Small RNAs Asserting Big Roles in Mycobacteria
Abstract
1. Introduction
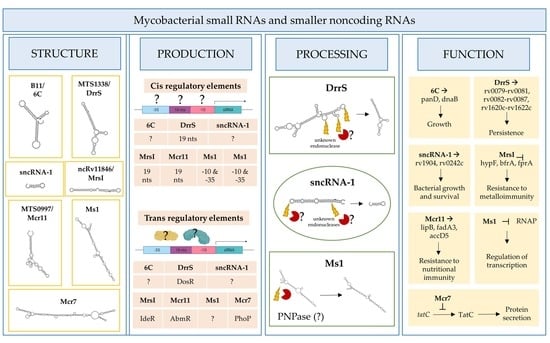
2. Identification and Designation of Mycobacterial Small RNAs
3. Functional Roles of Mycobacterial sRNAs and sncRNAs
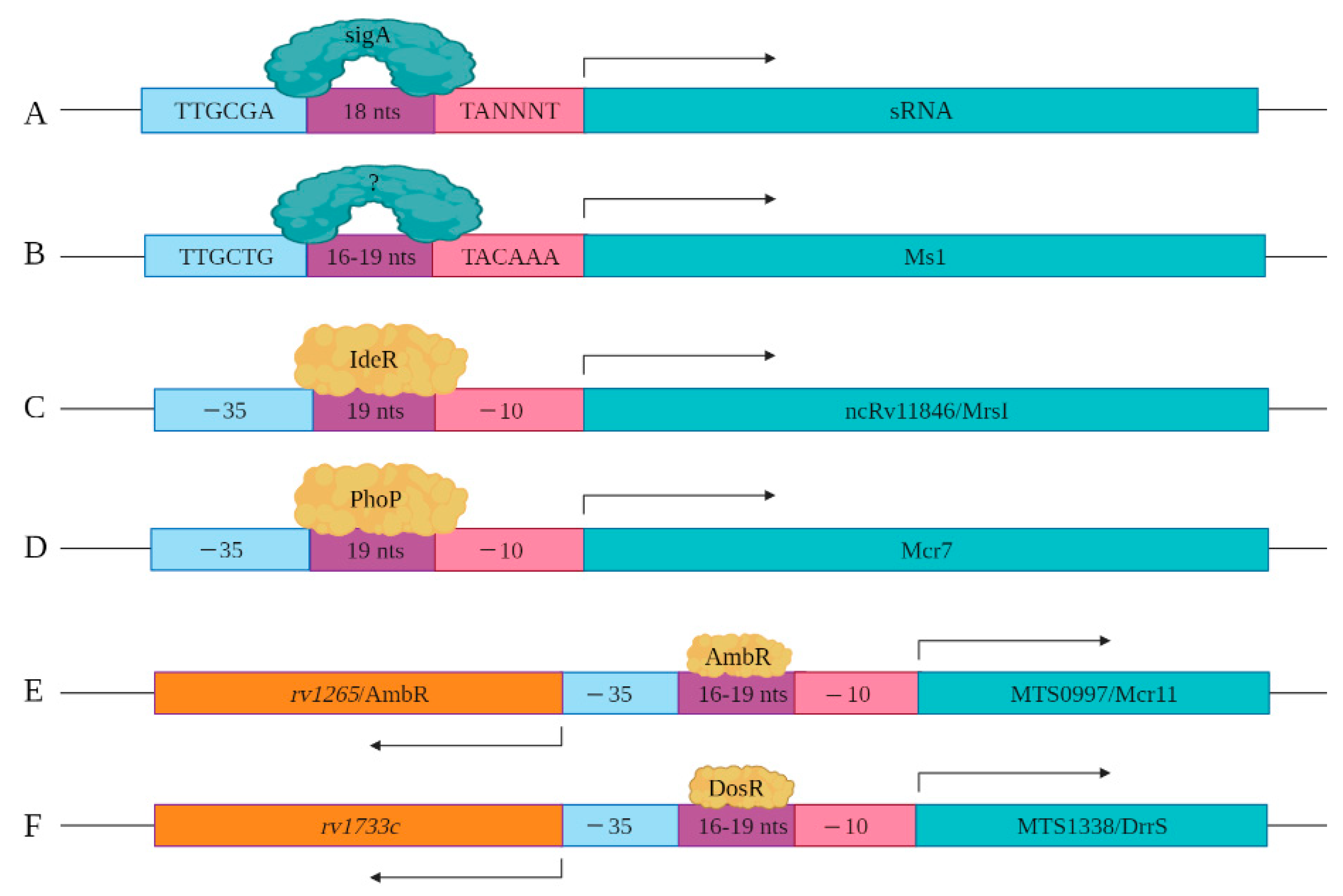
4. Regulation of Mycobacterial sRNAs’/sncRNAs’ Expression
5. Processing of Mycobacterial sRNAs and sncRNAs
6. tRNA Processing Enzymes as Potential Players for sRNA Maturation
7. The Hunt for the Mycobacterial Hfq Equivalent
8. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Supply, P.; Marceau, M.; Mangenot, S.; Roche, D.; Rouanet, C.; Khanna, V.; Majlessi, L.; Criscuolo, A.; Tap, J.; Pawlik, A.; et al. Genomic analysis of smooth tubercle bacilli provides insights into ancestry and pathoadaptation of Mycobacterium tuberculosis. Nat. Genet. 2013, 45, 172–179. [Google Scholar] [CrossRef]
- Cole, S.T.; Brosch, R.; Parkhill, J.; Garnier, T.; Churcher, C.; Harris, D.; Gordon, S.V.; Eiglmeier, K.; Gas, S.; Barry, C.E., 3rd; et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 1998, 393, 537–544. [Google Scholar] [CrossRef]
- Ohol, Y.M.; Goetz, D.H.; Chan, K.; Shiloh, M.U.; Craik, C.S.; Cox, J.S. Mycobacterium tuberculosis MycP1 protease plays a dual role in regulation of ESX-1 secretion and virulence. Cell Host Microbe 2010, 7, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Garces, A.; Atmakuri, K.; Chase, M.R.; Woodworth, J.S.; Krastins, B.; Rothchild, A.C.; Ramsdell, T.L.; Lopez, M.F.; Behar, S.M.; Sarracino, D.A.; et al. EspA acts as a critical mediator of ESX1-dependent virulence in Mycobacterium tuberculosis by affecting bacterial cell wall integrity. PLoS Pathog. 2010, 6, e1000957. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Singh, A.; Tyagi, A.K. Deciphering the genes involved in pathogenesis of Mycobacterium tuberculosis. Tuberculosis 2005, 85, 325–335. [Google Scholar] [CrossRef]
- Sassetti, C.M.; Rubin, E.J. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. USA 2003, 100, 12989–12994. [Google Scholar] [CrossRef] [PubMed]
- DeJesus, M.A.; Gerrick, E.R.; Xu, W.; Park, S.W.; Long, J.E.; Boutte, C.C.; Rubin, E.J.; Schnappinger, D.; Ehrt, S.; Fortune, S.M.; et al. Comprehensive Essentiality Analysis of the Mycobacterium tuberculosis Genome via Saturating Transposon Mutagenesis. mBio 2017, 8, e02133‐16. [Google Scholar] [CrossRef] [PubMed]
- Pellin, D.; Miotto, P.; Ambrosi, A.; Cirillo, D.M.; Di Serio, C. A genome-wide identification analysis of small regulatory RNAs in Mycobacterium tuberculosis by RNA-Seq and conservation analysis. PLoS ONE 2012, 7, e32723. [Google Scholar] [CrossRef]
- Arnvig, K.B.; Young, D.B. Identification of small RNAs in Mycobacterium tuberculosis. Mol. Microbiol. 2009, 73, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Arnvig, K.B.; Comas, I.; Thomson, N.R.; Houghton, J.; Boshoff, H.I.; Croucher, N.J.; Rose, G.; Perkins, T.T.; Parkhill, J.; Dougan, G.; et al. Sequence-based analysis uncovers an abundance of non-coding RNA in the total transcriptome of Mycobacterium tuberculosis. PLoS Pathog. 2011, 7, e1002342. [Google Scholar] [CrossRef]
- DiChiara, J.M.; Contreras-Martinez, L.M.; Livny, J.; Smith, D.; McDonough, K.A.; Belfort, M. Multiple small RNAs identified in Mycobacterium bovis BCG are also expressed in Mycobacterium tuberculosis and Mycobacterium smegmatis. Nucleic Acids Res. 2010, 38, 4067–4078. [Google Scholar] [CrossRef]
- Gerrick, E.R.; Barbier, T.; Chase, M.R.; Xu, R.; Francois, J.; Lin, V.H.; Szucs, M.J.; Rock, J.M.; Ahmad, R.; Tjaden, B.; et al. Small RNA profiling in Mycobacterium tuberculosis identifies MrsI as necessary for an anticipatory iron sparing response. Proc. Natl. Acad. Sci. USA 2018, 115, 6464–6469. [Google Scholar] [CrossRef]
- Coskun, F.S.; Srivastava, S.; Raj, P.; Dozmorov, I.; Belkaya, S.; Mehra, S.; Golden, N.A.; Bucsan, A.N.; Chapagain, M.L.; Wakeland, E.K.; et al. sncRNA-1 Is a Small Noncoding RNA Produced by Mycobacterium tuberculosis in Infected Cells That Positively Regulates Genes Coupled to Oleic Acid Biosynthesis. Front. Microbiol. 2020, 11, 1631. [Google Scholar] [CrossRef] [PubMed]
- Girardin, R.C.; Bai, G.; He, J.; Sui, H.; McDonough, K.A. AbmR (Rv1265) is a novel transcription factor of Mycobacterium tuberculosis that regulates host cell association and expression of the non-coding small RNA Mcr11. Mol. Microbiol. 2018, 110, 811–830. [Google Scholar] [CrossRef]
- Wassarman, K.M.; Zhang, A.; Storz, G. Small RNAs in Escherichia coli. Trends Microbiol. 1999, 7, 37–45. [Google Scholar] [CrossRef]
- Gottesman, S. The small RNA regulators of Escherichia coli: Roles and mechanisms. Annu Rev. Microbiol. 2004, 58, 303–328. [Google Scholar] [CrossRef]
- Waters, L.S.; Storz, G. Regulatory RNAs in bacteria. Cell 2009, 136, 615–628. [Google Scholar] [CrossRef]
- Gottesman, S.; Storz, G. Bacterial small RNA regulators: Versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect. Biol. 2011, 3, a003798. [Google Scholar] [CrossRef] [PubMed]
- Ignatov, D.V.; Salina, E.G.; Fursov, M.V.; Skvortsov, T.A.; Azhikina, T.L.; Kaprelyants, A.S. Dormant non-culturable Mycobacterium tuberculosis retains stable low-abundant mRNA. BMC Genom. 2015, 16, 954. [Google Scholar] [CrossRef] [PubMed]
- Afonyushkin, T.; Vecerek, B.; Moll, I.; Blasi, U.; Kaberdin, V.R. Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB. Nucleic Acids Res. 2005, 33, 1678–1689. [Google Scholar] [CrossRef]
- Morita, T.; Maki, K.; Aiba, H. RNase E-based ribonucleoprotein complexes: Mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005, 19, 2176–2186. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Li, W.; Wu, Z.; Tao, Y.; Wang, X.; Wei, J.; Jiang, P.; Wu, J.; Zhang, Z.; Zhang, W.; et al. Detection of mycobacterial small RNA in the bacterial culture supernatant and plasma of patients with active tuberculosis. Biochem. Biophys. Res. Commun. 2018, 503, 490–494. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Jiang, X.; Wu, A.; Zhou, J.; Liu, H.; He, F.; Zhang, Q.; Zen, K.; Gu, S.; Wang, J. Two Small Extracellular Vesicle sRNAs Derived from Mycobacterium tuberculosis Serve as Diagnostic Biomarkers for Active Pulmonary Tuberculosis. Front. Microbiol. 2021, 12, 642559. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Li, T.; Fan, Y.; Wang, X.; Gu, W.; Lu, W.; Yin, Y.; Meng, Q.; Zhang, W.; Zhao, J.; et al. Screening of 20 Mycobacterium tuberculosis sRNAs in plasma for detection of active pulmonary tuberculosis. Tuberculosis 2021, 129, 102086. [Google Scholar] [CrossRef]
- Hartkoorn, R.C.; Sala, C.; Uplekar, S.; Busso, P.; Rougemont, J.; Cole, S.T. Genome-wide definition of the SigF regulon in Mycobacterium tuberculosis. J. Bacteriol. 2012, 194, 2001–2009. [Google Scholar] [CrossRef] [PubMed]
- Hnilicova, J.; Jirat Matejckova, J.; Sikova, M.; Pospisil, J.; Halada, P.; Panek, J.; Krasny, L. Ms1, a novel sRNA interacting with the RNA polymerase core in mycobacteria. Nucleic Acids Res. 2014, 42, 11763–11776. [Google Scholar] [CrossRef]
- Sikova, M.; Janouskova, M.; Ramaniuk, O.; Palenikova, P.; Pospisil, J.; Bartl, P.; Suder, A.; Pajer, P.; Kubickova, P.; Pavlis, O.; et al. Ms1 RNA increases the amount of RNA polymerase in Mycobacterium smegmatis. Mol. Microbiol. 2019, 111, 354–372. [Google Scholar] [PubMed]
- Moores, A.; Riesco, A.B.; Schwenk, S.; Arnvig, K.B. Expression, maturation and turnover of DrrS, an unusually stable, DosR regulated small RNA in Mycobacterium tuberculosis. PLoS ONE 2017, 12, e0174079. [Google Scholar] [CrossRef]
- Salina, E.G.; Grigorov, A.; Skvortsova, Y.; Majorov, K.; Bychenko, O.; Ostrik, A.; Logunova, N.; Ignatov, D.; Kaprelyants, A.; Apt, A.; et al. MTS1338, A Small Mycobacterium tuberculosis RNA, Regulates Transcriptional Shifts Consistent With Bacterial Adaptation for Entering Into Dormancy and Survival Within Host Macrophages. Front. Cell Infect. Microbiol. 2019, 9, 405. [Google Scholar] [CrossRef]
- Girardin, R.C.; McDonough, K.A. Small RNA Mcr11 requires the transcription factor AbmR for stable expression and regulates genes involved in the central metabolism of Mycobacterium tuberculosis. Mol. Microbiol. 2020, 113, 504–520. [Google Scholar] [CrossRef]
- Solans, L.; Gonzalo-Asensio, J.; Sala, C.; Benjak, A.; Uplekar, S.; Rougemont, J.; Guilhot, C.; Malaga, W.; Martin, C.; Cole, S.T. The PhoP-dependent ncRNA Mcr7 modulates the TAT secretion system in Mycobacterium tuberculosis. PLoS Pathog. 2014, 10, e1004183. [Google Scholar] [CrossRef] [PubMed]
- Lamichhane, G.; Arnvig, K.B.; McDonough, K.A. Definition and annotation of (myco)bacterial non-coding RNA. Tuberculosis 2013, 93, 26–29. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; Zhao, C.; Zhang, T.; Liang, H.; Wang, X.M.; Pan, Y.; Chen, X.; Zhao, Q.; Li, D.; Liu, F.; et al. Salmonella produce microRNA-like RNA fragment Sal-1 in the infected cells to facilitate intracellular survival. Sci. Rep. 2017, 7, 2392. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.M.; Choi, J.W.; Lee, Y.; Hong, S.H.; Lee, H.J. Identification of microRNA-size, small RNAs in Escherichia coli. Curr. Microbiol. 2013, 67, 609–613. [Google Scholar] [CrossRef]
- Dang, T.H.Y.; Tyagi, S.; D’Cunha, G.; Bhave, M.; Crawford, R.; Ivanova, E.P. Computational prediction of microRNAs in marine bacteria of the genus Thalassospira. PLoS ONE 2019, 14, e0212996. [Google Scholar] [CrossRef]
- Shmaryahu, A.; Carrasco, M.; Valenzuela, P.D. Prediction of bacterial microRNAs and possible targets in human cell transcriptome. J. Microbiol. 2014, 52, 482–489. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Guo, H.; Ingolia, N.T.; Weissman, J.S.; Bartel, D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010, 466, 835–840. [Google Scholar] [CrossRef] [PubMed]
- Furuse, Y.; Finethy, R.; Saka, H.A.; Xet-Mull, A.M.; Sisk, D.M.; Smith, K.L.; Lee, S.; Coers, J.; Valdivia, R.H.; Tobin, D.M.; et al. Search for microRNAs expressed by intracellular bacterial pathogens in infected mammalian cells. PLoS ONE 2014, 9, e106434. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarty, S.; Kumar, A.; Raviprasad, K.; Mallya, S.; Satyamoorthy, K.; Chawla, K. Host and MTB genome encoded miRNA markers for diagnosis of tuberculosis. Tuberculosis 2019, 116, 37–43. [Google Scholar] [CrossRef]
- Ostrik, A.A.; Azhikina, T.L.; Salina, E.G. Small Noncoding RNAs and Their Role in the Pathogenesis of Mycobacterium tuberculosis Infection. Biochemistry 2021, 86, S109–S119. [Google Scholar] [PubMed]
- Weinberg, Z.; Barrick, J.E.; Yao, Z.; Roth, A.; Kim, J.N.; Gore, J.; Wang, J.X.; Lee, E.R.; Block, K.F.; Sudarsan, N.; et al. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res. 2007, 35, 4809–4819. [Google Scholar] [CrossRef] [PubMed]
- Mai, J.; Rao, C.; Watt, J.; Sun, X.; Lin, C.; Zhang, L.; Liu, J. Mycobacterium tuberculosis 6C sRNA binds multiple mRNA targets via C-rich loops independent of RNA chaperones. Nucleic Acids Res. 2019, 47, 4292–4307. [Google Scholar] [CrossRef] [PubMed]
- Grunweller, A.; Hartmann, R.K. Locked nucleic acid oligonucleotides: The next generation of antisense agents? BioDrugs 2007, 21, 235–243. [Google Scholar] [CrossRef]
- Manier, S.; Powers, J.T.; Sacco, A.; Glavey, S.V.; Huynh, D.; Reagan, M.R.; Salem, K.Z.; Moschetta, M.; Shi, J.; Mishima, Y.; et al. The LIN28B/let-7 axis is a novel therapeutic pathway in multiple myeloma. Leukemia 2017, 31, 853–860. [Google Scholar] [CrossRef]
- Cavanagh, A.T.; Wassarman, K.M. 6S RNA, a global regulator of transcription in Escherichia coli, Bacillus subtilis, and beyond. Annu. Rev. Microbiol. 2014, 68, 45–60. [Google Scholar] [CrossRef]
- McDonough, J.A.; Hacker, K.E.; Flores, A.R.; Pavelka, M.S., Jr.; Braunstein, M. The twin-arginine translocation pathway of Mycobacterium smegmatis is functional and required for the export of mycobacterial beta-lactamases. J. Bacteriol. 2005, 187, 7667–7679. [Google Scholar] [CrossRef]
- Gomez, M.; Doukhan, L.; Nair, G.; Smith, I. sigA is an essential gene in Mycobacterium smegmatis. Mol. Microbiol. 1998, 29, 617–628. [Google Scholar] [CrossRef]
- Hu, Y.; Coates, A.R. Transcription of two sigma 70 homologue genes, sigA and sigB, in stationary-phase Mycobacterium tuberculosis. J. Bacteriol. 1999, 181, 469–476. [Google Scholar] [CrossRef]
- Sachdeva, P.; Misra, R.; Tyagi, A.K.; Singh, Y. The sigma factors of Mycobacterium tuberculosis: Regulation of the regulators. FEBS J. 2010, 277, 605–626. [Google Scholar] [CrossRef]
- Wu, S.; Howard, S.T.; Lakey, D.L.; Kipnis, A.; Samten, B.; Safi, H.; Gruppo, V.; Wizel, B.; Shams, H.; Basaraba, R.J.; et al. The principal sigma factor sigA mediates enhanced growth of Mycobacterium tuberculosis in vivo. Mol. Microbiol. 2004, 51, 1551–1562. [Google Scholar] [CrossRef]
- Miotto, P.; Forti, F.; Ambrosi, A.; Pellin, D.; Veiga, D.F.; Balazsi, G.; Gennaro, M.L.; Di Serio, C.; Ghisotti, D.; Cirillo, D.M. Genome-wide discovery of small RNAs in Mycobacterium tuberculosis. PLoS ONE 2012, 7, e51950. [Google Scholar] [CrossRef]
- Schmitt, M.P.; Predich, M.; Doukhan, L.; Smith, I.; Holmes, R.K. Characterization of an iron-dependent regulatory protein (IdeR) of Mycobacterium tuberculosis as a functional homolog of the diphtheria toxin repressor (DtxR) from Corynebacterium diphtheriae. Infect. Immun. 1995, 63, 4284–4289. [Google Scholar] [CrossRef] [PubMed]
- Perez, E.; Samper, S.; Bordas, Y.; Guilhot, C.; Gicquel, B.; Martin, C. An essential role for phoP in Mycobacterium tuberculosis virulence. Mol. Microbiol. 2001, 41, 179–187. [Google Scholar] [CrossRef]
- Cortes, T.; Schubert, O.T.; Rose, G.; Arnvig, K.B.; Comas, I.; Aebersold, R.; Young, D.B. Genome-wide mapping of transcriptional start sites defines an extensive leaderless transcriptome in Mycobacterium tuberculosis. Cell Rep. 2013, 5, 1121–1131. [Google Scholar] [CrossRef] [PubMed]
- Park, H.D.; Guinn, K.M.; Harrell, M.I.; Liao, R.; Voskuil, M.I.; Tompa, M.; Schoolnik, G.K.; Sherman, D.R. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 2003, 48, 833–843. [Google Scholar] [CrossRef] [PubMed]
- Wang, B. Base Composition Characteristics of Mammalian miRNAs. J. Nucleic Acids 2013, 2013, 951570. [Google Scholar] [CrossRef]
- Plocinski, P.; Macios, M.; Houghton, J.; Niemiec, E.; Plocinska, R.; Brzostek, A.; Slomka, M.; Dziadek, J.; Young, D.; Dziembowski, A. Proteomic and transcriptomic experiments reveal an essential role of RNA degradosome complexes in shaping the transcriptome of Mycobacterium tuberculosis. Nucleic Acids Res. 2019, 47, 5892–5905. [Google Scholar] [CrossRef]
- Baek, Y.M.; Jang, K.J.; Lee, H.; Yoon, S.; Baek, A.; Lee, K.; Kim, D.E. The bacterial endoribonuclease RNase E can cleave RNA in the absence of the RNA chaperone Hfq. J. Biol. Chem. 2019, 294, 16465–16478. [Google Scholar] [CrossRef] [PubMed]
- Jester, B.C.; Romby, P.; Lioliou, E. When ribonucleases come into play in pathogens: A survey of gram-positive bacteria. Int. J. Microbiol. 2012, 2012, 592196. [Google Scholar] [CrossRef]
- Rock, J.M.; Hopkins, F.F.; Chavez, A.; Diallo, M.; Chase, M.R.; Gerrick, E.R.; Pritchard, J.R.; Church, G.M.; Rubin, E.J.; Sassetti, C.M.; et al. Programmable transcriptional repression in mycobacteria using an orthogonal CRISPR interference platform. Nat. Microbiol. 2017, 2, 16274. [Google Scholar] [CrossRef]
- Hopper, A.K.; Nostramo, R.T. tRNA Processing and Subcellular Trafficking Proteins Multitask in Pathways for Other RNAs. Front. Genet. 2019, 10, 96. [Google Scholar] [CrossRef]
- Oberbauer, V.; Schaefer, M.R. tRNA-Derived Small RNAs: Biogenesis, Modification, Function and Potential Impact on Human Disease Development. Genes 2018, 9, 607. [Google Scholar] [CrossRef]
- Singh, A.; Batra, J.K. Insight into the functional role of unique determinants in RNA component of RNase P of Mycobacterium tuberculosis. Int. J. Biol. Macromol. 2018, 119, 937–944. [Google Scholar] [CrossRef]
- Taverniti, V.; Forti, F.; Ghisotti, D.; Putzer, H. Mycobacterium smegmatis RNase J is a 5′-3′ exo-/endoribonuclease and both RNase J and RNase E are involved in ribosomal RNA maturation. Mol. Microbiol. 2011, 82, 1260–1276. [Google Scholar] [CrossRef] [PubMed]
- Abendroth, J.; Ollodart, A.; Andrews, E.S.; Myler, P.J.; Staker, B.L.; Edwards, T.E.; Arcus, V.L.; Grundner, C. Mycobacterium tuberculosis Rv2179c protein establishes a new exoribonuclease family with broad phylogenetic distribution. J. Biol. Chem. 2014, 289, 2139–2147. [Google Scholar] [CrossRef]
- Jain, C. Novel role for RNase PH in the degradation of structured RNA. J. Bacteriol. 2012, 194, 3883–3890. [Google Scholar] [CrossRef]
- Li, Z.; Pandit, S.; Deutscher, M.P. 3′ exoribonucleolytic trimming is a common feature of the maturation of small, stable RNAs in Escherichia coli. Proc. Natl. Acad. Sci. USA 1998, 95, 2856–2861. [Google Scholar] [CrossRef] [PubMed]
- Haiser, H.J.; Karginov, F.V.; Hannon, G.J.; Elliot, M.A. Developmentally regulated cleavage of tRNAs in the bacterium Streptomyces coelicolor. Nucleic Acids Res. 2008, 36, 732–741. [Google Scholar] [CrossRef]
- Cintron, M.; Zeng, J.M.; Barth, V.C.; Cruz, J.W.; Husson, R.N.; Woychik, N.A. Accurate target identification for Mycobacterium tuberculosis endoribonuclease toxins requires expression in their native host. Sci. Rep. 2019, 9, 5949. [Google Scholar] [CrossRef] [PubMed]
- Stanger, S.J.; Bernstein, I.R.; Anderson, A.L.; Hutcheon, K.; Dun, M.D.; Eamens, A.L.; Nixon, B. The abundance of a transfer RNA-derived RNA fragment small RNA subpopulation is enriched in cauda spermatozoa. ExRNA 2020, 2, 17. [Google Scholar] [CrossRef]
- Quendera, A.P.; Seixas, A.F.; Dos Santos, R.F.; Santos, I.; Silva, J.P.N.; Arraiano, C.M.; Andrade, J.M. RNA-Binding Proteins Driving the Regulatory Activity of Small Non-coding RNAs in Bacteria. Front. Mol. Biosci. 2020, 7, 78. [Google Scholar] [CrossRef] [PubMed]
- Muller, P.; Gimpel, M.; Wildenhain, T.; Brantl, S. A new role for CsrA: Promotion of complex formation between an sRNA and its mRNA target in Bacillus subtilis. RNA Biol. 2019, 16, 972–987. [Google Scholar] [CrossRef] [PubMed]
- Bruce, H.A.; Du, D.; Matak-Vinkovic, D.; Bandyra, K.J.; Broadhurst, R.W.; Martin, E.; Sobott, F.; Shkumatov, A.V.; Luisi, B.F. Analysis of the natively unstructured RNA/protein-recognition core in the Escherichia coli RNA degradosome and its interactions with regulatory RNA/Hfq complexes. Nucleic Acids Res. 2018, 46, 387–402. [Google Scholar] [CrossRef]
- Bloch, S.; Wegrzyn, A.; Wegrzyn, G.; Nejman-Falenczyk, B. Small and Smaller-sRNAs and MicroRNAs in the Regulation of Toxin Gene Expression in Prokaryotic Cells: A Mini-Review. Toxins 2017, 9, 181. [Google Scholar] [CrossRef]
- Caballero, C.J.; Menendez-Gil, P.; Catalan-Moreno, A.; Vergara-Irigaray, M.; Garcia, B.; Segura, V.; Irurzun, N.; Villanueva, M.; Ruiz de Los Mozos, I.; Solano, C.; et al. The regulon of the RNA chaperone CspA and its auto-regulation in Staphylococcus aureus. Nucleic Acids Res. 2018, 46, 1345–1361. [Google Scholar] [CrossRef] [PubMed]
- Rennella, E.; Sara, T.; Juen, M.; Wunderlich, C.; Imbert, L.; Solyom, Z.; Favier, A.; Ayala, I.; Weinhaupl, K.; Schanda, P.; et al. RNA binding and chaperone activity of the E. coli cold-shock protein CspA. Nucleic Acids Res. 2017, 45, 4255–4268. [Google Scholar]
- Mazzon, R.R.; Lang, E.A.; Silva, C.A.; Marques, M.V. Cold shock genes cspA and cspB from Caulobacter crescentus are posttranscriptionally regulated and important for cold adaptation. J. Bacteriol. 2012, 194, 6507–6517. [Google Scholar] [CrossRef]
- Resch, A.; Vecerek, B.; Palavra, K.; Blasi, U. Requirement of the CsdA DEAD-box helicase for low temperature riboregulation of rpoS mRNA. RNA Biol. 2010, 7, 796–802. [Google Scholar] [CrossRef][Green Version]
- Huang, X.; Miller, W. A Time-Efficient, Linear-Space Local Similarity Algorithm. Adv. Appl. Math. 1991, 12, 337–357. [Google Scholar] [CrossRef]
- Hogg, J.R.; Collins, K. Human Y5 RNA specializes a Ro ribonucleoprotein for 5S ribosomal RNA quality control. Genes Dev. 2007, 21, 3067–3072. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Smith, J.D.; Shi, H.; Yang, D.D.; Flavell, R.A.; Wolin, S.L. The Ro autoantigen binds misfolded U2 small nuclear RNAs and assists mammalian cell survival after UV irradiation. Curr. Biol. 2003, 13, 2206–2211. [Google Scholar] [CrossRef] [PubMed]



| Name | Northern or PCR Size | Location | Surrounding Genes | Expression |
|---|---|---|---|---|
| B11/6C (Candidate_1603) [8,9] | 93 | 4099386-4099478 (−) | rv3660c- rv3661 | H2O2 and pH = 5 |
| B55 (Candidate_84) [8,9] | 61 | 704187-704247 (+) | rv0609A- rv0610c | H2O2 and Mitomycin C |
| C8 (Mcr6, candidate_1621) [8,9,11] | 58, 70, 128 | 4168154-4168281 (−) | rv3722c- rv3723 | TBD a |
| F6 (Mcr14, candidate_29) [8,9,25] | 38, 58, 102 | 293604-293705 (+) | fadA2-fadE5 | H2O2 and pH = 5 |
| G2 (Candidate_1269) [8,9] | 67, 214, 229 | 1914962-1915190 (−) | tyrS-IprJ | TBD |
| ASdes (candidate_121) [8,9,22] | 48, 63, 68, 83, 94, 109, 149, 169, 195 | 918264-918458 (+) | within desA1 | TBD |
| ASpks [9] | 78, 89, 91, 102, 129, 142, 162 | 2299745-2299886 (+) | within pks12 | H202 |
| AS1726 [9] | 61, 77, 85, 110, 213 | 1952291-1952503 (−) | within Rv1726 | TBD |
| AS1890 [9] | 63, 109, 191, 238 | 2139419-2139656 (+) | within Rv1890 | TBD |
| MTS2823 or Ms1 [10,26,27] | 250, 300 | 4100669-4100968 (+) | rv3661- rv3662c | in vivo |
| MTS1338/DrrS [10,28,29] | 108, 109, ~160, 273 | 1960667-1960783 (+) | rv1733c- rv1735c | NO, stationary phase, in vivo |
| MTS0997/Mcr11 (Candidate_1693) [8,10,11,14,30] | 115 | 1413094-1413224 (−) | rv1264- rv1265 | in vivo, stationary phase, low pH, or hypoxia |
| Mcr1 [11] | >300 | 2029043-2029087 (TBD) | ppe26-ppe27 | TBD |
| Mcr2 [11] | 120 | 1108857-1108824 (TBD) | rv0967- rv0968 | TBD |
| Mcr3 (candidate_190) [8,11] | 118 | 1471619-1471742 (+) | murA-rrs | TBD |
| Mcr4 (candidate_1314) [8,11] | 200–250 | 2137148-2137103 (TBD) | fbpB-rv1887 | TBD |
| Mcr5 [11] | 80 | 2437823-2437866 (−) | within rv2175c | TBD |
| Mcr7 [11,31] | 350–400 | 2692172-2692521 (+) | rv2395-pe_PGRS41 | TBD |
| Mcr8 (candidate_1935) [8,11] | 200 | 4073966-4073908 (TBD) | rv3661–rv3662c | TBD |
| Mcr9 (candidate_1502) [8,11] | 66–82 | 3317634-3317517 (TBD) | ilvB1-cfp6 | TBD |
| Mcr10 [11] | 120 | 1283693-1283815 (+) | within rv1157c | TBD |
| Mcr12 [11] | 118 | 1228436-1228381 (TBD) | rv1072- rv1073 | TBD |
| Mcr13 [11] | 311 | 4315154-4315215 (TBD) | rv3866- rv3867 | TBD |
| Mcr15 [11] | >300 | 1535417-1535716 (−) | rv1363c- rv1364c | TBD |
| Mcr16 [11] | 100 | 2517032-2517134 (−) | within fabD | TBD |
| Mcr17 [11] | 82–90 | 2905457-2905402 (TBD) | within rv2613c | TBD |
| Mcr18 [11] | 82 | 3466287-3466332 (TBD) | within nuoC | TBD |
| Mcr19 [11] | 66–82 | 575033-575069 (+) | within rv0485 | TBD |
| ncRv11846/MrsI [12] | 100 | 2096766-2096867 (+) | blal-rv1847 | iron starvation, oxidative stress, and membrane stress |
| sncRNA-1 [13] | 25 | 4352927-4352951 | esxA-rv3876 | inside macrophages |
| sncRNA-6 [13] | 21 | 786003-786083 | rv0685- rv0686 | inside macrophages |
| sncRNA-8 [13] | 24 | 1471701-1471724 | murA-rrs | inside macrophages |
| Gene ID | Name | Species | Putative Function |
|---|---|---|---|
| MTB000026 | RnpB | M. bovis, Mtb, M. haemophilum | RNA component of RNase P: RNase P catalyzes the removal of the 5′-leader sequence from pre-tRNA to produce the mature 5′ terminus. |
| rv1340 | RphA | M. marinum, M. leprae, M. bovis, Mtb | Probable ribonuclease RphA (RNase PH). |
| rv2092c | HelY | M. marinum, M. leprae, M. bovis, Mtb, M. abscessus | DNA helicase activity. |
| rv2179c | Rnt | Mtb, M. smegmatis, M. leprae, M. marinum | Conserved hypothetical protein. |
| rv2228c | Rv2228c | N/A | Multifunctional protein. Has RNASE H, alpha-ribazole phosphatase, and acid phosphatase activities. |
| rv2407 | Rnz | Mtb, M. smegmatis, M. leprae, M. marinum, M. bovis | Endonucleolytic cleavage of RNA, removing extra 3′ nucleotides from tRNA precursor, generating 3′ termini of tRNAs. |
| rv2444c | Rne | M. bovis, Mtb, M. leprae, M. marinum, M. smegmatis | Putative RNase E. Plays a central role in the maturation of 5S and 16S rRNAs and the majority of tRNAs. Also involved in the degradation of most mRNAs. |
| rv2511 | Orn | Mtb, M. smegmatis, M. leprae, M. marinum, M. bovis | Involved in RNA degradation: 3′-to-5′ exoribonuclease specific for small oligoribonucleotides. |
| rv2681 | Rnd | Mtb, M. smegmatis, M. leprae, M. marinum, M. bovis | Conserved hypothetical protein. |
| rv2752c | Rnj | M. bovis, Mtb, M. leprae, M. marinum, M. smegmatis | Conserved hypothetical protein. |
| rv2783c | GpsI (Pnp) | M. marinum, M. leprae, M. bovis, Mtb, M. smegmatis, M. abscessus | Involved in mRNA degradation. Hydrolyses single-stranded polyribonucleotides processively in the 3′ to 5′ direction. |
| rv2902c | RnhB | M. marinum, M. leprae, M.bovis, Mtb, M. abscessus | Probable ribonuclease HII protein RnhB. |
| rv2907c | RimM | M. marinum, M. leprae, M. bovis, Mtb, M. smegmatis, M. abscessus | Essential for efficient processing of 16S rRNA. Probably part of the 30S subunit prior to or during the final step in the processing of 16S free 30S ribosomal subunits. It could be some accessory protein needed for efficient assembly of the 30S subunit. |
| rv2925c | Rnc | M. marinum, M. leprae, M. bovis, Mtb, M. smegmatis, M. abscessus | Digests double-stranded RNA. Involved in the processing of ribosomal RNA precursors and of some mRNAs. |
| rv3853 | RraA | M. leprae, M. bovis, Mtb | Regulator of RNase E activity a RraA. |
| rv3923c | RnpA | M. marinum, M. leprae, M. bovis, Mtb, M. smegmatis, M. abscessus | Ribonuclease P protein component RnpA. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coskun, F.S.; Płociński, P.; van Oers, N.S.C. Small RNAs Asserting Big Roles in Mycobacteria. Non-Coding RNA 2021, 7, 69. https://doi.org/10.3390/ncrna7040069
Coskun FS, Płociński P, van Oers NSC. Small RNAs Asserting Big Roles in Mycobacteria. Non-Coding RNA. 2021; 7(4):69. https://doi.org/10.3390/ncrna7040069
Chicago/Turabian StyleCoskun, Fatma S., Przemysław Płociński, and Nicolai S. C. van Oers. 2021. "Small RNAs Asserting Big Roles in Mycobacteria" Non-Coding RNA 7, no. 4: 69. https://doi.org/10.3390/ncrna7040069
APA StyleCoskun, F. S., Płociński, P., & van Oers, N. S. C. (2021). Small RNAs Asserting Big Roles in Mycobacteria. Non-Coding RNA, 7(4), 69. https://doi.org/10.3390/ncrna7040069