin Expression in Prostate Adenocarcinoma and Neuroendocrine Tumors. Int. J. Radiat. Oncol. Biol. Phys. 2020, 108, 936–940. [Google Scholar] [CrossRef]
- Park, J.W.; Lee, J.K.; Witte, O.N.; Huang, J. FOXA2 is a sensitive and specific marker for small cell neuroendocrine carcinoma of the prostate. Mod. Pathol. 2017, 30, 1262–1272. [Google Scholar] [CrossRef]
- Connelly, Z.M.; Yang, S.; Chen, F.; Yeh, Y.; Khater, N.; Jin, R.; Matusik, R.; Yu, X. Foxa2 activates the transcription of androgen receptor target genes in castrate resistant prostatic tumors. Am. J. Clin. Exp. Urol. 2018, 6, 172–181. [Google Scholar]
- Bishop, J.L.; Thaper, D.; Vahid, S.; Davies, A.; Ketola, K.; Kuruma, H.; Jama, R.; Nip, K.M.; Angeles, A.; Johnson, F.; et al. The Master Neural Transcription Factor BRN2 Is an Androgen Receptor-Suppressed Driver of Neuroendocrine Differentiation in Prostate Cancer. Cancer Discov. 2017, 7, 54–71. [Google Scholar] [CrossRef]
- Bhagirath, D.; Yang, T.L.; Tabatabai, Z.L.; Majid, S.; Dahiya, R.; Tanaka, Y.; Saini, S. BRN4 Is a Novel Driver of Neuroendocrine Differentiation in Castration-Resistant Prostate Cancer and Is Selectively Released in Extracellular Vesicles with BRN2. Clin. Cancer Res. 2019, 25, 6532–6545. [Google Scholar] [CrossRef]
- Lee, J.K.; Phillips, J.W.; Smith, B.A.; Park, J.W.; Stoyanova, T.; McCaffrey, E.F.; Baertsch, R.; Sokolov, A.; Meyerowitz, J.G.; Mathis, C.; et al. N-Myc Drives Neuroendocrine Prostate Cancer Initiated from Human Prostate Epithelial Cells. Cancer Cell 2016, 29, 536–547. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, B.; Wu, W.; Li, L.; Broom, B.M.; Basourakos, S.P.; Korentzelos, D.; Luan, Y.; Wang, J.; Yang, G.; et al. Targeting the MYCN-PARP-DNA Damage Response Pathway in Neuroendocrine Prostate Cancer. Clin. Cancer Res. 2018, 24, 696–707. [Google Scholar] [CrossRef]
- Guo, H.; Ci, X.; Ahmed, M.; Hua, J.T.; Soares, F.; Lin, D.; Puca, L.; Vosoughi, A.; Xue, H.; Li, E.; et al. ONECUT2 is a driver of neuroendocrine prostate cancer. Nat. Commun. 2019, 10, 278. [Google Scholar] [CrossRef]
- Varambally, S.; Dhanasekaran, S.M.; Zhou, M.; Barrette, T.R.; Kumar-Sinha, C.; Sanda, M.G.; Ghosh, D.; Pienta, K.J.; Sewalt, R.G.; Otte, A.P.; et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002, 419, 624–629. [Google Scholar] [CrossRef]
- Zhang, Y.; Zheng, D.; Zhou, T.; Song, H.; Hulsurkar, M.; Su, N.; Liu, Y.; Wang, Z.; Shao, L.; Ittmann, M.; et al. Androgen deprivation promotes neuroendocrine differentiation and angiogenesis through CREB-EZH2-TSP1 pathway in prostate cancers. Nat. Commun. 2018, 9, 4080. [Google Scholar] [CrossRef]
- Li, Y.; Donmez, N.; Sahinalp, C.; Xie, N.; Wang, Y.; Xue, H.; Mo, F.; Beltran, H.; Gleave, M.; Collins, C.; et al. SRRM4 Drives Neuroendocrine Transdifferentiation of Prostate Adenocarcinoma Under Androgen Receptor Pathway Inhibition. Eur. Urol. 2017, 71, 68–78. [Google Scholar] [CrossRef]
- Lee, A.R.; Gan, Y.; Tang, Y.; Dong, X. A novel mechanism of SRRM4 in promoting neuroendocrine prostate cancer development via a pluripotency gene network. EBioMedicine 2018, 35, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Akamatsu, S.; Wyatt, A.W.; Lin, D.; Lysakowski, S.; Zhang, F.; Kim, S.; Tse, C.; Wang, K.; Mo, F.; Haegert, A.; et al. The Placental Gene PEG10 Promotes Progression of Neuroendocrine Prostate Cancer. Cell Rep. 2015, 12, 922–936. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Thaper, D.; Bidnur, S.; Toren, P.; Akamatsu, S.; Bishop, J.L.; Colins, C.; Vahid, S.; Zoubeidi, A. PEG10 is associated with treatment-induced neuroendocrine prostate cancer. J. Mol. Endocrinol. 2019, 63, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Svensson, C.; Ceder, J.; Iglesias-Gato, D.; Chuan, Y.C.; Pang, S.T.; Bjartell, A.; Martinez, R.M.; Bott, L.; Helczynski, L.; Ulmert, D.; et al. REST mediates androgen receptor actions on gene repression and predicts early recurrence of prostate cancer. Nucleic Acids Res. 2014, 42, 999–1015. [Google Scholar] [CrossRef]
- Ballas, N.; Grunseich, C.; Lu, D.D.; Speh, J.C.; Mandel, G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell 2005, 121, 645–657. [Google Scholar] [CrossRef]
- Gao, Z.; Ure, K.; Ding, P.; Nashaat, M.; Yuan, L.; Ma, J.; Hammer, R.E.; Hsieh, J. The master negative regulator REST/NRSF controls adult neurogenesis by restraining the neurogenic program in quiescent stem cells. J. Neurosci. 2011, 31, 9772–9786. [Google Scholar] [CrossRef]
- Liu, K.; Wang, S.; Liu, Y.; Gu, J.; Gu, S.; Xu, Z.; Zhang, R.; Wang, Z.; Ma, H.; Chen, Y.; et al. Overexpression of MYCN promotes proliferation of non-small cell lung cancer. Tumour Biol. 2016, 37, 12855–12866. [Google Scholar] [CrossRef]
- Metz, E.P.; Wilder, P.J.; Dong, J.; Datta, K.; Rizzino, A. Elevating SOX2 in prostate tumor cells upregulates expression of neuroendocrine genes, but does not reduce the inhibitory effects of enzalutamide. J. Cell Physiol. 2020, 235, 3731–3740. [Google Scholar] [CrossRef]
- Niu, W.; Zang, T.; Smith, D.K.; Vue, T.Y.; Zou, Y.; Bachoo, R.; Johnson, J.E.; Zhang, C.L. SOX2 reprograms resident astrocytes into neural progenitors in the adult brain. Stem. Cell Rep. 2015, 4, 780–794. [Google Scholar] [CrossRef]
- Mirosevich, J.; Gao, N.; Gupta, A.; Shappell, S.B.; Jove, R.; Matusik, R.J. Expression and role of Foxa proteins in prostate cancer. Prostate 2006, 66, 1013–1028. [Google Scholar] [CrossRef]
- Baca, S.C.; Takeda, D.Y.; Seo, J.H.; Hwang, J.; Ku, S.Y.; Arafeh, R.; Arnoff, T.; Agarwal, S.; Bell, C.; O’Connor, E.; et al. Reprogramming of the FOXA1 cistrome in treatment-emergent neuroendocrine prostate cancer. Nat. Commun. 2021, 12, 1979. [Google Scholar] [CrossRef]
- Rotinen, M.; You, S.; Yang, J.; Coetzee, S.G.; Reis-Sobreiro, M.; Huang, W.C.; Huang, F.; Pan, X.; Yanez, A.; Hazelett, D.J.; et al. ONECUT2 is a targetable master regulator of lethal prostate cancer that suppresses the androgen axis. Nat. Med. 2018, 24, 1887–1898. [Google Scholar] [CrossRef]
- Burke, P.A.; Gregg, J.P.; Bakhtiar, B.; Beckett, L.A.; Denardo, G.L.; Albrecht, H.; De Vere White, R.W.; De Nardo, S.J. Characterization of MUC1 glycoprotein on prostate cancer for selection of targeting molecules. Int. J. Oncol. 2006, 29, 49–55. [Google Scholar] [CrossRef]
- Rajabi, H.; Kufe, D. MUC1-C Oncoprotein Integrates a Program of EMT, Epigenetic Reprogramming and Immune Evasion in Human Carcinomas. Biochim. Biophys Acta Rev. Cancer 2017, 1868, 117–122. [Google Scholar] [CrossRef]
- Yasumizu, Y.; Rajabi, H.; Jin, C.; Hata, T.; Pitroda, S.; Long, M.D.; Hagiwara, M.; Li, W.; Hu, Q.; Liu, S.; et al. MUC1-C regulates lineage plasticity driving progression to neuroendocrine prostate cancer. Nat. Commun. 2020, 11, 338. [Google Scholar] [CrossRef]
- Hagiwara, M.; Yasumizu, Y.; Yamashita, N.; Rajabi, H.; Fushimi, A.; Long, M.D.; Li, W.; Bhattacharya, A.; Ahmad, R.; Oya, M.; et al. MUC1-C Activates the BAF (mSWI/SNF) Complex in Prostate Cancer Stem Cells. Cancer Res. 2021, 81, 1111–1122. [Google Scholar] [CrossRef]
- Kwon, O.J.; Zhang, L.; Jia, D.; Zhou, Z.; Li, Z.; Haffner, M.; Lee, J.K.; True, L.; Morrissey, C.; Xin, L. De novo induction of lineage plasticity from human prostate luminal epithelial cells by activated AKT1 and c-Myc. Oncogene 2020, 39, 7142–7151. [Google Scholar] [CrossRef]
- Hsu, E.C.; Rice, M.A.; Bermudez, A.; Marques, F.J.G.; Aslan, M.; Liu, S.; Ghoochani, A.; Zhang, C.A.; Chen, Y.S.; Zlitni, A.; et al. Trop2 is a driver of metastatic prostate cancer with neuroendocrine phenotype via PARP1. Proc. Natl. Acad. Sci. USA 2020, 117, 2032–2042. [Google Scholar] [CrossRef]
- Abramovic, I.; Ulamec, M.; Katusic Bojanac, A.; Bulic-Jakus, F.; Jezek, D.; Sincic, N. miRNA in prostate cancer: Challenges toward translation. Epigenomics 2020, 12, 543–558. [Google Scholar] [CrossRef]
- Alles, J.; Fehlmann, T.; Fischer, U.; Backes, C.; Galata, V.; Minet, M.; Hart, M.; Abu-Halima, M.; Grasser, F.A.; Lenhof, H.P.; et al. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019, 47, 3353–3364. [Google Scholar] [CrossRef]
- Kabekkodu, S.P.; Shukla, V.; Varghese, V.K.; Jeevitha, D.S.; Chakrabarty, S.; Satyamoorthy, K. Clustered miRNAs and their role in biological functions and diseases. Biol. Rev. Camb. Philos. Soc. 2018, 93, 1955–1986. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. (Lausanne) 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, S.; Tong, Y.; Steitz, J.A. Switching from repression to activation: MicroRNAs can up-regulate translation. Science 2007, 318, 1931–1934. [Google Scholar] [CrossRef] [PubMed]
- Bayraktar, R.; Bertilaccio, M.T.S.; Calin, G.A. The Interaction Between Two Worlds: MicroRNAs and Toll-Like Receptors. Front. Immunol. 2019, 10, 1053. [Google Scholar] [CrossRef]
- Quillet, A.; Saad, C.; Ferry, G.; Anouar, Y.; Vergne, N.; Lecroq, T.; Dubessy, C. Improving Bioinformatics Prediction of microRNA Targets by Ranks Aggregation. Front. Genet. 2019, 10, 1330. [Google Scholar] [CrossRef]
- Huang, H.Y.; Lin, Y.C.; Li, J.; Huang, K.Y.; Shrestha, S.; Hong, H.C.; Tang, Y.; Chen, Y.G.; Jin, C.N.; Yu, Y.; et al. miRTarBase 2020: Updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 2020, 48, D148–D154. [Google Scholar] [CrossRef]
- Jiang, X.; Guo, S.; Zhang, Y.; Zhao, Y.; Li, X.; Jia, Y.; Xu, Y.; Ma, B. LncRNA NEAT1 promotes docetaxel resistance in prostate cancer by regulating ACSL4 via sponging miR-34a-5p and miR-204-5p. Cell Signal. 2020, 65, 109422. [Google Scholar] [CrossRef]
- Thomson, D.W.; Dinger, M.E. Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 2016, 17, 272–283. [Google Scholar] [CrossRef]
- Lo, U.G.; Pong, R.C.; Yang, D.; Gandee, L.; Hernandez, E.; Dang, A.; Lin, C.J.; Santoyo, J.; Ma, S.; Sonavane, R.; et al. IFNgamma-Induced IFIT5 Promotes Epithelial-to-Mesenchymal Transition in Prostate Cancer via miRNA Processing. Cancer Res. 2019, 79, 1098–1112. [Google Scholar] [CrossRef]
- Fernandes, R.C.; Toubia, J.; Townley, S.; Hanson, A.R.; Dredge, B.K.; Pillman, K.A.; Bert, A.G.; Winter, J.M.; Iggo, R.; Das, R.; et al. Post-transcriptional Gene Regulation by MicroRNA-194 Promotes Neuroendocrine Transdifferentiation in Prostate Cancer. Cell Rep. 2021, 34, 108585. [Google Scholar] [CrossRef]
- Das, R.; Gregory, P.A.; Fernandes, R.C.; Denis, I.; Wang, Q.; Townley, S.L.; Zhao, S.G.; Hanson, A.R.; Pickering, M.A.; Armstrong, H.K.; et al. MicroRNA-194 Promotes Prostate Cancer Metastasis by Inhibiting SOCS2. Cancer Res. 2017, 77, 1021–1034. [Google Scholar] [CrossRef]
- Selth, L.A.; Townley, S.L.; Bert, A.G.; Stricker, P.D.; Sutherland, P.D.; Horvath, L.G.; Goodall, G.J.; Butler, L.M.; Tilley, W.D. Circulating microRNAs predict biochemical recurrence in prostate cancer patients. Br. J. Cancer 2013, 109, 641–650. [Google Scholar] [CrossRef]
- Tong, A.W.; Fulgham, P.; Jay, C.; Chen, P.; Khalil, I.; Liu, S.; Senzer, N.; Eklund, A.C.; Han, J.; Nemunaitis, J. MicroRNA profile analysis of human prostate cancers. Cancer Gene Ther. 2009, 16, 206–216. [Google Scholar] [CrossRef]
- Bhagirath, D.; Liston, M.; Patel, N.; Akoto, T.; Lui, B.; Yang, T.L.; To, D.M.; Majid, S.; Dahiya, R.; Tabatabai, Z.L.; et al. MicroRNA determinants of neuroendocrine differentiation in metastatic castration-resistant prostate cancer. Oncogene 2020, 39, 7209–7223. [Google Scholar] [CrossRef]
- Abdelmohsen, K.; Hutchison, E.R.; Lee, E.K.; Kuwano, Y.; Kim, M.M.; Masuda, K.; Srikantan, S.; Subaran, S.S.; Marasa, B.S.; Mattson, M.P.; et al. miR-375 inhibits differentiation of neurites by lowering HuD levels. Mol. Cell Biol. 2010, 30, 4197–4210. [Google Scholar] [CrossRef]
- Liu, Y.; Xing, R.; Zhang, X.; Dong, W.; Zhang, J.; Yan, Z.; Li, W.; Cui, J.; Lu, Y. miR-375 targets the p53 gene to regulate cellular response to ionizing radiation and etoposide in gastric cancer cells. DNA Repair (Amst) 2013, 12, 741–750. [Google Scholar] [CrossRef]
- Wang, Y.; Lieberman, R.; Pan, J.; Zhang, Q.; Du, M.; Zhang, P.; Nevalainen, M.; Kohli, M.; Shenoy, N.K.; Meng, H.; et al. miR-375 induces docetaxel resistance in prostate cancer by targeting SEC23A and YAP1. Mol. Cancer 2016, 15, 70. [Google Scholar] [CrossRef]
- He, S.; Shi, J.; Mao, J.; Luo, X.; Liu, W.; Liu, R.; Yang, F. The expression of miR-375 in prostate cancer: A study based on GEO, TCGA data and bioinformatics analysis. Pathol. Res. Pr. 2019, 215, 152375. [Google Scholar] [CrossRef]
- Valera, V.A.; Parra-Medina, R.; Walter, B.A.; Pinto, P.; Merino, M.J. microRNA Expression Profiling in Young Prostate Cancer Patients. J. Cancer 2020, 11, 4106–4114. [Google Scholar] [CrossRef]
- Costa-Pinheiro, P.; Ramalho-Carvalho, J.; Vieira, F.Q.; Torres-Ferreira, J.; Oliveira, J.; Goncalves, C.S.; Costa, B.M.; Henrique, R.; Jeronimo, C. MicroRNA-375 plays a dual role in prostate carcinogenesis. Clin. Epigenetics 2015, 7, 42. [Google Scholar] [CrossRef]
- Benoist, G.E.; van Oort, I.M.; Boerrigter, E.; Verhaegh, G.W.; van Hooij, O.; Groen, L.; Smit, F.; de Mol, P.; Hamberg, P.; Dezentje, V.O.; et al. Prognostic Value of Novel Liquid Biomarkers in Patients with Metastatic Castration-Resistant Prostate Cancer Treated with Enzalutamide: A Prospective Observational Study. Clin. Chem. 2020, 66, 842–851. [Google Scholar] [CrossRef] [PubMed]
- Selth, L.A.; Das, R.; Townley, S.L.; Coutinho, I.; Hanson, A.R.; Centenera, M.M.; Stylianou, N.; Sweeney, K.; Soekmadji, C.; Jovanovic, L.; et al. A ZEB1-miR-375-YAP1 pathway regulates epithelial plasticity in prostate cancer. Oncogene 2017, 36, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Pickl, J.M.; Tichy, D.; Kuryshev, V.Y.; Tolstov, Y.; Falkenstein, M.; Schuler, J.; Reidenbach, D.; Hotz-Wagenblatt, A.; Kristiansen, G.; Roth, W.; et al. Ago-RIP-Seq identifies Polycomb repressive complex I member CBX7 as a major target of miR-375 in prostate cancer progression. Oncotarget 2016, 7, 59589–59603. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yuan, T.; Liang, M.; Du, M.; Xia, S.S.; Dittmar, R.; Wang, D.; See, W.; Costello, B.A.; Quevedo, F.; et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur. Urol. 2015, 67, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Zedan, A.H.; Osther, P.J.S.; Assenholt, J.; Madsen, J.S.; Hansen, T.F. Circulating miR-141 and miR-375 are associated with treatment outcome in metastatic castration resistant prostate cancer. Sci. Rep. 2020, 10, 227. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.H.; Plets, M.; Li, H.; Higano, C.S.; Tangen, C.M.; Agarwal, N.; Vogelzang, N.J.; Hussain, M.; Thompson, I.M., Jr.; Tewari, M.; et al. Circulating microRNAs and treatment response in the Phase II SWOG S0925 study for patients with new metastatic hormone-sensitive prostate cancer. Prostate 2018, 78, 121–127. [Google Scholar] [CrossRef]
- Foj, L.; Ferrer, F.; Serra, M.; Arevalo, A.; Gavagnach, M.; Gimenez, N.; Filella, X. Exosomal and Non-Exosomal Urinary miRNAs in Prostate Cancer Detection and Prognosis. Prostate 2017, 77, 573–583. [Google Scholar] [CrossRef]
- Ciszkowicz, E.; Porzycki, P.; Semik, M.; Kaznowska, E.; Tyrka, M. MiR-93/miR-375: Diagnostic Potential, Aggressiveness Correlation and Common Target Genes in Prostate Cancer. Int. J. Mol. Sci. 2020, 21, 5667. [Google Scholar] [CrossRef]
- Jin, W.; Fei, X.; Wang, X.; Chen, F.; Song, Y. Circulating miRNAs as Biomarkers for Prostate Cancer Diagnosis in Subjects with Benign Prostatic Hyperplasia. J. Immunol. Res. 2020, 2020, 5873056. [Google Scholar] [CrossRef]
- Haldrup, C.; Kosaka, N.; Ochiya, T.; Borre, M.; Hoyer, S.; Orntoft, T.F.; Sorensen, K.D. Profiling of circulating microRNAs for prostate cancer biomarker discovery. Drug Deliv. Transl. Res. 2014, 4, 19–30. [Google Scholar] [CrossRef]
- Fredsoe, J.; Rasmussen, A.K.I.; Mouritzen, P.; Bjerre, M.T.; Ostergren, P.; Fode, M.; Borre, M.; Sorensen, K.D. Profiling of Circulating microRNAs in Prostate Cancer Reveals Diagnostic Biomarker Potential. Diagnostics 2020, 10, 188. [Google Scholar] [CrossRef]
- Bidarra, D.; Constancio, V.; Barros-Silva, D.; Ramalho-Carvalho, J.; Moreira-Barbosa, C.; Antunes, L.; Mauricio, J.; Oliveira, J.; Henrique, R.; Jeronimo, C. Circulating MicroRNAs as Biomarkers for Prostate Cancer Detection and Metastasis Development Prediction. Front. Oncol. 2019, 9, 900. [Google Scholar] [CrossRef]
- Paiva, R.M.; Zauli, D.A.G.; Neto, B.S.; Brum, I.S. Urinary microRNAs expression in prostate cancer diagnosis: A systematic review. Clin. Transl. Oncol. 2020, 22, 2061–2073. [Google Scholar] [CrossRef]
- Wach, S.; Al-Janabi, O.; Weigelt, K.; Fischer, K.; Greither, T.; Marcou, M.; Theil, G.; Nolte, E.; Holzhausen, H.J.; Stohr, R.; et al. The combined serum levels of miR-375 and urokinase plasminogen activator receptor are suggested as diagnostic and prognostic biomarkers in prostate cancer. Int. J. Cancer 2015, 137, 1406–1416. [Google Scholar] [CrossRef]
- Chu, M.; Chang, Y.; Li, P.; Guo, Y.; Zhang, K.; Gao, W. Androgen receptor is negatively correlated with the methylation-mediated transcriptional repression of miR-375 in human prostate cancer cells. Oncol. Rep. 2014, 31, 34–40. [Google Scholar] [CrossRef]
- Pillman, K.A.; Phillips, C.A.; Roslan, S.; Toubia, J.; Dredge, B.K.; Bert, A.G.; Lumb, R.; Neumann, D.P.; Li, X.; Conn, S.J.; et al. miR-200/375 control epithelial plasticity-associated alternative splicing by repressing the RNA-binding protein Quaking. EMBO J. 2018, 37, e99016. [Google Scholar] [CrossRef]
- Bhagirath, D.; Liston, M.; Akoto, T.; Lui, B.; Bensing, B.A.; Sharma, A.; Saini, S. Novel, non-invasive markers for detecting therapy induced neuroendocrine differentiation in castration-resistant prostate cancer patients. Sci. Rep. 2021, 11, 8279. [Google Scholar] [CrossRef]
- Ma, F.; Zhang, J.; Zhong, L.; Wang, L.; Liu, Y.; Wang, Y.; Peng, L.; Guo, B. Upregulated microRNA-301a in breast cancer promotes tumor metastasis by targeting PTEN and activating Wnt/beta-catenin signaling. Gene 2014, 535, 191–197. [Google Scholar] [CrossRef]
- Xie, H.; Li, L.; Zhu, G.; Dang, Q.; Ma, Z.; He, D.; Chang, L.; Song, W.; Chang, H.C.; Krolewski, J.J.; et al. Infiltrated pre-adipocytes increase prostate cancer metastasis via modulation of the miR-301a/androgen receptor (AR)/TGF-beta1/Smad/MMP9 signals. Oncotarget 2015, 6, 12326–12339. [Google Scholar] [CrossRef]
- Fan, L.; Wang, Y.; Huo, W.; Wang, W.H. MicroRNA301a3p overexpression promotes cell invasion and proliferation by targeting runtrelated transcription factor 3 in prostate cancer. Mol. Med. Rep. 2019, 20, 3755–3763. [Google Scholar] [CrossRef]
- Li, X.; Li, J.; Cai, Y.; Peng, S.; Wang, J.; Xiao, Z.; Wang, Y.; Tao, Y.; Leng, Q.; Wu, D.; et al. Hyperglycaemia-induced miR-301a promotes cell proliferation by repressing p21 and Smad4 in prostate cancer. Cancer Lett. 2018, 418, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liu, M.; Guan, Y.; Wu, Q. Hypoxia-Responsive Mir-301a and Mir-301b Promote Radioresistance of Prostate Cancer Cells via Downregulating NDRG2. Med. Sci. Monit. 2016, 22, 2126–2132. [Google Scholar] [CrossRef] [PubMed]
- Nam, R.K.; Benatar, T.; Wallis, C.J.; Amemiya, Y.; Yang, W.; Garbens, A.; Naeim, M.; Sherman, C.; Sugar, L.; Seth, A. MiR-301a regulates E-cadherin expression and is predictive of prostate cancer recurrence. Prostate 2016, 76, 869–884. [Google Scholar] [CrossRef] [PubMed]
- Damodaran, C.; Das, T.P.; Papu John, A.M.; Suman, S.; Kolluru, V.; Morris, T.J.; Faber, E.N.; Rai, S.N.; Messer, J.C.; Alatassi, H.; et al. miR-301a expression: A prognostic marker for prostate cancer. Urol. Oncol. 2016, 34, 336.e13–336.e20. [Google Scholar] [CrossRef]
- Nam, R.K.; Amemiya, Y.; Benatar, T.; Wallis, C.J.; Stojcic-Bendavid, J.; Bacopulos, S.; Sherman, C.; Sugar, L.; Naeim, M.; Yang, W.; et al. Identification and Validation of a Five MicroRNA Signature Predictive of Prostate Cancer Recurrence and Metastasis: A Cohort Study. J. Cancer 2015, 6, 1160–1171. [Google Scholar] [CrossRef]
- Kolluru, V.; Chandrasekaran, B.; Tyagi, A.; Dervishi, A.; Ankem, M.; Yan, X.; Maiying, K.; Alatassi, H.; Shaheen, S.P.; Messer, J.C.; et al. miR-301a expression: Diagnostic and prognostic marker for prostate cancer. Urol. Oncol. 2018, 36, 503.e509–503.e515. [Google Scholar] [CrossRef]
- Saran, U.; Chandrasekaran, B.; Kolluru, V.; Tyagi, A.; Nguyen, K.D.; Valadon, C.L.; Shaheen, S.P.; Kong, M.; Poddar, T.; Ankem, M.K.; et al. Diagnostic molecular markers predicting aggressive potential in low-grade prostate cancer. Transl. Res. 2021, 231, 92–101. [Google Scholar] [CrossRef]
- Dankert, J.T.; Wiesehofer, M.; Czyrnik, E.D.; Singer, B.B.; von Ostau, N.; Wennemuth, G. The deregulation of miR-17/CCND1 axis during neuroendocrine transdifferentiation of LNCaP prostate cancer cells. PLoS ONE 2018, 13, e0200472. [Google Scholar] [CrossRef]
- Arabi, L.; Gsponer, J.R.; Smida, J.; Nathrath, M.; Perrina, V.; Jundt, G.; Ruiz, C.; Quagliata, L.; Baumhoer, D. Upregulation of the miR-17-92 cluster and its two paraloga in osteosarcoma—Reasons and consequences. Genes Cancer 2014, 5, 56–63. [Google Scholar] [CrossRef]
- Fang, Y.; Shen, H.; Li, H.; Cao, Y.; Qin, R.; Long, L.; Zhu, X.; Xie, C.; Xu, W. miR-106a confers cisplatin resistance by regulating PTEN/Akt pathway in gastric cancer cells. Acta Biochim. Biophys Sin. (Shanghai) 2013, 45, 963–972. [Google Scholar] [CrossRef]
- Jiang, Y.; Wu, Y.; Greenlee, A.R.; Wu, J.; Han, Z.; Li, X.; Zhao, Y. miR-106a-mediated malignant transformation of cells induced by anti-benzo[a]pyrene-trans-7,8-diol-9,10-epoxide. Toxicol. Sci. 2011, 119, 50–60. [Google Scholar] [CrossRef]
- Hoey, C.; Ray, J.; Jeon, J.; Huang, X.; Taeb, S.; Ylanko, J.; Andrews, D.W.; Boutros, P.C.; Liu, S.K. miRNA-106a and prostate cancer radioresistance: A novel role for LITAF in ATM regulation. Mol. Oncol. 2018, 12, 1324–1341. [Google Scholar] [CrossRef]
- Cochetti, G.; Poli, G.; Guelfi, G.; Boni, A.; Egidi, M.G.; Mearini, E. Different levels of serum microRNAs in prostate cancer and benign prostatic hyperplasia: Evaluation of potential diagnostic and prognostic role. Onco Targets Ther. 2016, 9, 7545–7553. [Google Scholar] [CrossRef]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef]
- Luo, B.; Kang, N.; Chen, Y.; Liu, L.; Zhang, Y. Oncogene miR-106a promotes proliferation and metastasis of prostate cancer cells by directly targeting PTEN in vivo and in vitro. Minerva Med. 2018, 109, 24–30. [Google Scholar] [CrossRef]
- Hoey, C.; Ahmed, M.; Fotouhi Ghiam, A.; Vesprini, D.; Huang, X.; Commisso, K.; Commisso, A.; Ray, J.; Fokas, E.; Loblaw, D.A.; et al. Circulating miRNAs as non-invasive biomarkers to predict aggressive prostate cancer after radical prostatectomy. J. Transl. Med. 2019, 17, 173. [Google Scholar] [CrossRef]
- Sharova, E.; Grassi, A.; Marcer, A.; Ruggero, K.; Pinto, F.; Bassi, P.; Zanovello, P.; Zattoni, F.; D’Agostino, D.M.; Iafrate, M.; et al. A circulating miRNA assay as a first-line test for prostate cancer screening. Br. J. Cancer 2016, 114, 1362–1366. [Google Scholar] [CrossRef]
- Alhasan, A.H.; Scott, A.W.; Wu, J.J.; Feng, G.; Meeks, J.J.; Thaxton, C.S.; Mirkin, C.A. Circulating microRNA signature for the diagnosis of very high-risk prostate cancer. Proc. Natl. Acad. Sci. USA 2016, 113, 10655–10660. [Google Scholar] [CrossRef]
- Wei, P.; Yang, J.; Zhang, D.; Cui, M.; Li, L. lncRNA HAND2-AS1 Regulates Prostate Cancer Cell Growth Through Targeting the miR-106a-5p/RBM24 Axis. Onco Targets Ther. 2020, 13, 4523–4531. [Google Scholar] [CrossRef]
- Xia, T.; Liao, Q.; Jiang, X.; Shao, Y.; Xiao, B.; Xi, Y.; Guo, J. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci. Rep. 2014, 4, 6088. [Google Scholar] [CrossRef]
- Zhang, G.; Zhou, H.; Xiao, H.; Liu, Z.; Tian, H.; Zhou, T. MicroRNA-92a functions as an oncogene in colorectal cancer by targeting PTEN. Dig. Dis. Sci. 2014, 59, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Liao, G.; Xiong, H.; Tang, J.; Li, Y.; Liu, Y. MicroRNA-92a Inhibits the Cell Viability and Metastasis of Prostate Cancer by Targeting SOX4. Technol. Cancer Res. Treat 2020, 19, 1533033820959354. [Google Scholar] [CrossRef] [PubMed]
- Xiaoli, Z.; Yawei, W.; Lianna, L.; Haifeng, L.; Hui, Z. Screening of Target Genes and Regulatory Function of miRNAs as Prognostic Indicators for Prostate Cancer. Med. Sci. Monit. 2015, 21, 3748–3759. [Google Scholar] [CrossRef][Green Version]
- Zhang, R.; Li, F.; Wang, Y.; Yao, M.; Chi, C. Prognostic value of microRNA-20b expression level in patients with prostate cancer. Histol. Histopathol. 2020, 35, 827–831. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Fang, Y.X.; Xue, J.L.; Chen, J.Z. Four microRNAs promote prostate cell proliferation with regulation of PTEN and its downstream signals in vitro. PLoS ONE 2013, 8, e75885. [Google Scholar] [CrossRef]
- Konoshenko, M.Y.; Lekchnov, E.A.; Bryzgunova, O.E.; Zaporozhchenko, I.A.; Yarmoschuk, S.V.; Pashkovskaya, O.A.; Pak, S.V.; Laktionov, P.P. The Panel of 12 Cell-Free MicroRNAs as Potential Biomarkers in Prostate Neoplasms. Diagnostics 2020, 10, 38. [Google Scholar] [CrossRef]
- Huo, W.; Qi, F.; Wang, K. Long non-coding RNA FER1L4 inhibits prostate cancer progression via sponging miR-92a-3p and upregulation of FBXW7. Cancer Cell Int. 2020, 20, 64. [Google Scholar] [CrossRef]
- Buechner, J.; Tomte, E.; Haug, B.H.; Henriksen, J.R.; Lokke, C.; Flaegstad, T.; Einvik, C. Tumour-suppressor microRNAs let-7 and mir-101 target the proto-oncogene MYCN and inhibit cell proliferation in MYCN-amplified neuroblastoma. Br. J. Cancer 2011, 105, 296–303. [Google Scholar] [CrossRef]
- Olive, V.; Bennett, M.J.; Walker, J.C.; Ma, C.; Jiang, I.; Cordon-Cardo, C.; Li, Q.J.; Lowe, S.W.; Hannon, G.J.; He, L. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009, 23, 2839–2849. [Google Scholar] [CrossRef]
- Fan, Y.; Yin, S.; Hao, Y.; Yang, J.; Zhang, H.; Sun, C.; Ma, M.; Chang, Q.; Xi, J.J. miR-19b promotes tumor growth and metastasis via targeting TP53. Rna 2014, 20, 765–772. [Google Scholar] [CrossRef]
- Osip’yants, A.I.; Knyazev, E.N.; Galatenko, A.V.; Nyushko, K.M.; Galatenko, V.V.; Shkurnikov, M.Y.; Alekseev, B.Y. Changes in the Level of Circulating hsa-miR-297 and hsa-miR-19b-3p miRNA Are Associated with Generalization of Prostate Cancer. Bull. Exp. Biol. Med. 2017, 162, 379–382. [Google Scholar] [CrossRef]
- Stuopelyte, K.; Daniunaite, K.; Jankevicius, F.; Jarmalaite, S. Detection of miRNAs in urine of prostate cancer patients. Medicina 2016, 52, 116–124. [Google Scholar] [CrossRef]
- Duca, R.B.; Massillo, C.; Dalton, G.N.; Farré, P.L.; Graña, K.D.; Gardner, K.; De Siervi, A. MiR-19b-3p and miR-101-3p as potential biomarkers for prostate cancer diagnosis and prognosis. Am. J. Cancer Res. 2021, 11, 2802–2820. [Google Scholar]
- Li, D.; Ilnytskyy, Y.; Kovalchuk, A.; Khachigian, L.M.; Bronson, R.T.; Wang, B.; Kovalchuk, O. Crucial role for early growth response-1 in the transcriptional regulation of miR-20b in breast cancer. Oncotarget 2013, 4, 1373–1387. [Google Scholar] [CrossRef]
- Guo, J.; Xiao, Z.; Yu, X.; Cao, R. miR-20b promotes cellular proliferation and migration by directly regulating phosphatase and tensin homolog in prostate cancer. Oncol. Lett. 2017, 14, 6895–6900. [Google Scholar] [CrossRef]
- Qi, J.C.; Yang, Z.; Zhang, Y.P.; Lu, B.S.; Yin, Y.W.; Liu, K.L.; Xue, W.Y.; Qu, C.B.; Li, W. miR-20b-5p, TGFBR2, and E2F1 Form a Regulatory Loop to Participate in Epithelial to Mesenchymal Transition in Prostate Cancer. Front. Oncol. 2019, 9, 1535. [Google Scholar] [CrossRef]
- Pan, Z.; Mo, F.; Liu, H.; Zeng, J.; Huang, K.; Huang, S.; Cao, Z.; Xu, X.; Xu, J.; Liu, T.; et al. LncRNA prostate androgen-regulated transcript 1 (PART 1) functions as an oncogene in osteosarcoma via sponging miR-20b-5p to upregulate BAMBI. Ann. Transl. Med. 2021, 9, 488. [Google Scholar] [CrossRef]
- Pashaei, E.; Ahmady, M.; Ozen, M.; Aydin, N. Meta-analysis of miRNA expression profiles for prostate cancer recurrence following radical prostatectomy. PLoS ONE 2017, 12, e0179543. [Google Scholar] [CrossRef]
- Chen, Y.; Lu, X.; Wu, B.; Su, Y.; Li, J.; Wang, H. MicroRNA 363 mediated positive regulation of c-myc translation affect prostate cancer development and progress. Neoplasma 2015, 62, 191–198. [Google Scholar] [CrossRef]
- Liang, H.; Studach, L.; Hullinger, R.L.; Xie, J.; Andrisani, O.M. Down-regulation of RE-1 silencing transcription factor (REST) in advanced prostate cancer by hypoxia-induced miR-106b~25. Exp. Cell Res. 2014, 320, 188–199. [Google Scholar] [CrossRef]
- Cai, K.; Wang, Y.; Bao, X. MiR-106b promotes cell proliferation via targeting RB in laryngeal carcinoma. J. Exp. Clin. Cancer Res. 2011, 30, 73. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.S.; Yang, X.H.; Chen, X.; Wang, X.D.; Hua, J.; Zhou, D.L.; Zhou, B.; Song, Z.S. MicroRNA-106b in cancer-associated fibroblasts from gastric cancer promotes cell migration and invasion by targeting PTEN. FEBS Lett. 2014, 588, 2162–2169. [Google Scholar] [CrossRef] [PubMed]
- Song, C.J.; Chen, H.; Chen, L.Z.; Ru, G.M.; Guo, J.J.; Ding, Q.N. The potential of microRNAs as human prostate cancer biomarkers: A meta-analysis of related studies. J. Cell Biochem. 2018, 119, 2763–2786. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Chen, J.; Wang, G.; Zhang, D. MicroRNA106b functions as an oncogene and regulates tumor viability and metastasis by targeting LARP4B in prostate cancer. Mol. Med. Rep. 2019, 20, 951–958. [Google Scholar] [CrossRef]
- Poliseno, L.; Salmena, L.; Riccardi, L.; Fornari, A.; Song, M.S.; Hobbs, R.M.; Sportoletti, P.; Varmeh, S.; Egia, A.; Fedele, G.; et al. Identification of the miR-106b~25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci. Signal. 2010, 3, ra29. [Google Scholar] [CrossRef]
- Hudson, R.S.; Yi, M.; Esposito, D.; Glynn, S.A.; Starks, A.M.; Yang, Y.; Schetter, A.J.; Watkins, S.K.; Hurwitz, A.A.; Dorsey, T.H.; et al. MicroRNA-106b-25 cluster expression is associated with early disease recurrence and targets caspase-7 and focal adhesion in human prostate cancer. Oncogene 2013, 32, 4139–4147. [Google Scholar] [CrossRef]
- Li, B.; Shi, X.B.; Nori, D.; Chao, C.K.; Chen, A.M.; Valicenti, R.; White Rde, V. Down-regulation of microRNA 106b is involved in p21-mediated cell cycle arrest in response to radiation in prostate cancer cells. Prostate 2011, 71, 567–574. [Google Scholar] [CrossRef]
- Fu, X.; Tian, J.; Zhang, L.; Chen, Y.; Hao, Q. Involvement of microRNA-93, a new regulator of PTEN/Akt signaling pathway, in regulation of chemotherapeutic drug cisplatin chemosensitivity in ovarian cancer cells. FEBS Lett. 2012, 586, 1279–1286. [Google Scholar] [CrossRef]
- Pudova, E.A.; Krasnov, G.S.; Nyushko, K.M.; Kobelyatskaya, A.A.; Savvateeva, M.V.; Poloznikov, A.A.; Dolotkazin, D.R.; Klimina, K.M.; Guvatova, Z.G.; Simanovsky, S.A.; et al. miRNAs expression signature potentially associated with lymphatic dissemination in locally advanced prostate cancer. BMC Med. Genom. 2020, 13, 129. [Google Scholar] [CrossRef]
- Liu, J.-J.; Zhang, X.; Wu, X.-H. miR-93 Promotes the Growth and Invasion of Prostate Cancer by Upregulating Its Target Genes TGFBR2, ITGB8, and LATS2. Mol. Ther.-Oncolytics 2018, 11, 14–19. [Google Scholar] [CrossRef]
- Choi, N.; Park, J.; Lee, J.-S.; Yoe, J.; Park, G.Y.; Kim, E.; Jeon, H.; Cho, Y.M.; Roh, T.-Y.; Lee, Y. miR-93/miR-106b/miR-375-CIC-CRABP1: A novel regulatory axis in prostate cancer progression. Oncotarget 2015, 6, 23533. [Google Scholar] [CrossRef]
- Wang, C.; Tian, S.; Zhang, D.; Deng, J.; Cai, H.; Shi, C.; Yang, W. Increased expression of microRNA-93 correlates with progression and prognosis of prostate cancer. Medicine (Baltimore) 2020, 99, e18432. [Google Scholar] [CrossRef]
- Barceló, M.; Castells, M.; Pérez-Riba, M.; Bassas, L.; Vigués, F.; Larriba, S. Seminal plasma microRNAs improve diagnosis/prognosis of prostate cancer in men with moderately altered prostate-specific antigen. Am. J. Transl. Res. 2020, 12, 2041–2051. [Google Scholar]
- Martínez-González, L.J.; Sánchez-Conde, V.; González-Cabezuelo, J.M.; Antunez-Rodríguez, A.; Andrés-León, E.; Robles-Fernandez, I.; Lorente, J.A.; Vázquez-Alonso, F.; Alvarez-Cubero, M.J. Identification of MicroRNAs as Viable Aggressiveness Biomarkers for Prostate Cancer. Biomedicines 2021, 9, 646. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, C.; Zou, X.; Geng, X.; Zhou, X.; Fan, X.; Zhu, D.; Zhang, H.; Zhu, W. MicroRNA panel in serum reveals novel diagnostic biomarkers for prostate cancer. PeerJ 2021, 9, e11441. [Google Scholar] [CrossRef]
- Li, Z.; Sun, Y.; Chen, X.; Squires, J.; Nowroozizadeh, B.; Liang, C.; Huang, J. p53 Mutation Directs AURKA Overexpression via miR-25 and FBXW7 in Prostatic Small Cell Neuroendocrine Carcinoma. Mol. Cancer Res. 2015, 13, 584–591. [Google Scholar] [CrossRef]
- Esposito, F.; Tornincasa, M.; Pallante, P.; Federico, A.; Borbone, E.; Pierantoni, G.M.; Fusco, A. Down-regulation of the miR-25 and miR-30d contributes to the development of anaplastic thyroid carcinoma targeting the polycomb protein EZH2. J. Clin. Endocrinol. Metab. 2012, 97, E710–E718. [Google Scholar] [CrossRef]
- Kumar, M.; Lu, Z.; Takwi, A.A.; Chen, W.; Callander, N.S.; Ramos, K.S.; Young, K.H.; Li, Y. Negative regulation of the tumor suppressor p53 gene by microRNAs. Oncogene 2011, 30, 843–853. [Google Scholar] [CrossRef]
- Zoni, E.; van der Horst, G.; van de Merbel, A.F.; Chen, L.; Rane, J.K.; Pelger, R.C.M.; Collins, A.T.; Visakorpi, T.; Snaar-Jagalska, B.E.; Maitland, N.J.; et al. miR-25 Modulates Invasiveness and Dissemination of Human Prostate Cancer Cells via Regulation of αv- and α6- Integrin Expression. Cancer Res. 2015, 75, 2326–2336. [Google Scholar] [CrossRef]
- Srivastava, A.; Goldberger, H.; Dimtchev, A.; Marian, C.; Soldin, O.; Li, X.; Collins, S.P.; Suy, S.; Kumar, D. Circulatory miR-628-5p is downregulated in prostate cancer patients. Tumour. Biol. 2014, 35, 4867–4873. [Google Scholar] [CrossRef]
- Cai, K.; Wan, Y.; Sun, G.; Shi, L.; Bao, X.; Wang, Z. Let-7a inhibits proliferation and induces apoptosis by targeting EZH2 in nasopharyngeal carcinoma cells. Oncol. Rep. 2012, 28, 2101–2106. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Yang, C.; Yang, Q.; Ding, H.; Jia, J.; Guo, J.; Wang, J.; Wang, Z. Deregulation of let-7e in epithelial ovarian cancer promotes the development of resistance to cisplatin. Oncogenesis 2013, 2, e75. [Google Scholar] [CrossRef] [PubMed]
- Murray, M.J.; Saini, H.K.; Siegler, C.A.; Hanning, J.E.; Barker, E.M.; van Dongen, S.; Ward, D.M.; Raby, K.L.; Groves, I.J.; Scarpini, C.G.; et al. LIN28 Expression in malignant germ cell tumors downregulates let-7 and increases oncogene levels. Cancer Res. 2013, 73, 4872–4884. [Google Scholar] [CrossRef] [PubMed]
- Cimadamore, F.; Amador-Arjona, A.; Chen, C.; Huang, C.-T.; Terskikh, A.V. SOX2–LIN28/let-7 pathway regulates proliferation and neurogenesis in neural precursors. Proc. Natl. Acad. Sci. USA 2013, 110, E3017–E3026. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.; Heath, E.; Chen, W.; Cher, M.L.; Powell, I.; Heilbrun, L.; Li, Y.; Ali, S.; Sethi, S.; Hassan, O.; et al. Loss of Let-7 Up-Regulates EZH2 in Prostate Cancer Consistent with the Acquisition of Cancer Stem Cell Signatures That Are Attenuated by BR-DIM. PLoS ONE 2012, 7, e33729. [Google Scholar] [CrossRef]
- Schubert, M.; Spahn, M.; Kneitz, S.; Scholz, C.J.; Joniau, S.; Stroebel, P.; Riedmiller, H.; Kneitz, B. Distinct microRNA Expression Profile in Prostate Cancer Patients with Early Clinical Failure and the Impact of let-7 as Prognostic Marker in High-Risk Prostate Cancer. PLoS ONE 2013, 8, e65064. [Google Scholar] [CrossRef]
- Costanzi, E.; Romani, R.; Scarpelli, P.; Bellezza, I. Extracellular Vesicles-Mediated Transfer of miRNA Let-7b from PC3 Cells to Macrophages. Genes 2020, 11, 1495. [Google Scholar] [CrossRef]
- Guelfi, G.; Cochetti, G.; Stefanetti, V.; Zampini, D.; Diverio, S.; Boni, A.; Mearini, E. Next Generation Sequencing of urine exfoliated cells: An approach of prostate cancer microRNAs research. Sci. Rep. 2018, 8, 7111. [Google Scholar] [CrossRef]
- Xiao, G.a.; Yao, J.; Kong, D.; Ye, C.; Chen, R.; Li, L.; Zeng, T.; Wang, L.; Zhang, W.; Shi, X.; et al. The Long Noncoding RNA TTTY15, Which Is Located on the Y Chromosome, Promotes Prostate Cancer Progression by Sponging let-7. Eur. Urol. 2019, 76, 315–326. [Google Scholar] [CrossRef]
- Nadiminty, N.; Tummala, R.; Lou, W.; Zhu, Y.; Shi, X.-B.; Zou, J.X.; Chen, H.; Zhang, J.; Chen, X.; Luo, J.; et al. MicroRNA let-7c Is Downregulated in Prostate Cancer and Suppresses Prostate Cancer Growth. PLoS ONE 2012, 7, e32832. [Google Scholar] [CrossRef]
- Pernicová, Z.; Slabáková, E.; Fedr, R.; Šimečková, Š.; Jaroš, J.; Suchánková, T.; Bouchal, J.; Kharaishvili, G.; Král, M.; Kozubík, A.; et al. The role of high cell density in the promotion of neuroendocrine transdifferentiation of prostate cancer cells. Mol. Cancer 2014, 13, 113. [Google Scholar] [CrossRef]
- Akoto, T.; Bhagirath, D.; Saini, S. MicroRNAs in treatment-induced neuroendocrine differentiation in prostate cancer. Cancer Drug Resist. 2020, 3, 804–818. [Google Scholar] [CrossRef]
- Chen, J.; Bardes, E.E.; Aronow, B.J.; Jegga, A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009, 37, W305–W311. [Google Scholar] [CrossRef]
- Kong, Q.; Chen, X.S.; Tian, T.; Xia, X.Y.; Xu, P. MicroRNA-194 suppresses prostate cancer migration and invasion by downregulating human nuclear distribution protein. Oncol. Rep. 2017, 37, 803–812. [Google Scholar] [CrossRef]
- Gao, S.; Zhao, Z.; Wu, R.; Wu, L.; Tian, X.; Zhang, Z. MicroRNA-194 regulates cell viability and apoptosis by targeting CDH2 in prostatic cancer. Onco Targets Ther. 2018, 11, 4837–4844. [Google Scholar] [CrossRef]
- Li, Z.; Li, L.X.; Diao, Y.J.; Wang, J.; Ye, Y.; Hao, X.K. Identification of Urinary Exosomal miRNAs for the Non-Invasive Diagnosis of Prostate Cancer. Cancer Manag. Res. 2021, 13, 25–35. [Google Scholar] [CrossRef]
- Konoshenko, M.Y.; Bryzgunova, O.E.; Lekchnov, E.A.; Amelina, E.V.; Yarmoschuk, S.V.; Pak, S.V.; Laktionov, P.P. The Influence of Radical Prostatectomy on the Expression of Cell-Free MiRNA. Diagnostics 2020, 10, 600. [Google Scholar] [CrossRef]
- Mosquera, J.M.; Beltran, H.; Park, K.; MacDonald, T.Y.; Robinson, B.D.; Tagawa, S.T.; Perner, S.; Bismar, T.A.; Erbersdobler, A.; Dhir, R.; et al. Concurrent AURKA and MYCN gene amplifications are harbingers of lethal treatment-related neuroendocrine prostate cancer. Neoplasia 2013, 15, 1–10. [Google Scholar] [CrossRef]
- Kishore, S.; Jaskiewicz, L.; Burger, L.; Hausser, J.; Khorshid, M.; Zavolan, M. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat. Methods 2011, 8, 559–564. [Google Scholar] [CrossRef]
- Riley, K.J.; Rabinowitz, G.S.; Yario, T.A.; Luna, J.M.; Darnell, R.B.; Steitz, J.A. EBV and human microRNAs co-target oncogenic and apoptotic viral and human genes during latency. Embo J 2012, 31, 2207–2221. [Google Scholar] [CrossRef]
- Heo, I.; Joo, C.; Kim, Y.K.; Ha, M.; Yoon, M.J.; Cho, J.; Yeom, K.H.; Han, J.; Kim, V.N. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 2009, 138, 696–708. [Google Scholar] [CrossRef] [PubMed]
- Mogilyansky, E.; Rigoutsos, I. The miR-17/92 cluster: A comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013, 20, 1603–1614. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wu, Z.; Zhou, H.; Cai, W.; Li, X.; Hu, J.; Gao, L.; Feng, T.; Wang, L.; Peng, X.; et al. The SOX4/miR-17-92/RB1 Axis Promotes Prostate Cancer Progression. Neoplasia 2019, 21, 765–776. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.L.; Sood, A.; Rahimi, H.A.; Wang, W.; Gupta, N.; Hicks, J.; Mosier, S.; Gocke, C.D.; Epstein, J.I.; Netto, G.J.; et al. Rb loss is characteristic of prostatic small cell neuroendocrine carcinoma. Clin. Cancer Res. 2014, 20, 890–903. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Li, H.; He, Y. MicroRNA-17 downregulates expression of the PTEN gene to promote the occurrence and development of adenomyosis. Exp. Ther. Med. 2017, 14, 3805–3811. [Google Scholar] [CrossRef] [PubMed]
- Dong, P.; Xiong, Y.; Yu, J.; Chen, L.; Tao, T.; Yi, S.; Hanley, S.J.B.; Yue, J.; Watari, H.; Sakuragi, N. Control of PD-L1 expression by miR-140/142/340/383 and oncogenic activation of the OCT4-miR-18a pathway in cervical cancer. Oncogene 2018, 37, 5257–5268. [Google Scholar] [CrossRef]
- Liang, Z.; Li, Y.; Huang, K.; Wagar, N.; Shim, H. Regulation of miR-19 to breast cancer chemoresistance through targeting PTEN. Pharm Res. 2011, 28, 3091–3100. [Google Scholar] [CrossRef]
- Wang, F.; Li, T.; Zhang, B.; Li, H.; Wu, Q.; Yang, L.; Nie, Y.; Wu, K.; Shi, Y.; Fan, D. MicroRNA-19a/b regulates multidrug resistance in human gastric cancer cells by targeting PTEN. Biochem. Biophys Res. Commun. 2013, 434, 688–694. [Google Scholar] [CrossRef]
- Trompeter, H.I.; Abbad, H.; Iwaniuk, K.M.; Hafner, M.; Renwick, N.; Tuschl, T.; Schira, J.; Muller, H.W.; Wernet, P. MicroRNAs MiR-17, MiR-20a, and MiR-106b act in concert to modulate E2F activity on cell cycle arrest during neuronal lineage differentiation of USSC. PLoS ONE 2011, 6, e16138. [Google Scholar] [CrossRef]
- Jiang, Y.; Chang, H.; Chen, G. Effects of microRNA-20a on the proliferation, migration and apoptosis of multiple myeloma via the PTEN/PI3K/AKT signaling pathway. Oncol. Lett. 2018, 15, 10001–10007. [Google Scholar] [CrossRef]
- Lewis, B.P.; Shih, I.H.; Jones-Rhoades, M.W.; Bartel, D.P.; Burge, C.B. Prediction of mammalian microRNA targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef]
- Lu, W.D.; Zuo, Y.; Xu, Z.; Zhang, M. MiR-19a promotes epithelial-mesenchymal transition through PI3K/AKT pathway in gastric cancer. World J. Gastroenterol. 2015, 21, 4564–4573. [Google Scholar] [CrossRef]
- Wiesehofer, M.; Czyrnik, E.D.; Spahn, M.; Ting, S.; Reis, H.; Dankert, J.T.; Wennemuth, G. Increased Expression of AKT3 in Neuroendocrine Differentiated Prostate Cancer Cells Alters the Response Towards Anti-Androgen Treatment. Cancers 2021, 13, 578. [Google Scholar] [CrossRef]
- Jiao, L.; Deng, Z.; Xu, C.; Yu, Y.; Li, Y.; Yang, C.; Chen, J.; Liu, Z.; Huang, G.; Li, L.C.; et al. miR-663 induces castration-resistant prostate cancer transformation and predicts clinical recurrence. J. Cell Physiol. 2014, 229, 834–844. [Google Scholar] [CrossRef]
- Cho, J.G.; Park, S.; Lim, C.H.; Kim, H.S.; Song, S.Y.; Roh, T.Y.; Sung, J.H.; Suh, W.; Ham, S.J.; Lim, K.H.; et al. ZNF224, Kruppel like zinc finger protein, induces cell growth and apoptosis-resistance by down-regulation of p21 and p53 via miR-663a. Oncotarget 2016, 7, 31177–31190. [Google Scholar] [CrossRef]
- Sadeghi, M.; Ranjbar, B.; Ganjalikhany, M.R.; Khan, F.M.; Schmitz, U.; Wolkenhauer, O.; Gupta, S.K. MicroRNA and Transcription Factor Gene Regulatory Network Analysis Reveals Key Regulatory Elements Associated with Prostate Cancer Progression. PLoS ONE 2016, 11, e0168760. [Google Scholar] [CrossRef]
- Dang, Q.; Li, L.; Xie, H.; He, D.; Chen, J.; Song, W.; Chang, L.S.; Chang, H.C.; Yeh, S.; Chang, C. Anti-androgen enzalutamide enhances prostate cancer neuroendocrine (NE) differentiation via altering the infiltrated mast cells → androgen receptor (AR) → miRNA32 signals. Mol. Oncol. 2015, 9, 1241–1251. [Google Scholar] [CrossRef]
- Jalava, S.E.; Urbanucci, A.; Latonen, L.; Waltering, K.K.; Sahu, B.; Janne, O.A.; Seppala, J.; Lahdesmaki, H.; Tammela, T.L.; Visakorpi, T. Androgen-regulated miR-32 targets BTG2 and is overexpressed in castration-resistant prostate cancer. Oncogene 2012, 31, 4460–4471. [Google Scholar] [CrossRef]
- Zhang, L.; Li, X.; Chao, Y.; He, R.; Liu, J.; Yuan, Y.; Zhao, W.; Han, C.; Song, X. KLF4, a miR-32-5p targeted gene, promotes cisplatin-induced apoptosis by upregulating BIK expression in prostate cancer. Cell Commun. Signal. 2018, 16, 53. [Google Scholar] [CrossRef]
- Latonen, L.; Scaravilli, M.; Gillen, A.; Hartikainen, S.; Zhang, F.P.; Ruusuvuori, P.; Kujala, P.; Poutanen, M.; Visakorpi, T. In Vivo Expression of miR-32 Induces Proliferation in Prostate Epithelium. Am. J. Pathol. 2017, 187, 2546–2557. [Google Scholar] [CrossRef]
- Liao, H.; Xiao, Y.; Hu, Y.; Yin, Z.; Liu, L. microRNA-32 induces radioresistance by targeting DAB2IP and regulating autophagy in prostate cancer cells. Oncol. Lett. 2015, 10, 2055–2062. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhu, G.; Chai, J.; Ma, L.; Duan, H.; Zhang, H. Downregulated microRNA-32 expression induced by high glucose inhibits cell cycle progression via PTEN upregulation and Akt inactivation in bone marrow-derived mesenchymal stem cells. Biochem. Biophys Res. Commun. 2013, 433, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.; Heath, E.; Chen, W.; Cher, M.; Powell, I.; Heilbrun, L.; Li, Y.; Ali, S.; Sethi, S.; Hassan, O.; et al. Epigenetic silencing of miR-34a in human prostate cancer cells and tumor tissue specimens can be reversed by BR-DIM treatment. Am. J. Transl. Res. 2012, 4, 14–23. [Google Scholar] [PubMed]
- Chen, W.Y.; Liu, S.Y.; Chang, Y.S.; Yin, J.J.; Yeh, H.L.; Mouhieddine, T.H.; Hadadeh, O.; Abou-Kheir, W.; Liu, Y.N. MicroRNA-34a regulates WNT/TCF7 signaling and inhibits bone metastasis in Ras-activated prostate cancer. Oncotarget 2015, 6, 441–457. [Google Scholar] [CrossRef]
- Shu, Y.; Ren, L.; Xie, B.; Liang, Z.; Chen, J.M. MiR-204 enhances mitochondrial apoptosis in doxorubicin-treated prostate cancer cells by targeting SIRT1/p53 pathway. Oncotarget 2017, 8, 97313–97322. [Google Scholar] [CrossRef]
- Wa, Q.; Huang, S.; Pan, J.; Tang, Y.; He, S.; Fu, X.; Peng, X.; Chen, X.; Yang, C.; Ren, D.; et al. miR-204-5p Represses Bone Metastasis via Inactivating NF-kappaB Signaling in Prostate Cancer. Mol. Ther. Nucleic Acids 2019, 18, 567–579. [Google Scholar] [CrossRef]
- Kashat, M.; Azzouz, L.; Sarkar, S.H.; Kong, D.; Li, Y.; Sarkar, F.H. Inactivation of AR and Notch-1 signaling by miR-34a attenuates prostate cancer aggressiveness. Am. J. Transl. Res. 2012, 4, 432–442. [Google Scholar]
- Wei, J.S.; Song, Y.K.; Durinck, S.; Chen, Q.R.; Cheuk, A.T.C.; Tsang, P.; Zhang, Q.; Thiele, C.J.; Slack, A.; Shohet, J.; et al. The MYCN oncogene is a direct target of miR-34a. Oncogene 2008, 27, 5204–5213. [Google Scholar] [CrossRef]
- Choi, Y.J.; Lin, C.P.; Ho, J.J.; He, X.; Okada, N.; Bu, P.; Zhong, Y.; Kim, S.Y.; Bennett, M.J.; Chen, C.; et al. miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat. Cell Biol. 2011, 13, 1353–1360. [Google Scholar] [CrossRef]
- Javeri, A.; Ghaffarpour, M.; Taha, M.F.; Houshmand, M. Downregulation of miR-34a in breast tumors is not associated with either p53 mutations or promoter hypermethylation while it correlates with metastasis. Med. Oncol 2013, 30, 413. [Google Scholar] [CrossRef]
- Zheng, C.; Yinghao, S.; Li, J. MiR-221 expression affects invasion potential of human prostate carcinoma cell lines by targeting DVL2. Med. Oncol. 2012, 29, 815–822. [Google Scholar] [CrossRef]
- Kiener, M.; Chen, L.; Krebs, M.; Grosjean, J.; Klima, I.; Kalogirou, C.; Riedmiller, H.; Kneitz, B.; Thalmann, G.N.; Snaar-Jagalska, E.; et al. miR-221-5p regulates proliferation and migration in human prostate cancer cells and reduces tumor growth in vivo. BMC Cancer 2019, 19, 627. [Google Scholar] [CrossRef]
- Sun, T.; Du, S.-Y.; Armenia, J.; Qu, F.; Fan, J.; Wang, X.; Fei, T.; Komura, K.; Liu, S.X.; Lee, G.-S.M.; et al. Expression of lncRNA MIR222HG co-transcribed from the miR-221/222 gene promoter facilitates the development of castration-resistant prostate cancer. Oncogenesis 2018, 7, 30. [Google Scholar] [CrossRef]
- Xuan, H.; Xue, W.; Pan, J.; Sha, J.; Dong, B.; Huang, Y. Downregulation of miR-221, -30d, and -15a contributes to pathogenesis of prostate cancer by targeting Bmi-1. Biochemistry (Moscow) 2015, 80, 276–283. [Google Scholar] [CrossRef]
- Lupini, L.; Bassi, C.; Ferracin, M.; Bartonicek, N.; D’Abundo, L.; Zagatti, B.; Callegari, E.; Musa, G.; Moshiri, F.; Gramantieri, L.; et al. miR-221 affects multiple cancer pathways by modulating the level of hundreds messenger RNAs. Front. Genet. 2013, 4, 64. [Google Scholar] [CrossRef]
- Garofalo, M.; Di Leva, G.; Romano, G.; Nuovo, G.; Suh, S.-S.; Ngankeu, A.; Taccioli, C.; Pichiorri, F.; Alder, H.; Secchiero, P.; et al. miR-221&222 Regulate TRAIL Resistance and Enhance Tumorigenicity through PTEN and TIMP3 Downregulation. Cancer Cell 2009, 16, 498–509. [Google Scholar] [CrossRef]
- Boyle, G.M.; Woods, S.L.; Bonazzi, V.F.; Stark, M.S.; Hacker, E.; Aoude, L.G.; Dutton-Regester, K.; Cook, A.L.; Sturm, R.A.; Hayward, N.K. Melanoma cell invasiveness is regulated by miR-211 suppression of the BRN2 transcription factor. Pigment. Cell Melanoma Res. 2011, 24, 525–537. [Google Scholar] [CrossRef]
- Kumar, B.; Khaleghzadegan, S.; Mears, B.; Hatano, K.; Kudrolli, T.A.; Chowdhury, W.H.; Yeater, D.B.; Ewing, C.M.; Luo, J.; Isaacs, W.B.; et al. Identification of miR-30b-3p and miR-30d-5p as direct regulators of androgen receptor signaling in prostate cancer by complementary functional microRNA library screening. Oncotarget 2016, 7, 72593–72607. [Google Scholar] [CrossRef]
- Lin, P.C.; Chiu, Y.L.; Banerjee, S.; Park, K.; Mosquera, J.M.; Giannopoulou, E.; Alves, P.; Tewari, A.K.; Gerstein, M.B.; Beltran, H.; et al. Epigenetic repression of miR-31 disrupts androgen receptor homeostasis and contributes to prostate cancer progression. Cancer Res. 2013, 73, 1232–1244. [Google Scholar] [CrossRef]
- Ostano, P.; Mello-Grand, M.; Sesia, D.; Gregnanin, I.; Peraldo-Neia, C.; Guana, F.; Jachetti, E.; Farsetti, A.; Chiorino, G. Gene Expression Signature Predictive of Neuroendocrine Transformation in Prostate Adenocarcinoma. Int. J. Mol. Sci. 2020, 21, 1078. [Google Scholar] [CrossRef]
- Coarfa, C.; Fiskus, W.; Eedunuri, V.K.; Rajapakshe, K.; Foley, C.; Chew, S.A.; Shah, S.S.; Geng, C.; Shou, J.; Mohamed, J.S.; et al. Comprehensive proteomic profiling identifies the androgen receptor axis and other signaling pathways as targets of microRNAs suppressed in metastatic prostate cancer. Oncogene 2016, 35, 2345–2356. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, C.E.; Sulpice, E.; Combe, S.; Shibakawa, A.; Leach, D.A.; Hamilton, M.P.; Chrysostomou, S.L.; Sharp, A.; Welti, J.; Yuan, W.; et al. Androgen receptor-modulatory microRNAs provide insight into therapy resistance and therapeutic targets in advanced prostate cancer. Oncogene 2019, 38, 5700–5724. [Google Scholar] [CrossRef] [PubMed]
- Ebron, J.S.; Shankar, E.; Singh, J.; Sikand, K.; Weyman, C.M.; Gupta, S.; Lindner, D.J.; Liu, X.; Campbell, M.J.; Shukla, G.C. MiR-644a Disrupts Oncogenic Transformation and Warburg Effect by Direct Modulation of Multiple Genes of Tumor-Promoting Pathways. Cancer Res. 2019, 79, 1844–1856. [Google Scholar] [CrossRef] [PubMed]
- Qu, H.W.; Jin, Y.; Cui, Z.L.; Jin, X.B. MicroRNA-373-3p inhibits prostate cancer progression by targeting AKT1. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 6252–6259. [Google Scholar] [CrossRef]
- Josson, S.; Gururajan, M.; Hu, P.; Shao, C.; Chu, G.Y.; Zhau, H.E.; Liu, C.; Lao, K.; Lu, C.L.; Lu, Y.T.; et al. miR-409-3p/-5p promotes tumorigenesis, epithelial-to-mesenchymal transition, and bone metastasis of human prostate cancer. Clin. Cancer Res. 2014, 20, 4636–4646. [Google Scholar] [CrossRef]
- Josson, S.; Gururajan, M.; Sung, S.Y.; Hu, P.; Shao, C.; Zhau, H.E.; Liu, C.; Lichterman, J.; Duan, P.; Li, Q.; et al. Stromal fibroblast-derived miR-409 promotes epithelial-to-mesenchymal transition and prostate tumorigenesis. Oncogene 2015, 34, 2690–2699. [Google Scholar] [CrossRef]
- Yang, K.; Handorean, A.M.; Iczkowski, K.A. MicroRNAs 373 and 520c are downregulated in prostate cancer, suppress CD44 translation and enhance invasion of prostate cancer cells in vitro. Int. J. Clin. Exp. Pathol. 2009, 2, 361–369. [Google Scholar]
- Huang, Q.; Ma, B.; Su, Y.; Chan, K.; Qu, H.; Huang, J.; Wang, D.; Qiu, J.; Liu, H.; Yang, X.; et al. miR-197-3p Represses the Proliferation of Prostate Cancer by Regulating the VDAC1/AKT/beta-catenin Signaling Axis. Int. J. Biol. Sci. 2020, 16, 1417–1426. [Google Scholar] [CrossRef]
- Yang, J.; Song, Q.; Cai, Y.; Wang, P.; Wang, M.; Zhang, D. RLIP76-dependent suppression of PI3K/AKT/Bcl-2 pathway by miR-101 induces apoptosis in prostate cancer. Biochem. Biophys Res. Commun. 2015, 463, 900–906. [Google Scholar] [CrossRef]
- Wu, X.; Bhayani, M.K.; Dodge, C.T.; Nicoloso, M.S.; Chen, Y.; Yan, X.; Adachi, M.; Thomas, L.; Galer, C.E.; Jiffar, T.; et al. Coordinated targeting of the EGFR signaling axis by microRNA-27a *. Oncotarget 2013, 4, 1388–1398. [Google Scholar] [CrossRef]
- Fletcher, C.E.; Dart, D.A.; Sita-Lumsden, A.; Cheng, H.; Rennie, P.S.; Bevan, C.L. Androgen-regulated processing of the oncomir miR-27a, which targets Prohibitin in prostate cancer. Hum. Mol. Genet. 2012, 21, 3112–3127. [Google Scholar] [CrossRef]
- Wan, X.; Huang, W.; Yang, S.; Zhang, Y.; Zhang, P.; Kong, Z.; Li, T.; Wu, H.; Jing, F.; Li, Y. Androgen-induced miR-27A acted as a tumor suppressor by targeting MAP2K4 and mediated prostate cancer progression. Int. J. Biochem. Cell Biol. 2016, 79, 249–260. [Google Scholar] [CrossRef]
- Mo, W.; Zhang, J.; Li, X.; Meng, D.; Gao, Y.; Yang, S.; Wan, X.; Zhou, C.; Guo, F.; Huang, Y.; et al. Identification of Novel AR-Targeted MicroRNAs Mediating Androgen Signalling through Critical Pathways to Regulate Cell Viability in Prostate Cancer. PLoS ONE 2013, 8, e56592. [Google Scholar] [CrossRef]
- Barros-Silva, D.; Costa-Pinheiro, P.; Duarte, H.; Sousa, E.J.; Evangelista, A.F.; Graça, I.; Carneiro, I.; Martins, A.T.; Oliveira, J.; Carvalho, A.L.; et al. MicroRNA-27a-5p regulation by promoter methylation and MYC signaling in prostate carcinogenesis. Cell Death Dis. 2018, 9, 167. [Google Scholar] [CrossRef]
- Lin, S.-C.; Kao, C.-Y.; Lee, H.-J.; Creighton, C.J.; Ittmann, M.M.; Tsai, S.-J.; Tsai, S.Y.; Tsai, M.-J. Dysregulation of miRNAs-COUP-TFII-FOXM1-CENPF axis contributes to the metastasis of prostate cancer. Nat. Commun. 2016, 7, 11418. [Google Scholar] [CrossRef]
- Gao, W.; Hong, Z.; Huang, H.; Zhu, A.; Lin, S.; Cheng, C.; Zhang, X.; Zou, G.; Shi, Z. miR-27a in serum acts as biomarker for prostate cancer detection and promotes cell proliferation by targeting Sprouty2. Oncol. Lett. 2018, 16, 5291–5298. [Google Scholar] [CrossRef]
- Ku, A.; Fredsøe, J.; Sørensen, K.D.; Borre, M.; Evander, M.; Laurell, T.; Lilja, H.; Ceder, Y. High-Throughput and Automated Acoustic Trapping of Extracellular Vesicles to Identify microRNAs with Diagnostic Potential for Prostate Cancer. Front. Oncol. 2021, 11, 386. [Google Scholar] [CrossRef]
- Nam, R.K.; Wallis, C.J.D.; Amemiya, Y.; Benatar, T.; Seth, A. Identification of a Novel MicroRNA Panel Associated with Metastasis Following Radical Prostatectomy for Prostate Cancer. Anticancer Res. 2018, 38, 5027–5034. [Google Scholar] [CrossRef]
- Varambally, S.; Cao, Q.; Mani, R.S.; Shankar, S.; Wang, X.; Ateeq, B.; Laxman, B.; Cao, X.; Jing, X.; Ramnarayanan, K.; et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 2008, 322, 1695–1699. [Google Scholar] [CrossRef]
- Li, K.; Liu, C.; Zhou, B.; Bi, L.; Huang, H.; Lin, T.; Xu, K. Role of EZH2 in the Growth of Prostate Cancer Stem Cells Isolated from LNCaP Cells. Int. J. Mol. Sci. 2013, 14, 11981–11993. [Google Scholar] [CrossRef]
- Gu, Z.; You, Z.; Yang, Y.; Ding, R.; Wang, M.; Pu, J.; Chen, J. Inhibition of MicroRNA miR-101-3p on prostate cancer progression by regulating Cullin 4B (CUL4B) and PI3K/AKT/mTOR signaling pathways. Bioengineered 2021, 12, 4719–4735. [Google Scholar] [CrossRef] [PubMed]
- Antognelli, C.; Cecchetti, R.; Riuzzi, F.; Peirce, M.J.; Talesa, V.N. Glyoxalase 1 sustains the metastatic phenotype of prostate cancer cells via EMT control. J. Cell. Mol. Med. 2018, 22, 2865–2883. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Gu, X.; Zhao, Y.; Greene, S.; Sha, W.; Smoot, D.T.; Califano, J.; Wu, T.C.; Pang, X. Enforced expression of miR-101 inhibits prostate cancer cell growth by modulating the COX-2 pathway in vivo. Cancer Prev. Res. 2011, 4, 1073–1083. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; You, S.; Nguyen, C.; Wang, Y.; Kim, J.; Sirohi, D.; Ziembiec, A.; Luthringer, D.; Lin, S.C.; Daskivich, T.; et al. Genes involved in prostate cancer progression determine MRI visibility. Theranostics 2018, 8, 1752–1765. [Google Scholar] [CrossRef]
- Lin, Y.; Chen, F.; Shen, L.; Tang, X.; Du, C.; Sun, Z.; Ding, H.; Chen, J.; Shen, B. Biomarker microRNAs for prostate cancer metastasis: Screened with a network vulnerability analysis model. J. Transl. Med. 2018, 16, 134. [Google Scholar] [CrossRef]
- Watahiki, A.; Wang, Y.; Morris, J.; Dennis, K.; O’Dwyer, H.M.; Gleave, M.; Gout, P.W.; Wang, Y. MicroRNAs Associated with Metastatic Prostate Cancer. PLoS ONE 2011, 6, e24950. [Google Scholar] [CrossRef]
- Chen, J.H.; Tong, W.; Pu, X.F.; Wang, J.Z. Long noncoding RNA CRNDE promotes proliferation, migration and invasion in prostate cancer through miR-101/Rap1A. Neoplasma 2020, 67, 584–594. [Google Scholar] [CrossRef]
- Cao, P.; Deng, Z.; Wan, M.; Huang, W.; Cramer, S.D.; Xu, J.; Lei, M.; Sui, G. MicroRNA-101 negatively regulates Ezh2 and its expression is modulated by androgen recept or and HIF-1α/HIF-1β. Mol. Cancer 2010, 9, 108. [Google Scholar] [CrossRef]
- Ramnarine, V.R.; Alshalalfa, M.; Mo, F.; Nabavi, N.; Erho, N.; Takhar, M.; Shukin, R.; Brahmbhatt, S.; Gawronski, A.; Kobelev, M.; et al. The long noncoding RNA landscape of neuroendocrine prostate cancer and its clinical implications. GigaScience 2018, 7, 1–23. [Google Scholar] [CrossRef]
- Luo, J.; Wang, K.; Yeh, S.; Sun, Y.; Liang, L.; Xiao, Y.; Xu, W.; Niu, Y.; Cheng, L.; Maity, S.N.; et al. LncRNA-p21 alters the antiandrogen enzalutamide-induced prostate cancer neuroendocrine differentiation via modulating the EZH2/STAT3 signaling. Nat. Commun. 2019, 10, 2571. [Google Scholar] [CrossRef]
- Işın, M.; Uysaler, E.; Özgür, E.; Köseoğlu, H.; Şanlı, Ö.; Yücel, Ö.B.; Gezer, U.; Dalay, N. Exosomal lncRNA-p21 levels may help to distinguish prostate cancer from benign disease. Front. Genet. 2015, 6, 168. [Google Scholar] [CrossRef]
- Liu, B.; Jiang, H.-Y.; Yuan, T.; Luo, J.; Zhou, W.-D.; Jiang, Q.-Q.; Wu, D. Enzalutamide-Induced Upregulation of PCAT6 Promotes Prostate Cancer Neuroendocrine Differentiation by Regulating miR-326/HNRNPA2B1 Axis. Front. Oncol. 2021, 11, 1803. [Google Scholar] [CrossRef]
- Lang, C.; Yin, C.; Lin, K.; Li, Y.; Yang, Q.; Wu, Z.; Du, H.; Ren, D.; Dai, Y.; Peng, X. m6A modification of lncRNA PCAT6 promotes bone metastasis in prostate cancer through IGF2BP2-mediated IGF1R mRNA stabilization. Clin. Transl. Med. 2021, 11, e426. [Google Scholar] [CrossRef]
- Li, Y.; Li, H.; Wei, X. Long noncoding RNA LINC00261 suppresses prostate cancer tumorigenesis through upregulation of GATA6-mediated DKK3. Cancer Cell Int. 2020, 20, 474. [Google Scholar] [CrossRef]
- Chang, Y.-T.; Lin, T.-P.; Tang, J.-T.; Campbell, M.; Luo, Y.-L.; Lu, S.-Y.; Yang, C.-P.; Cheng, T.-Y.; Chang, C.-H.; Liu, T.-T.; et al. HOTAIR is a REST-regulated lncRNA that promotes neuroendocrine differentiation in castration resistant prostate cancer. Cancer Lett. 2018, 433, 43–52. [Google Scholar] [CrossRef]
- Mather, R.L.; Wang, Y.; Crea, F. Is HOTAIR really involved in neuroendocrine prostate cancer differentiation? Epigenomics 2018, 10, 1259–1261. [Google Scholar] [CrossRef]
- Zhang, A.; Zhao, J.C.; Kim, J.; Fong, K.W.; Yang, Y.A.; Chakravarti, D.; Mo, Y.Y.; Yu, J. LncRNA HOTAIR Enhances the Androgen-Receptor-Mediated Transcriptional Program and Drives Castration-Resistant Prostate Cancer. Cell Rep. 2015, 13, 209–221. [Google Scholar] [CrossRef]
- Sowalsky, A.G.; Xia, Z.; Wang, L.; Zhao, H.; Chen, S.; Bubley, G.J.; Balk, S.P.; Li, W. Whole transcriptome sequencing reveals extensive unspliced mRNA in metastatic castration-resistant prostate cancer. Mol. Cancer Res. 2015, 13, 98–106. [Google Scholar] [CrossRef]
- Wang, R.; Sun, Y.; Li, L.; Niu, Y.; Lin, W.; Lin, C.; Antonarakis, E.S.; Luo, J.; Yeh, S.; Chang, C. Preclinical Study using Malat1 Small Interfering RNA or Androgen Receptor. Splicing Variant 7 Degradation Enhancer ASC-J9(®) to Suppress Enzalutamide-resistant Prostate Cancer Progression. Eur. Urol. 2017, 72, 835–844. [Google Scholar] [CrossRef]
- Cai, S.; Pataillot-Meakin, T.; Shibakawa, A.; Ren, R.; Bevan, C.L.; Ladame, S.; Ivanov, A.P.; Edel, J.B. Single-molecule amplification-free multiplexed detection of circulating microRNA cancer biomarkers from serum. Nat. Commun. 2021, 12, 3515. [Google Scholar] [CrossRef]
- Lakshmanan, V.K.; Ojha, S.; Jung, Y.D. A modern era of personalized medicine in the diagnosis, prognosis, and treatment of prostate cancer. Comput. Biol. Med. 2020, 126, 104020. [Google Scholar] [CrossRef] [PubMed]
- Ahadi, A.; Brennan, S.; Kennedy, P.J.; Hutvagner, G.; Tran, N. Long non-coding RNAs harboring miRNA seed regions are enriched in prostate cancer exosomes. Sci. Rep. 2016, 6, 24922. [Google Scholar] [CrossRef] [PubMed]
- Reda El Sayed, S.; Cristante, J.; Guyon, L.; Denis, J.; Chabre, O.; Cherradi, N. MicroRNA Therapeutics in Cancer: Current Advances and Challenges. Cancers 2021, 13, 2680. [Google Scholar] [CrossRef] [PubMed]
- Velagapudi, S.P.; Luo, Y.; Tran, T.; Haniff, H.S.; Nakai, Y.; Fallahi, M.; Martinez, G.J.; Childs-Disney, J.L.; Disney, M.D. Defining RNA-Small Molecule Affinity Landscapes Enables Design of a Small Molecule Inhibitor of an Oncogenic Noncoding RNA. ACS Cent. Sci. 2017, 3, 205–216. [Google Scholar] [CrossRef]
- Liu, X.; Haniff, H.S.; Childs-Disney, J.L.; Shuster, A.; Aikawa, H.; Adibekian, A.; Disney, M.D. Targeted Degradation of the Oncogenic MicroRNA 17-92 Cluster by Structure-Targeting Ligands. J. Am. Chem. Soc. 2020, 142, 6970–6982. [Google Scholar] [CrossRef]
- Ottman, R.; Levy, J.; Grizzle, W.E.; Chakrabarti, R. The other face of miR-17-92a cluster, exhibiting tumor suppressor effects in prostate cancer. Oncotarget 2016, 7, 73739–73753. [Google Scholar] [CrossRef]
- Zhou, P.; Ma, L.; Zhou, J.; Jiang, M.; Rao, E.; Zhao, Y.; Guo, F. miR-17-92 plays an oncogenic role and conveys chemo-resistance to cisplatin in human prostate cancer cells. Int. J. Oncol. 2016, 48, 1737–1748. [Google Scholar] [CrossRef]
- Feng, S.; Qian, X.; Li, H.; Zhang, X. Combinations of elevated tissue miRNA-17-92 cluster expression and serum prostate-specific antigen as potential diagnostic biomarkers for prostate cancer. Oncol. Lett. 2017, 14, 6943–6949. [Google Scholar] [CrossRef][Green Version]
- Zhang, X.; Ladd, A.; Dragoescu, E.; Budd, W.T.; Ware, J.L.; Zehner, Z.E. MicroRNA-17-3p is a prostate tumor suppressor in vitro and in vivo, and is decreased in high grade prostate tumors analyzed by laser capture microdissection. Clin. Exp. Metastasis 2009, 26, 965–979. [Google Scholar] [CrossRef]
- Dai, H.; Wang, C.; Yu, Z.; He, D.; Yu, K.; Liu, Y.; Wang, S. MiR-17 Regulates Prostate Cancer Cell Proliferation and Apoptosis Through Inhibiting JAK-STAT3 Signaling Pathway. Cancer Biother. Radiopharm. 2018, 33, 103–109. [Google Scholar] [CrossRef]
- Xu, Z.; Zhang, Y.; Ding, J.; Hu, W.; Tan, C.; Wang, M.; Tang, J.; Xu, Y. miR-17-3p Downregulates Mitochondrial Antioxidant Enzymes and Enhances the Radiosensitivity of Prostate Cancer Cells. Mol. Ther. Nucleic Acids 2018, 13, 64–77. [Google Scholar] [CrossRef]
- Yang, X.; Du, W.W.; Li, H.; Liu, F.; Khorshidi, A.; Rutnam, Z.J.; Yang, B.B. Both mature miR-17-5p and passenger strand miR-17-3p target TIMP3 and induce prostate tumor growth and invasion. Nucleic Acids Res. 2013, 41, 9688–9704. [Google Scholar] [CrossRef]
- Gong, A.Y.; Eischeid, A.N.; Xiao, J.; Zhao, J.; Chen, D.; Wang, Z.Y.; Young, C.Y.; Chen, X.M. miR-17-5p targets the p300/CBP-associated factor and modulates androgen receptor transcriptional activity in cultured prostate cancer cells. BMC Cancer 2012, 12, 492. [Google Scholar] [CrossRef]
- Dyson, G.; Farran, B.; Bolton, S.; Craig, D.B.; Dombkowski, A.; Beebe-Dimmer, J.L.; Powell, I.J.; Podgorski, I.; Heilbrun, L.K.; Bock, C.H. The extrema of circulating miR-17 are identified as biomarkers for aggressive prostate cancer. Am. J. Cancer Res. 2018, 8, 2088–2095. [Google Scholar]
- Urabe, F.; Matsuzaki, J.; Yamamoto, Y.; Kimura, T.; Hara, T.; Ichikawa, M.; Takizawa, S.; Aoki, Y.; Niida, S.; Sakamoto, H.; et al. Large-scale Circulating microRNA Profiling for the Liquid Biopsy of Prostate Cancer. Clin. Cancer Res. 2019, 25, 3016–3025. [Google Scholar] [CrossRef]
- Wang, X.; Wang, R.; Wu, Z.; Bai, P. Circular RNA ITCH suppressed prostate cancer progression by increasing HOXB13 expression via spongy miR-17-5p. Cancer Cell Int. 2019, 19, 328. [Google Scholar] [CrossRef]
- Hu, Y.; Guo, B. Circ-MTO1 correlates with favorable prognosis and inhibits cell proliferation, invasion as well as miR-17-5p expression in prostate cancer. J. Clin. Lab. Anal. 2020, 34, e23086. [Google Scholar] [CrossRef]
- Xu, Y.; Fang, F.; Zhang, J.; Josson, S.; St Clair, W.H.; St Clair, D.K. miR-17* suppresses tumorigenicity of prostate cancer by inhibiting mitochondrial antioxidant enzymes. PLoS ONE 2010, 5, e14356. [Google Scholar] [CrossRef]
- Liang, B.; Zhou, C.; Cui, S.; Lu, H.; Xu, R.; Xue, D.; Zou, S.; He, X. Upregulation of miR-18a-5p promotes the proliferation of prostate cancer via inhibiting the expression of SLC40A1. Pathol.-Res. Pract. 2021, 224, 153448. [Google Scholar] [CrossRef]
- Hsu, T.I.; Hsu, C.H.; Lee, K.H.; Lin, J.T.; Chen, C.S.; Chang, K.C.; Su, C.Y.; Hsiao, M.; Lu, P.J. MicroRNA-18a is elevated in prostate cancer and promotes tumorigenesis through suppressing STK4 in vitro and in vivo. Oncogenesis 2014, 3, e99. [Google Scholar] [CrossRef]
- Ibrahim, N.H.; Abdellateif, M.S.; Kassem, S.H.; Abd El Salam, M.A.; El Gammal, M.M. Diagnostic significance of miR-21, miR-141, miR-18a and miR-221 as novel biomarkers in prostate cancer among Egyptian patients. Andrologia 2019, 51, e13384. [Google Scholar] [CrossRef] [PubMed]
- Al-Kafaji, G.; Al-Naieb, Z.T.; Bakhiet, M. Increased oncogenic microRNA-18a expression in the peripheral blood of patients with prostate cancer: A potential novel non-invasive biomarker. Oncol. Lett. 2016, 11, 1201–1206. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Hao, T.; Sun, J.; Wei, P.; Zhang, H. Long noncoding RNA GAS5 modulates α-Solanine-induced radiosensitivity by negatively regulating miR-18a in human prostate cancer cells. Biomed. Pharm. 2019, 112, 108656. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Han, G.; Zhang, X.; Yu, Q.; Li, Z.; Li, Z.; Li, J. Long non-coding RNA FENDRR reduces prostate cancer malignancy by competitively binding miR-18a-5p with RUNX1. Biomarkers 2018, 23, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, Q.; Liu, H.; Wang, N.; Zhang, X.; Yang, S. lncRNA PART1, manipulated by transcriptional factor FOXP2, suppresses proliferation and invasion in ESCC by regulating the miR-18a-5p/SOX6 signaling axis. Oncol. Rep. 2021, 45, 1118–1132. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Zhu, X.; Fan, B.; Xie, D.; Li, T.; Zhang, X. miR-19a-3p targets PMEPA1 and induces prostate cancer cell proliferation, migration and invasion. Mol. Med. Rep. 2016, 13, 4030–4038. [Google Scholar] [CrossRef] [PubMed]
- Wa, Q.; Li, L.; Lin, H.; Peng, X.; Ren, D.; Huang, Y.; He, P.; Huang, S. Downregulation of miR-19a-3p promotes invasion, migration and bone metastasis via activating TGF-β signaling in prostate cancer. Oncol. Rep. 2018, 39, 81–90. [Google Scholar] [CrossRef]
- Fu, F.; Wan, X.; Wang, D.; Kong, Z.; Zhang, Y.; Huang, W.; Wang, C.; Wu, H.; Li, Y. MicroRNA-19a acts as a prognostic marker and promotes prostate cancer progression via inhibiting VPS37A expression. Oncotarget 2017, 9, 1931. [Google Scholar] [CrossRef]
- Lu, K.; Liu, C.; Tao, T.; Zhang, X.; Zhang, L.; Sun, C.; Wang, Y.; Chen, S.; Xu, B.; Chen, M. MicroRNA-19a regulates proliferation and apoptosis of castration-resistant prostate cancer cells by targeting BTG1. FEBS Lett. 2015, 589, 1485–1490. [Google Scholar] [CrossRef]
- Wang, S.-Y.; Shiboski, S.; Belair, C.D.; Cooperberg, M.R.; Simko, J.P.; Stoppler, H.; Cowan, J.; Carroll, P.R.; Blelloch, R. miR-19, miR-345, miR-519c-5p Serum Levels Predict Adverse Pathology in Prostate Cancer Patients Eligible for Active Surveillance. PLoS ONE 2014, 9, e98597. [Google Scholar] [CrossRef]
- Qiang, X.F.; Zhang, Z.W.; Liu, Q.; Sun, N.; Pan, L.L.; Shen, J.; Li, T.; Yun, C.; Li, H.; Shi, L.H. miR-20a promotes prostate cancer invasion and migration through targeting ABL2. J. Cell Biochem. 2014, 115, 1269–1276. [Google Scholar] [CrossRef]
- Pesta, M.; Klecka, J.; Kulda, V.; Topolcan, O.; Hora, M.; Eret, V.; Ludvikova, M.; Babjuk, M.; Novak, K.; Stolz, J.; et al. Importance of miR-20a expression in prostate cancer tissue. Anticancer Res. 2010, 30, 3579–3583. [Google Scholar]
- Shen, J.; Hruby, G.W.; McKiernan, J.M.; Gurvich, I.; Lipsky, M.J.; Benson, M.C.; Santella, R.M. Dysregulation of circulating microRNAs and prediction of aggressive prostate cancer. Prostate 2012, 72, 1469–1477. [Google Scholar] [CrossRef]
- Hart, M.; Nolte, E.; Wach, S.; Szczyrba, J.; Taubert, H.; Rau, T.T.; Hartmann, A.; Grasser, F.A.; Wullich, B. Comparative microRNA profiling of prostate carcinomas with increasing tumor stage by deep sequencing. Mol. Cancer Res. 2014, 12, 250–263. [Google Scholar] [CrossRef]
- Li, X.; Pan, J.H.; Song, B.; Xiong, E.Q.; Chen, Z.W.; Zhou, Z.S.; Su, Y.P. Suppression of CX43 expression by miR-20a in the progression of human prostate cancer. Cancer Biol. Ther. 2012, 13, 890–898. [Google Scholar] [CrossRef]
- Sylvestre, Y.; De Guire, V.; Querido, E.; Mukhopadhyay, U.K.; Bourdeau, V.; Major, F.; Ferbeyre, G.; Chartrand, P. An E2F/miR-20a autoregulatory feedback loop. J. Biol. Chem. 2007, 282, 2135–2143. [Google Scholar] [CrossRef]
- Lin, H.M.; Castillo, L.; Mahon, K.L.; Chiam, K.; Lee, B.Y.; Nguyen, Q.; Boyer, M.J.; Stockler, M.R.; Pavlakis, N.; Marx, G.; et al. Circulating microRNAs are associated with docetaxel chemotherapy outcome in castration-resistant prostate cancer. Br. J. Cancer 2014, 110, 2462–2471. [Google Scholar] [CrossRef]
- Mohammadi Torbati, P.; Asadi, F.; Fard-Esfahani, P. Circulating miR-20a and miR-26a as Biomarkers in Prostate Cancer. Asian Pac. J. Cancer Prev. 2019, 20, 1453–1456. [Google Scholar] [CrossRef]
- Daniel, R.; Wu, Q.; Williams, V.; Clark, G.; Guruli, G.; Zehner, Z. A Panel of MicroRNAs as Diagnostic Biomarkers for the Identification of Prostate Cancer. Int. J. Mol. Sci. 2017, 18, 1281. [Google Scholar] [CrossRef]
- Li, J.; Yang, X.; Guan, H.; Mizokami, A.; Keller, E.T.; Xu, X.; Liu, X.; Tan, J.; Hu, L.; Lu, Y.; et al. Exosome-derived microRNAs contribute to prostate cancer chemoresistance. Int. J. Oncol. 2016, 49, 838–846. [Google Scholar] [CrossRef]
- Ambrozkiewicz, F.; Karczmarski, J.; Kulecka, M.; Paziewska, A.; Cybulska, M.; Szymanski, M.; Dobruch, J.; Antoniewicz, A.; Mikula, M.; Ostrowski, J. Challenges in Cancer Biomarker Discovery Exemplified by the Identification of Diagnostic MicroRNAs in Prostate Tissues. Biomed. Res. Int. 2020, 2020, 9086829. [Google Scholar] [CrossRef] [PubMed]
- Paziewska, A.; Mikula, M.; Dabrowska, M.; Kulecka, M.; Goryca, K.; Antoniewicz, A.; Dobruch, J.; Borowka, A.; Rutkowski, P.; Ostrowski, J. Candidate diagnostic miRNAs that can detect cancer in prostate biopsy. Prostate 2018, 78, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Wang, J.; Chen, G.; Zhao, X. microRNA-204 modulates chemosensitivity and apoptosis of prostate cancer cells by targeting zinc-finger E-box-binding homeobox 1 (ZEB1). Am. J. Transl. Res. 2017, 9, 3599–3610. [Google Scholar] [PubMed]
- Lin, Y.C.; Lin, J.F.; Tsai, T.F.; Chou, K.Y.; Chen, H.E.; Hwang, T.I. Tumor suppressor miRNA-204-5p promotes apoptosis by targeting BCL2 in prostate cancer cells. Asian J. Surg. 2017, 40, 396–406. [Google Scholar] [CrossRef]
- Fredsoe, J.; Rasmussen, A.K.I.; Mouritzen, P.; Borre, M.; Orntoft, T.; Sorensen, K.D. A five-microRNA model (pCaP) for predicting prostate cancer aggressiveness using cell-free urine. Int. J. Cancer 2019, 145, 2558–2567. [Google Scholar] [CrossRef]
- Koppers-Lalic, D.; Hackenberg, M.; de Menezes, R.; Misovic, B.; Wachalska, M.; Geldof, A.; Zini, N.; de Reijke, T.; Wurdinger, T.; Vis, A.; et al. Noninvasive prostate cancer detection by measuring miRNA variants (isomiRs) in urine extracellular vesicles. Oncotarget 2016, 7, 22566–22578. [Google Scholar] [CrossRef]
- He, C.; Lu, X.; Yang, F.; Qin, L.; Guo, Z.; Sun, Y.; Wu, J. LncRNA UCA1 acts as a sponge of miR-204 to up-regulate CXCR4 expression and promote prostate cancer progression. Biosci. Rep. 2019, 39, BSR20181465. [Google Scholar] [CrossRef]
- Zhang, S.; Dong, X.; Ji, T.; Chen, G.; Shan, L. Long non-coding RNA UCA1 promotes cell progression by acting as a competing endogenous RNA of ATF2 in prostate cancer. Am. J. Transl. Res. 2017, 9, 366–375. [Google Scholar]
- Todorova, K.; Metodiev, M.V.; Metodieva, G.; Mincheff, M.; Fernandez, N.; Hayrabedyan, S. Micro-RNA-204 Participates in TMPRSS2/ERG Regulation and Androgen Receptor Reprogramming in Prostate Cancer. Horm. Cancer 2017, 8, 28–48. [Google Scholar] [CrossRef]
- Panigrahi, G.K.; Ramteke, A.; Birks, D.; Abouzeid Ali, H.E.; Venkataraman, S.; Agarwal, C.; Vibhakar, R.; Miller, L.D.; Agarwal, R.; Abd Elmageed, Z.Y.; et al. Exosomal microRNA profiling to identify hypoxia-related biomarkers in prostate cancer. Oncotarget 2018, 9, 13894–13910. [Google Scholar] [CrossRef]
- Zhang, G.; Tian, X.; Li, Y.; Wang, Z.; Li, X.; Zhu, C. miR-27b and miR-34a enhance docetaxel sensitivity of prostate cancer cells through inhibiting epithelial-to-mesenchymal transition by targeting ZEB1. Biomed. Pharmacother. 2018, 97, 736–744. [Google Scholar] [CrossRef]
- Rokavec, M.; Oner, M.G.; Li, H.; Jackstadt, R.; Jiang, L.; Lodygin, D.; Kaller, M.; Horst, D.; Ziegler, P.K.; Schwitalla, S.; et al. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J. Clin. Investig. 2014, 124, 1853–1867. [Google Scholar] [CrossRef]
- Duan, K.; Ge, Y.C.; Zhang, X.P.; Wu, S.Y.; Feng, J.S.; Chen, S.L.; Zhang, L.I.; Yuan, Z.H.; Fu, C.H. miR-34a inhibits cell proliferation in prostate cancer by downregulation of SIRT1 expression. Oncol. Lett. 2015, 10, 3223–3227. [Google Scholar] [CrossRef]
- Liang, J.; Li, Y.; Daniels, G.; Sfanos, K.; De Marzo, A.; Wei, J.; Li, X.; Chen, W.; Wang, J.; Zhong, X.; et al. LEF1 Targeting EMT in Prostate Cancer Invasion Is Regulated by miR-34a. Mol. Cancer Res. 2015, 13, 681–688. [Google Scholar] [CrossRef]
- Liao, H.; Xiao, Y.; Hu, Y.; Yin, Z.; Liu, L.; Kang, X.; Chen, Y. Methylation-induced silencing of miR-34a enhances chemoresistance by directly upregulating ATG4B-induced autophagy through AMPK/mTOR pathway in prostate cancer. Oncol. Rep. 2016, 35, 64–72. [Google Scholar] [CrossRef]
- Liu, X.; Luo, X.; Wu, Y.; Xia, D.; Chen, W.; Fang, Z.; Deng, J.; Hao, Y.; Yang, X.; Zhang, T.; et al. MicroRNA-34a Attenuates Paclitaxel Resistance in Prostate Cancer Cells via Direct Suppression of JAG1/Notch1 Axis. Cell Physiol. Biochem. 2018, 50, 261–276. [Google Scholar] [CrossRef]
- Corcoran, C.; Rani, S.; O’Driscoll, L. miR-34a is an intracellular and exosomal predictive biomarker for response to docetaxel with clinical relevance to prostate cancer progression. Prostate 2014, 74, 1320–1334. [Google Scholar] [CrossRef]
- Ma, Y.; Fan, B.; Ren, Z.; Liu, B.; Wang, Y. Long noncoding RNA DANCR contributes to docetaxel resistance in prostate cancer through targeting the miR-34a-5p/JAG1 pathway. Onco Targets Ther. 2019, 12, 5485–5497. [Google Scholar] [CrossRef]
- Ma, E.; Wang, Q.; Li, J.; Zhang, X.; Guo, Z.; Yang, X. LINC01006 facilitates cell proliferation, migration and invasion in prostate cancer through targeting miR-34a-5p to up-regulate DAAM1. Cancer Cell. Int. 2020, 20, 515. [Google Scholar] [CrossRef]
- Li, N.; Zhang, L.Y.; Qiao, Y.H.; Song, R.J. Long noncoding RNA LINC00662 functions as miRNA sponge to promote the prostate cancer tumorigenesis through targeting miR-34a. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3688–3698. [Google Scholar] [CrossRef]
- Lodygin, D.; Tarasov, V.; Epanchintsev, A.; Berking, C.; Knyazeva, T.; Körner, H.; Knyazev, P.; Diebold, J.; Hermeking, H. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle 2008, 7, 2591–2600. [Google Scholar] [CrossRef] [PubMed]
- Krebs, M.; Behrmann, C.; Kalogirou, C.; Sokolakis, I.; Kneitz, S.; Kruithof-de Julio, M.; Zoni, E.; Rech, A.; Schilling, B.; Kubler, H.; et al. miR-221 Augments TRAIL-Mediated Apoptosis in Prostate Cancer Cells by Inducing Endogenous TRAIL Expression and Targeting the Functional Repressors SOCS3 and PIK3R1. Biomed. Res. Int. 2019, 2019, 6392748. [Google Scholar] [CrossRef] [PubMed]
- Kurul, N.O.; Ates, F.; Yilmaz, I.; Narli, G.; Yesildal, C.; Senkul, T. The association of let-7c, miR-21, miR-145, miR-182, and miR-221 with clinicopathologic parameters of prostate cancer in patients diagnosed with low-risk disease. Prostate 2019, 79, 1125–1132. [Google Scholar] [CrossRef] [PubMed]
- Maqbool, R.; Lone, S.N.; Ul Hussain, M. Post-transcriptional regulation of the tumor suppressor p53 by a novel miR-27a, with implications during hypoxia and tumorigenesis. Biochem. J. 2016, 473, 3597–3610. [Google Scholar] [CrossRef]
- Cao, Z.; Xu, L.; Zhao, S. Exosome-derived miR-27a produced by PSC-27 cells contributes to prostate cancer chemoresistance through p53. Biochem. Biophys Res. Commun. 2019, 515, 345–351. [Google Scholar] [CrossRef]
- Lyu, J.; Zhao, L.; Wang, F.; Ji, J.; Cao, Z.; Xu, H.; Shi, X.; Zhu, Y.; Zhang, C.; Guo, F.; et al. Discovery and Validation of Serum MicroRNAs as Early Diagnostic Biomarkers for Prostate Cancer in Chinese Population. Biomed. Res. Int. 2019, 2019, 9306803. [Google Scholar] [CrossRef]
- Sun, F.; Wu, K.; Yao, Z.; Mu, X.; Zheng, Z.; Sun, M.; Wang, Y.; Liu, Z.; Zhu, Y. Long Noncoding RNA PVT1 Promotes Prostate Cancer Metastasis by Increasing NOP2 Expression via Targeting Tumor Suppressor MicroRNAs. Onco Targets Ther. 2020, 13, 6755–6765. [Google Scholar] [CrossRef]
- Cui, X.; Piao, C.; Lv, C.; Lin, X.; Zhang, Z.; Liu, X. ZNFX1 anti-sense RNA 1 promotes the tumorigenesis of prostate cancer by regulating c-Myc expression via a regulatory network of competing endogenous RNAs. Cell Mol. Life Sci. 2020, 77, 1135–1152. [Google Scholar] [CrossRef]
- Wu, J.; Zheng, C.; Fan, Y.; Zeng, C.; Chen, Z.; Qin, W.; Zhang, C.; Zhang, W.; Wang, X.; Zhu, X.; et al. Downregulation of microRNA-30 facilitates podocyte injury and is prevented by glucocorticoids. J. Am. Soc. Nephrol. 2014, 25, 92–104. [Google Scholar] [CrossRef]
- Ren, Q.; Liang, J.; Wei, J.; Basturk, O.; Wang, J.; Daniels, G.; Gellert, L.L.; Li, Y.; Shen, Y.; Osman, I.; et al. Epithelial and stromal expression of miRNAs during prostate cancer progression. Am. J. Transl. Res. 2014, 6, 329–339. [Google Scholar]
- Fredsøe, J.; Rasmussen, A.K.I.; Thomsen, A.R.; Mouritzen, P.; Høyer, S.; Borre, M.; Ørntoft, T.F.; Sørensen, K.D. Diagnostic and Prognostic MicroRNA Biomarkers for Prostate Cancer in Cell-free Urine. Eur. Urol. Focus 2018, 4, 825–833. [Google Scholar] [CrossRef]
- Zhao, Z.; Weickmann, S.; Jung, M.; Lein, M.; Kilic, E.; Stephan, C.; Erbersdobler, A.; Fendler, A.; Jung, K. A Novel Predictor Tool of Biochemical Recurrence after Radical Prostatectomy Based on a Five-MicroRNA Tissue Signature. Cancers 2019, 11, 1603. [Google Scholar] [CrossRef]
- Chen, W.; Yao, G.; Zhou, K. miR-103a-2-5p/miR-30c-1-3p inhibits the progression of prostate cancer resistance to androgen ablation therapy via targeting androgen receptor variant 7. J. Cell Biochem. 2019, 120, 14055–14064. [Google Scholar] [CrossRef]
- Song, Y.; Song, C.; Yang, S. Tumor-Suppressive Function of miR-30d-5p in Prostate Cancer Cell Proliferation and Migration by Targeting NT5E. Cancer Biother. Radiopharm. 2018, 33, 203–211. [Google Scholar] [CrossRef]
- Fuse, M.; Kojima, S.; Enokida, H.; Chiyomaru, T.; Yoshino, H.; Nohata, N.; Kinoshita, T.; Sakamoto, S.; Naya, Y.; Nakagawa, M.; et al. Tumor suppressive microRNAs (miR-222 and miR-31) regulate molecular pathways based on microRNA expression signature in prostate cancer. J. Hum. Genet. 2012, 57, 691–699. [Google Scholar] [CrossRef]
- Tsuchiyama, K.; Ito, H.; Taga, M.; Naganuma, S.; Oshinoya, Y.; Nagano, K.; Yokoyama, O.; Itoh, H. Expression of microRNAs associated with Gleason grading system in prostate cancer: miR-182-5p is a useful marker for high grade prostate cancer. Prostate 2013, 73, 827–834. [Google Scholar] [CrossRef]
- Bian, X.; Shen, Y.; Zhang, G.; Gu, C.; Cai, Y.; Wang, C.; Zhu, Y.; Zhu, Y.; Zhang, H.; Dai, B.; et al. Expression of Dicer and Its Related MiRNAs in the Progression of Prostate Cancer. PLoS ONE 2015, 10, e0120159. [Google Scholar] [CrossRef][Green Version]
- Zhao, J.; Xu, H.; Duan, Z.; Chen, X.; Ao, Z.; Chen, Y.; Ruan, Y.; Ni, M. miR-31-5p Regulates 14-3-3 ɛ to Inhibit Prostate Cancer 22RV1 Cell Survival and Proliferation via PI3K/AKT/Bcl-2 Signaling Pathway. Cancer Manag. Res. 2020, 12, 6679–6694. [Google Scholar] [CrossRef]
- Bhatnagar, N.; Li, X.; Padi, S.K.R.; Zhang, Q.; Tang, M.s.; Guo, B. Downregulation of miR-205 and miR-31 confers resistance to chemotherapy-induced apoptosis in prostate cancer cells. Cell Death. Dis. 2010, 1, e105. [Google Scholar] [CrossRef]
- Daniunaite, K.; Dubikaityte, M.; Gibas, P.; Bakavicius, A.; Rimantas Lazutka, J.; Ulys, A.; Jankevicius, F.; Jarmalaite, S. Clinical significance of miRNA host gene promoter methylation in prostate cancer. Hum. Mol. Genet. 2017, 26, 2451–2461. [Google Scholar] [CrossRef]
- Lekchnov, E.A.; Amelina, E.V.; Bryzgunova, O.E.; Zaporozhchenko, I.A.; Konoshenko, M.Y.; Yarmoschuk, S.V.; Murashov, I.S.; Pashkovskaya, O.A.; Gorizkii, A.M.; Zheravin, A.A.; et al. Searching for the Novel Specific Predictors of Prostate Cancer in Urine: The Analysis of 84 miRNA Expression. Int. J. Mol. Sci. 2018, 19, 4088. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Sun, Y.; Tang, M.; Liang, C.; Huang, C.P.; Niu, Y.; Wang, Z.; Chang, C. The miR-361-3p increases enzalutamide (Enz) sensitivity via targeting the ARv7 and MKNK2 to better suppress the Enz-resistant prostate cancer. Cell Death Dis. 2020, 11, 807. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Tao, T.; Xu, B.; Chen, S.; Liu, C.; Zhang, L.; Lu, K.; Huang, Y.; Jiang, L.; Zhang, X.; et al. MiR-361-5p acts as a tumor suppressor in prostate cancer by targeting signal transducer and activator of transcription-6(STAT6). Biochem. Biophys Res. Commun. 2014, 445, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Wang, S.; Zhang, W.; Qiu, J.; Shan, Y.; Yang, D.; Shen, B. Screening key microRNAs for castration-resistant prostate cancer based on miRNA/mRNA functional synergistic network. Oncotarget 2015, 6, 43819–43830. [Google Scholar] [CrossRef] [PubMed]
- Ju, G.; Zhu, Y.; Du, T.; Cao, W.; Lin, J.; Li, C.; Xu, D.; Wang, Z. MiR-197 Inhibitor Loaded AbCD133@MSNs@GNR Affects the Development of Prostate Cancer Through Targeting ITGAV. Front. Cell Dev. Biol. 2021, 9, 1289. [Google Scholar] [CrossRef]
- Walter, B.A.; Valera, V.A.; Pinto, P.A.; Merino, M.J. Comprehensive microRNA Profiling of Prostate Cancer. J. Cancer 2013, 4, 350–357. [Google Scholar] [CrossRef]
- Qiu, X.; Zhu, J.; Sun, Y.; Fan, K.; Yang, D.R.; Li, G.; Yang, G.; Chang, C. TR4 nuclear receptor increases prostate cancer invasion via decreasing the miR-373-3p expression to alter TGFβR2/p-Smad3 signals. Oncotarget 2015, 6, 15397–15409. [Google Scholar] [CrossRef]
- Zhang, G.; Liu, Z.; Xu, H.; Yang, Q. miR-409-3p suppresses breast cancer cell growth and invasion by targeting Akt1. Biochem. Biophys. Res. Commun. 2016, 469, 189–195. [Google Scholar] [CrossRef]
- Nguyen, H.C.; Xie, W.; Yang, M.; Hsieh, C.L.; Drouin, S.; Lee, G.S.; Kantoff, P.W. Expression differences of circulating microRNAs in metastatic castration resistant prostate cancer and low-risk, localized prostate cancer. Prostate 2013, 73, 346–354. [Google Scholar] [CrossRef]
- Yu, Q.; Li, P.; Weng, M.; Wu, S.; Zhang, Y.; Chen, X.; Zhang, Q.; Shen, G.; Ding, X.; Fu, S. Nano-Vesicles are a Potential Tool to Monitor Therapeutic Efficacy of Carbon Ion Radiotherapy in Prostate Cancer. J. Biomed. Nanotechnol. 2018, 14, 168–178. [Google Scholar] [CrossRef]
- Xiang, S.; Zou, P.; Tang, Q.; Zheng, F.; Wu, J.; Chen, Z.; Hann, S.S. HOTAIR-mediated reciprocal regulation of EZH2 and DNMT1 contribute to polyphyllin I-inhibited growth of castration-resistant prostate cancer cells in vitro and in vivo. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2018, 1862, 589–599. [Google Scholar] [CrossRef]
- Li, T.; Liu, N.; Gao, Y.; Quan, Z.; Hao, Y.; Yu, C.; Li, L.; Yuan, M.; Niu, L.; Luo, C.; et al. Long noncoding RNA HOTAIR regulates the invasion and metastasis of prostate cancer by targeting hepaCAM. Br. J. Cancer 2021, 124, 247–258. [Google Scholar] [CrossRef]
- Wang, N.; Jiang, Y.; Lv, S.; Wen, H.; Wu, D.; Wei, Q.; Dang, Q. HOTAIR expands the population of prostatic cancer stem-like cells and causes Docetaxel resistance via activating STAT3 signaling. Aging (Albany NY) 2020, 12, 12771–12782. [Google Scholar] [CrossRef]
- Ling, Z.; Wang, X.; Tao, T.; Zhang, L.; Guan, H.; You, Z.; Lu, K.; Zhang, G.; Chen, S.; Wu, J.; et al. Involvement of aberrantly activated HOTAIR/EZH2/miR-193a feedback loop in progression of prostate cancer. J. Exp. Clin. Cancer Res. 2017, 36, 159. [Google Scholar] [CrossRef]
- Wang, D.; Ding, L.; Wang, L.; Zhao, Y.; Sun, Z.; Karnes, R.J.; Zhang, J.; Huang, H. LncRNA MALAT1 enhances oncogenic activities of EZH2 in castration-resistant prostate cancer. Oncotarget 2015, 6, 41045. [Google Scholar] [CrossRef]
- Hao, T.; Wang, Z.; Yang, J.; Zhang, Y.; Shang, Y.; Sun, J. MALAT1 knockdown inhibits prostate cancer progression by regulating miR-140/BIRC6 axis. Biomed. Pharmacother. 2020, 123, 109666. [Google Scholar] [CrossRef]
- Chang, J.; Xu, W.; Du, X.; Hou, J. MALAT1 silencing suppresses prostate cancer progression by upregulating miR-1 and downregulating KRAS. Onco Targets Ther. 2018, 11, 3461–3473. [Google Scholar] [CrossRef]
- Xue, D.; Lu, H.; Xu, H.-Y.; Zhou, C.-X.; He, X.-Z. Long noncoding RNA MALAT1 enhances the docetaxel resistance of prostate cancer cells via miR-145-5p-mediated regulation of AKAP12. J. Cell. Mol. Med. 2018, 22, 3223–3237. [Google Scholar] [CrossRef]
- Li, Y.; Ji, J.; Lyu, J.; Jin, X.; He, X.; Mo, S.; Xu, H.; He, J.; Cao, Z.; Chen, X.; et al. A Novel Urine Exosomal lncRNA Assay to Improve the Detection of Prostate Cancer at Initial Biopsy: A Retrospective Multicenter Diagnostic Feasibility Study. Cancers 2021, 13, 4075. [Google Scholar] [CrossRef]
- Wang, F.; Ren, S.; Chen, R.; Lu, J.; Shi, X.; Zhu, Y.; Zhang, W.; Jing, T.; Zhang, C.; Shen, J.; et al. Development and prospective multicenter evaluation of the long noncoding RNA MALAT-1 as a diagnostic urinary biomarker for prostate cancer. Oncotarget 2014, 5, 11091. [Google Scholar] [CrossRef]
- Ren, S.; Wang, F.; Shen, J.; Sun, Y.; Xu, W.; Lu, J.; Wei, M.; Xu, C.; Wu, C.; Zhang, Z.; et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur. J. Cancer 2013, 49, 2949–2959. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Liang, Z.; Liu, L.; Guo, K.; Xu, S.; Wang, H. Silencing of MALAT1 inhibits migration and invasion by sponging miR-1-3p in prostate cancer cells. Mol. Med. Rep. 2019, 20, 3499–3508. [Google Scholar] [CrossRef] [PubMed]
- Eke, I.; Bylicky, M.A.; Sandfort, V.; Chopra, S.; Martello, S.; Graves, E.E.; Coleman, C.N.; Aryankalayil, M.J. The lncRNAs LINC00261 and LINC00665 are upregulated in long-term prostate cancer adaptation after radiotherapy. Mol. Ther. Nucleic Acids 2021, 24, 175–187. [Google Scholar] [CrossRef] [PubMed]
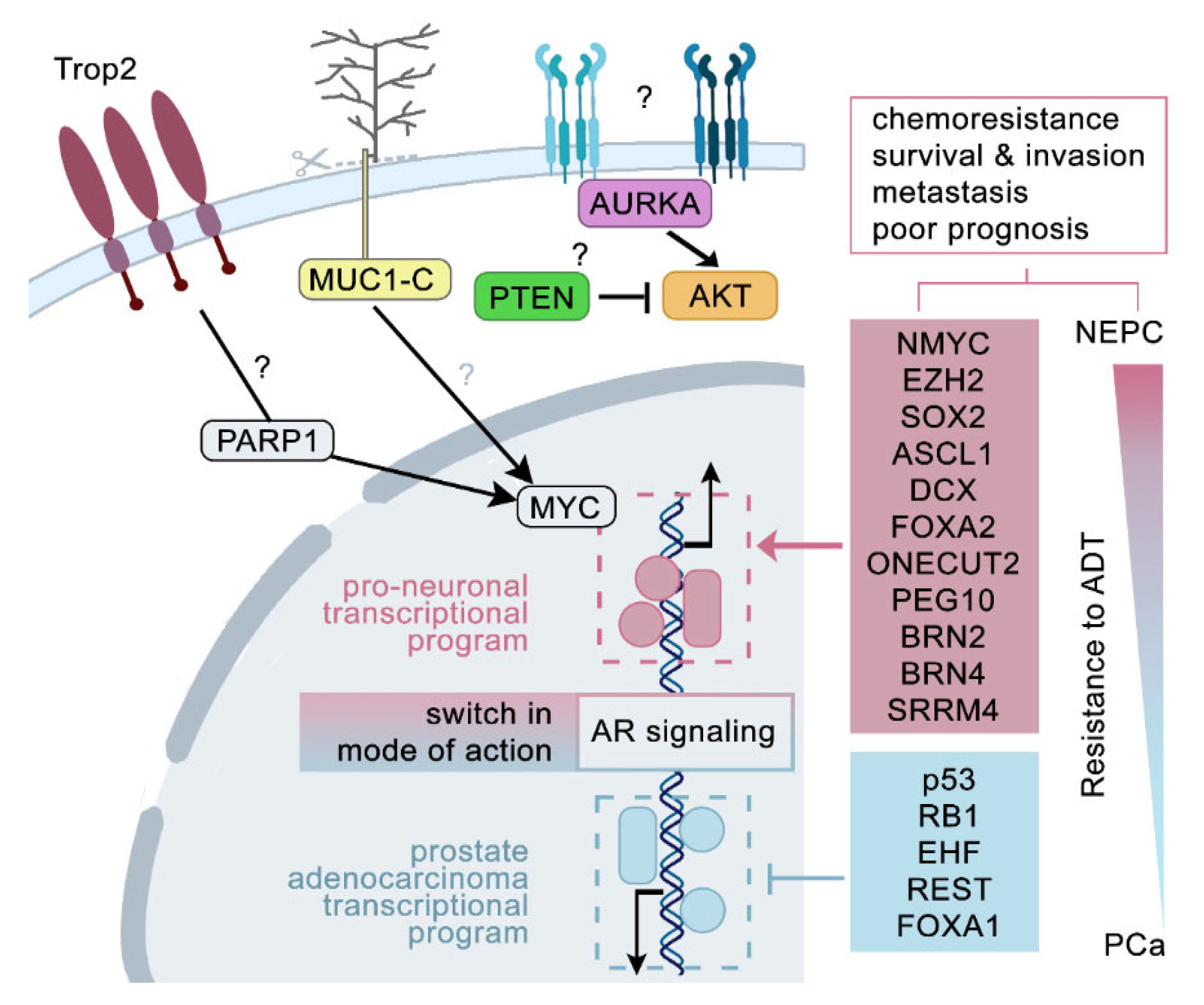
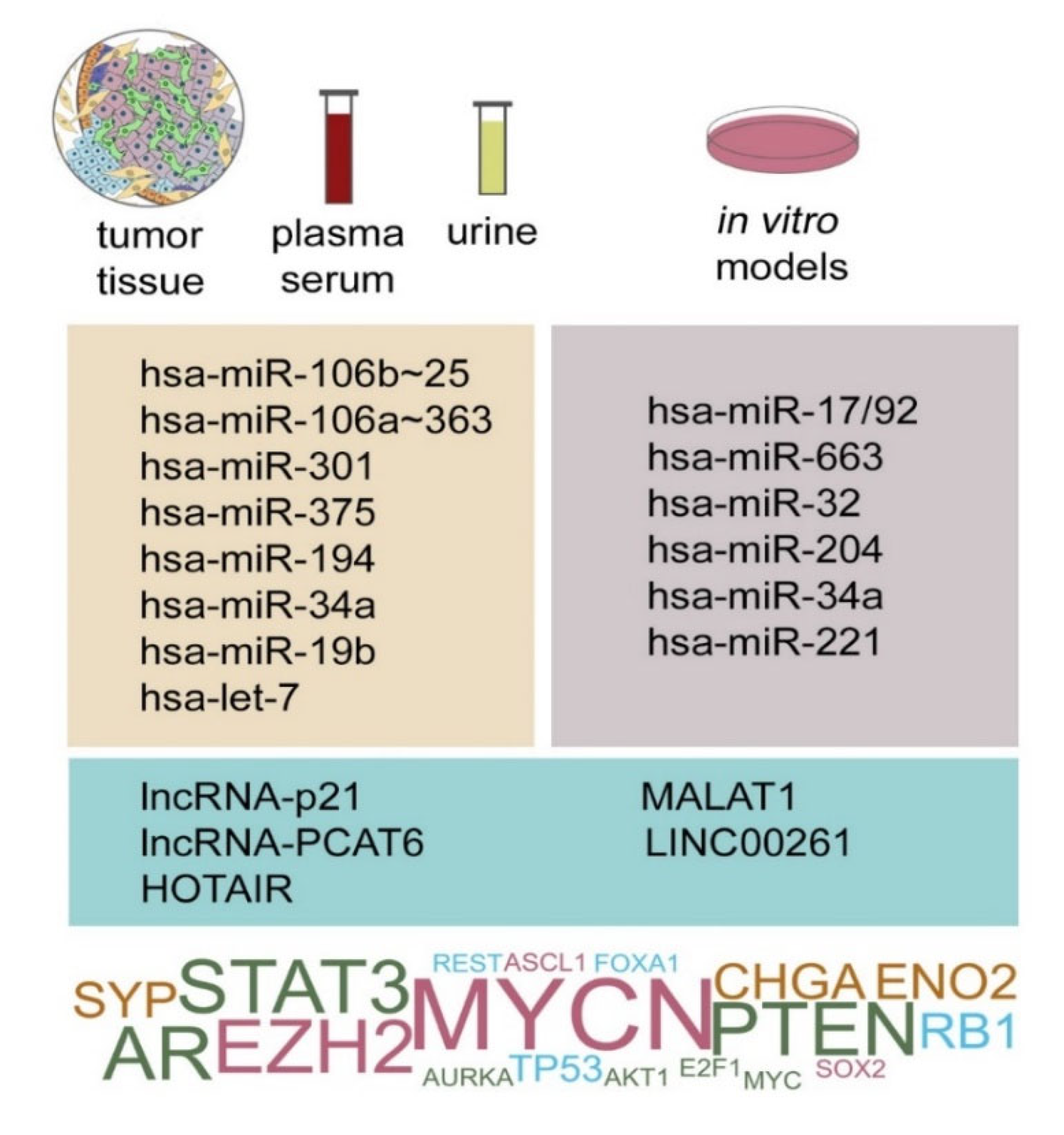
| Name | Biological Function | Ref | Role in PCa/NEPC | Ref |
|---|---|---|---|---|
| Chromogranin A (CgA) |
| [20,21] |
| [22,23,24,25] |
| Chromogranin B (CgB) |
| [21] |
| [26,27] |
| Neuron-specific enolase (γ-enolase, NSE) |
| [28] |
| [29,30,31] |
| Synaptophysin (Syn) |
| [32] |
| [33,34] |
| CD56 (N-CAM, neural cell adhesion molecule-1) |
| [35] |
| [36,37] |
| L-dopa decarboxylase (DDC) |
| [38] |
| [39,40,41] |
| Class III β-tubulin (TUBBIII) |
| [42] |
| [43,44] |
| Gastrin-releasing peptide (GRP) |
| [45,46] |
| [47,48,49] |
| Calcitonin gene-related peptide (CGRP) |
| [50] |
| [51,52,53] |
| Proadrenomedullin N-terminal 20-peptide (PAMP) |
| [54] |
| [55,56] |
| Adrenomedullin (AM) |
| [57] |
| [55,56,58,59] |
| Secretagogin |
| [60] |
| [61] |
| Parathyroid hormone-related peptide (PTHrP) |
| [62] |
| [27,63,64,65,66,67] |
| Neurotensin (NTS) |
| [68] |
| [27,69,70,71,72,73] |
| Vascular endothelial growth factor (VEGF) |
| [74] |
| [75,76,77] |
| Histamine |
| [78] |
| [78,79] |
| Serotonin (5-hydroxy-tryptamine, 5-HT) |
|
| [80,81] | |
| Carcinoembryonic antigen-related cell adhesion molecule 5 (CEACAM5) |
| [82,83,84] |
| [37,85] |
| Nerve growth factor (NGF) |
| [86] |
| [87,88,89] |
| Neuropeptide Y (NPY) |
| [90] |
| [91,92] |
| miRNA | Associationwith NED | Validated Target | Expression in PCa Clinical Samples | Cancer-Related Effect | Prognosis Correlation with Clinical Data | Biomarker | Other Findings | ||
|---|---|---|---|---|---|---|---|---|---|
| NED Marker | Positive NED Regulator | Negative NED Regulator | Experimental Findings | Source: Indication | |||||
| hsa-miR-194 |
| - | - | - |
|
|
| ||
| hsa-miR-375 |
| - |
| ||||||
| hsa-miR-301a | - | - |
| ||||||
| hsa-miR-106a | - | - |
| ||||||
| hsa-miR-92a | - | - | - |
| - |
|
| ||
| hsa-miR-19b |
| - |
| - |
| - | - | ||
| hsa-miR-20b |
| - | - |
|
|
|
| ||
| hsa-miR-363 |
| - | - | - |
| - | - |
| |
| hsa-miR-106b | - | - |
| - | - | ||||
| hsa-miR-93 |
| - | - |
|
| - | |||
| hsa-miR-25 | - |
|
| - | - | ||||
| hsa-let-7 |
| - | - |
|
|
| |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Slabáková, E.; Kahounová, Z.; Procházková, J.; Souček, K. Regulation of Neuroendocrine-like Differentiation in Prostate Cancer by Non-Coding RNAs. Non-Coding RNA 2021, 7, 75. https://doi.org/10.3390/ncrna7040075
Slabáková E, Kahounová Z, Procházková J, Souček K. Regulation of Neuroendocrine-like Differentiation in Prostate Cancer by Non-Coding RNAs. Non-Coding RNA. 2021; 7(4):75. https://doi.org/10.3390/ncrna7040075
Chicago/Turabian StyleSlabáková, Eva, Zuzana Kahounová, Jiřina Procházková, and Karel Souček. 2021. "Regulation of Neuroendocrine-like Differentiation in Prostate Cancer by Non-Coding RNAs" Non-Coding RNA 7, no. 4: 75. https://doi.org/10.3390/ncrna7040075
APA StyleSlabáková, E., Kahounová, Z., Procházková, J., & Souček, K. (2021). Regulation of Neuroendocrine-like Differentiation in Prostate Cancer by Non-Coding RNAs. Non-Coding RNA, 7(4), 75. https://doi.org/10.3390/ncrna7040075