Abstract
The cancer genome is characterized by extensive variability, in the form of Single Nucleotide Polymorphisms (SNPs) or structural variations such as Copy Number Alterations (CNAs) across wider genomic areas. At the molecular level, most SNPs and/or CNAs reside in non-coding sequences, ultimately affecting the regulation of oncogenes and/or tumor-suppressors in a cancer-specific manner. Notably, inherited non-coding variants can predispose for cancer decades prior to disease onset. Furthermore, accumulation of additional non-coding driver mutations during progression of the disease, gives rise to genomic instability, acting as the driving force of neoplastic development and malignant evolution. Therefore, detection and characterization of such mutations can improve risk assessment for healthy carriers and expand the diagnostic and therapeutic toolbox for the patient. This review focuses on functional variants that reside in transcribed or not transcribed non-coding regions of the cancer genome and presents a collection of appropriate state-of-the-art methodologies to study them.
1. Introduction
Cancer specific regulation of transcription is manifested through ectopic activity of proximal and/or distal Regulatory Elements (REs) [1,2,3,4,5]. REs are divided into proximal or cis-acting regulatory elements (CREs), such as promoters, and distal or trans-acting regulatory elements (TREs) comprising of enhancers that establish physical contact with the former via long range 3D chromatin loops [6,7]. Given their proximity to transcriptional start sites, promoters predominantly function in a directional manner with regards to transcript orientation [8]. In contrast, enhancers can be located upstream or downstream of the target gene as well as within intronic regions and they can operate from a distance and in a bidirectional fashion [9]. Promoter–enhancer communication is mainly established in the form of intrachromosomal chromatin loops, while in some rare occasions enhancers may establish interchromosomal interactions with promoters [10,11]. Distinct epigenetic marks for each regulatory element facilitate dynamic chromatin accessibility and nucleosomal repositioning, which in turn dictate transcriptional status of the target locus.
More specifically, nucleosomes in enhancer and promoter elements are decorated with histone acetylation H3K27Ac, which generally marks open chromatin, while histone methylation such as H3K4Me3 is indicative of active promoters [8]. Epigenetic modifications of enhancer loci can be subdivided in three main types according to their activity: nucleosomes of neutral enhancers carry H3K4Μe1 histone tag, poised/bivalent enhancers are decorated with histone methylation active (H3K4Me1) and repressive (H3K27Μe3) mark at the same time, while active enhancers carry both H3K4Μe1 and H3K27Ac histone marks [12,13,14]. Moreover, promoters along with enhancers are the main binding sites of the Mediator Complex, which specializes gene expression patterns, recruits general transcription factors and establishes transcriptional memory between tissues and across development [15]. At a chromatin architecture level, promoter–enhancer communication is achieved through the extrusion of chromatin loops leading to the stabilization of topologically associated domains (TADs), which serve as functional genomic boundaries that restrict RE interactions and specify gene expression in a spatiotemporal manner [16]. The adjacent genomic space of TADs hosts specific motifs that facilitate binding of the CCCTC-binding factor (CTCF), a key regulator of chromatin conformation as it interacts with the cohesin complex, which acts as a chromatin loop stabilizer that dictates TAD formation [17,18,19,20,21]. Apart from chromatin architecture, enhancer transcription by itself may generate enhancer-RNAs (eRNAs- referring to non-coding RNAs transcribed from enhancer loci) [22,23]. On many occasions, eRNAs have a regulatory role in the establishment or maintenance of enhancer–promoter loops. At the level of genomic organization, enhancers can exert their regulatory function either individually or through the formation of clusters, known as super-enhancers, which concentrate transcription factor binding, are characterized by extensive eRNA transcription and serve as organizational centers for complex TAD formation [24,25,26,27].
Distinctively, all this operational heterogeneity that underlies enhancer–promoter communication ensures acute yet precise transcriptional responses at a spatiotemporal level. Given the pivotal control of regulatory elements in diverse physiological processes, such as cell-lineage specialization, differentiation, organogenesis and morphogenesis, immune cell diversification and stroma cell interactions, deregulation of their physiological function by mutations often serves as the molecular basis of pathological conditions like cancer [26,28,29]. Identification of such variants in tumor progression can be used as a diagnostic or prognostic tool to predict the clinical outcome for the patient and/or tailor therapeutic strategy in a personalized manner [30,31]. It is therefore imperative to identify and most importantly, functionally dissect genetic variability in the cancer genome that is characterized by extensive variability, in the form of SNPs, or CNAs across wider genomic areas [32,33]. International consortia document and metanalyze functional genomic experiments, providing insights regarding the effect of tumor-specific driver mutations on regulatory elements during neoplastic progression [34,35,36,37]. Such consortia are the Encyclopedia of DNA Elements (ENCODE—a public research consortium focused in identifying all functional DNA regulatory elements) [38], NONCODE which is an integrated knowledge database dedicated to non-coding RNAs (excluding tRNAs and rRNAs) [39], and The Cancer Genome Atlas (TCGA—a landmark program of cancer genomics, containing genomic, epigenomic, transcriptomic, and proteomic data from tumors, along with the clinical profile of the patients) [40].
Although some genetic variants are stably inherited and occasionally predispose for hereditary forms of cancer, carcinogenesis itself relies on inactivation of DNA repair mechanisms, leading to genomic instability and extensive accumulation of a mutational burden on a global scale [41]. Genomic variants can be further classified based on structural (SNPs or CNAs), expression (transcribed or not transcribed sequences) or functional (coding or non-coding regions) criteria, all of which ultimately reflect the mechanism through which these genetic lesions are implicated in the development and progression of human malignancies [42,43,44,45]. This review focuses on the mechanisms though which non-coding regulatory variants in transcribed or non-transcribed parts of the genome control carcinogenesis, together with the appropriate state-of-the-art methodologies to identify and study them.
2. Genetic Variability in the Cancer Genome
2.1. Structural Classification of Mutations in Cancer
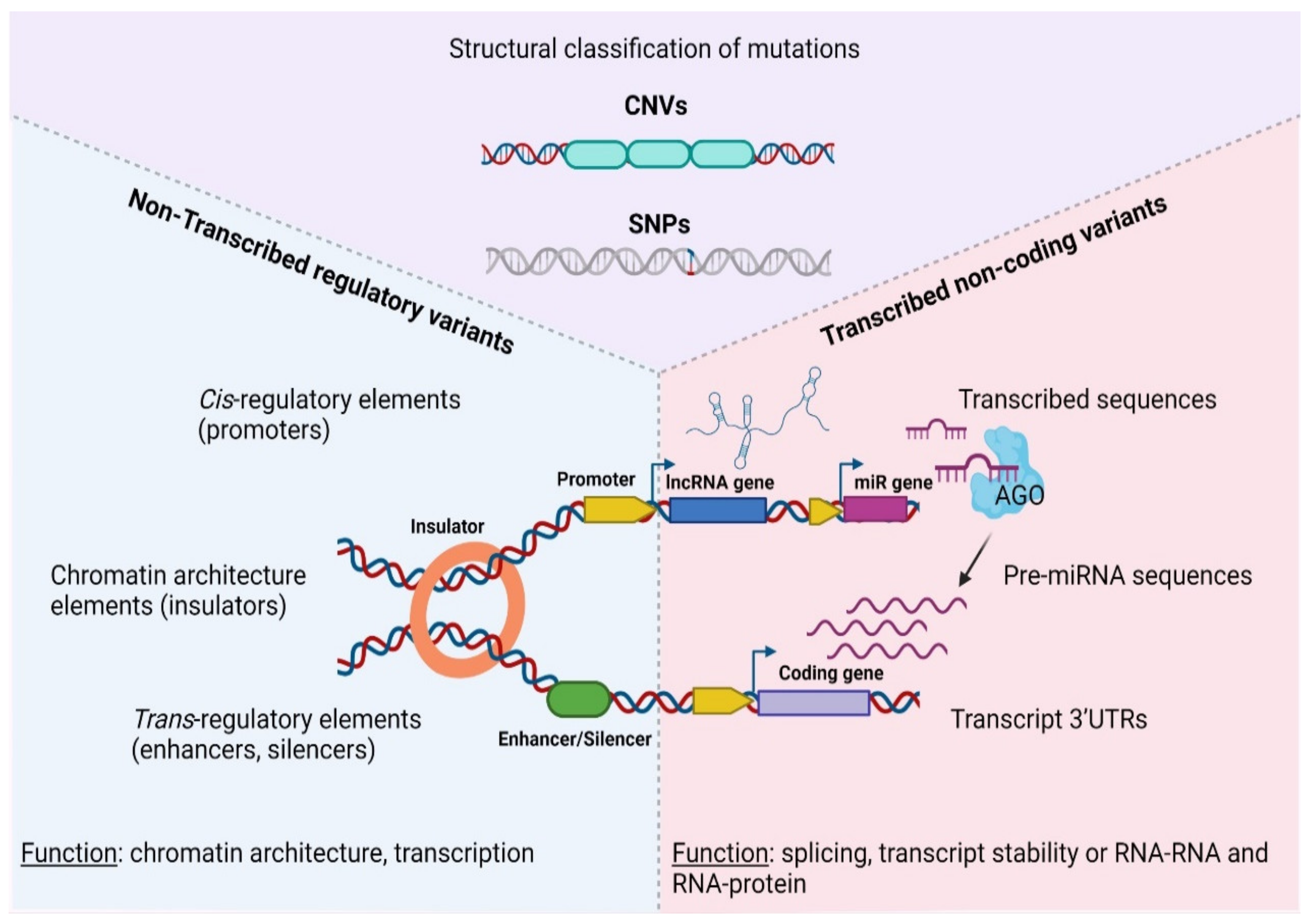
At the molecular level, SNPs and/or CNAs can reside both in coding, as well as non-coding sequences that are either transcribed or not inside tumors (Figure 1). Depending on the nature of the mutation and the function of the underlining sequence, genetic lesions fuel carcinogenesis through a diverse array of mechanisms, including but not limited to chromatin modification, transcriptional regulation and alternative splicing, to altered transcript/protein structure or activity due to premature stop codons, non-synonymous amino-acid changes and aberrant gene fusions [46,47,48,49]. Therefore, different types of mutations or affected sequences predispose for cancer via an array of distinct mechanisms that must be examined separately.
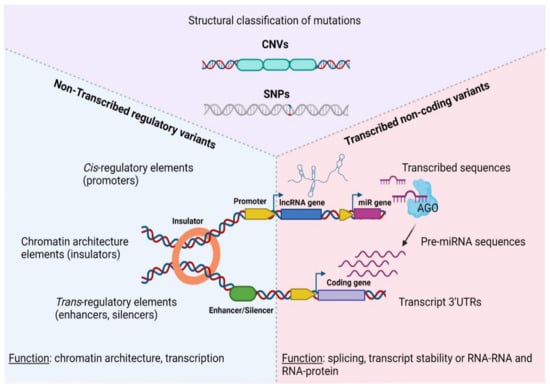
Figure 1.
Categorization of genetic variations in the non-coding coding cancer genome. AGO—Argonaute protein, CNV—Copy Number Variation, SNP—Single Nucleotide Polymorphism, UTR—Untranslated Region, lncRNA—long non—coding RNA, pre-miRNA, precursor microRNA. Created with BioRender.com, permission: 15 July 2021.
Focusing on single nucleotide polymorphisms, genome-wide association studies (GWAS) were successful in identifying their mechanistic interplay with normal development or pathology [50]. Depending on the strength of their alleles, SNPs can be characterized as high or low risk factors for complex traits, such as cancer [51]. Nevertheless, association between SNP alleles and phenotypic impact can be confounding due to linkage disequilibrium that segregates driver mutations (that directly control the trait of interest) together with passenger mutations (that are passively inherited with the former but are regulatory neutral) within populations. Therefore GWAS approaches should be accompanied by extensive and careful mechanistic characterization with the aim of determining which of the associated SNPs are the true causative factors of the disease [52].
At the level of CNAs, two main categories can be identified: (i) germline CNAs that include duplications or deletions and (ii) somatic copy number alterations of specific loci. Germline CNAs range from 50 bp up to 1 Mb in length and predispose for hereditary types of cancer, such as familiar breast cancer. Somatic CNAs are typically longer than 1 Kbp (100 Kbp on average [53,54]) and like germline CNAs, include duplications (known as Copy-Number Gains or CNGs), or deletions (representing Copy-Number Losses or CNLs) [55]. Both types of somatic CNVs (CNGs and CNLs) are prominent lesion types in tumors that are characterized by extreme Chromosomal INstability (CIN) [56].
Presence of CNAs is heavily linked with malignant manifestation through three main mechanisms: (i) alterations in gene dosage, in which the copy number of at least one gene locus is affected [57,58], (ii) gene fusions (mainly due to genomic deletions) [59] and (iii) alterations of cis and/or trans regulatory elements [60,61,62,63]. In some cases, a correlation between copy number variations and DNA methylation status of the CpG islets for a given locus has been reported to negatively affect target gene expression [64]. Apart from cancer, germline CNAs also predispose for various developmental disorders and diseases, such as autism, schizophrenia with a parental-specific pattern of inheritance, for which they can be also potent biomarkers for prenatal diagnosis [65,66].
2.2. Functional Classification of Mutations in Cancer
Regardless of their structure, genomic variants (including both SNPs or CNAs) can be further classified according to their occurrence in transcribed or non-transcribed sequences (Table 1, [67]). This type of classification underpins the function through which different variants associate with disease etiology.

Table 1.
Structural and functional genetic diversity in cancer.
For example, functional variants that occur in transcribed portions of the genome generally associate with altered transcript message or function (manifested as modified exonic sequences, alternative splicing, modified UTRs, altered ncRNA folding and/or gene fusions [96,97,98]). Interestingly, most GWAS/transcribed regulatory variants are not limited to protein coding genes but primarily localize in transcribed non-coding sequences that may generate regulatory transcripts with low or no protein coding potential [99,100,101,102]. Non-coding RNAs (ncRNAs) are categorized based on their processed length, with transcripts less than 200 nt referring to short non-coding RNAs (consisting mainly of microRNAs-miRNAs [103], small nucleolar RNAs-snoRNAs [104] and piwi-interacting RNAs [105]), in contrast to long non-coding RNAs (lncRNAs), which comprise transcripts with lengths larger than 200 nt [106].
3. Non-Transcribed Regulatory Variants
Variability in non-transcribed regulatory sequences (e.g., promoters, enhancers, CTCF sites) strongly associates with a mechanistic impact of non-coding variants during neoplastic development [107]. Genome-wide studies revealed an extensive correlation of these variants with conditionally deregulated spatiotemporal gene expression networks and disrupted genomic organization in various tumor forms, thus highlighting the importance of genetic non-coding variability in cancer onset and progression [67,108]. Rare SNP alleles, associated with increased risk of carcinogenesis (and/or other diseases), are enriched within expressed quantitative trait loci (eQTLs), with prominence in promoter regions of oncogenes and tumor-suppressors [109,110,111].
Apart from SNPs, somatic CNAs act as the driving force of the CIN subtype that is typical for various neoplasms. For example, 65% of gastric adenocarcinomas are categorized as CIN and since CNAs are one of the leading causes of extensive genomic and transcriptomic alterations, defining their functional role has a clinical interest [112]. Another cancer type with high percentage of CIN is colorectal cancer, in which CNVs contribute to loss of heterozygosity in TP53 and APC, or amplification in KRAS and FGFR1, leading to poor prognosis due to drug resistance [113,114]. Despite of their discovery and statistical association with diagnostic or prognostic markers, such variants often lack functional characterization due to the small effect that a single SNP may have in gene expression, together with tissue-specific restrictions in the expression of the target gene [115,116,117]. Therefore, it is crucial to first stratify and subsequently present some of the elucidated mechanisms through which non transcribed regulatory variants dictate neoplastic development.
3.1. Genetic Variability in Promoters
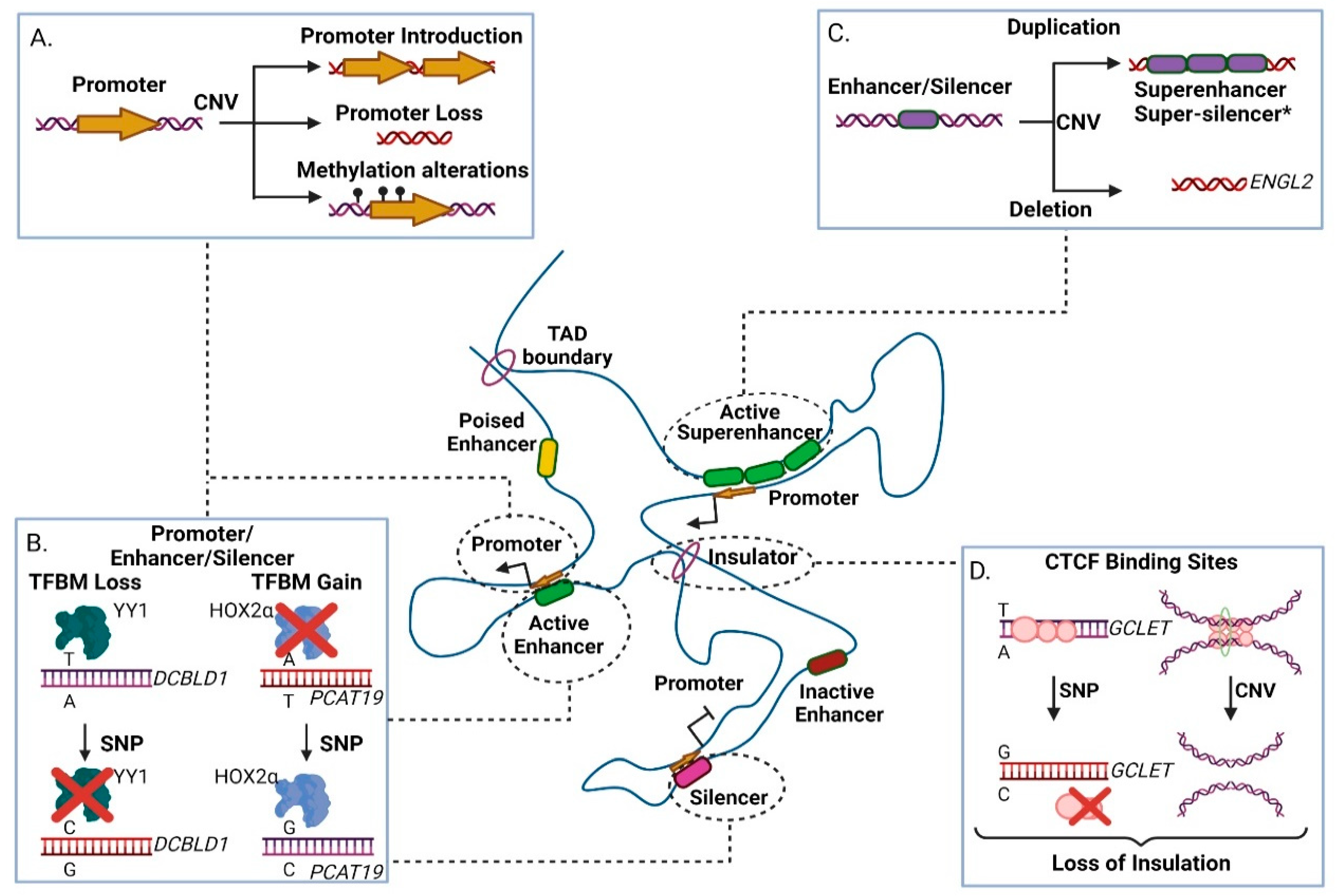
There are numerous examples of genomic variants in cis-regulatory regions that affect transcription of coding or non-coding target genes [118,119,120]. Promoters (especially the core promoter) are the prime regulatory units of transcription, as they embed transcription factor motifs that enable formation of the Pre-Initiation Complex (PIC) adjacent to the transcription start site of the gene [121]. In many cancer types of promoter activity is altered by inherited or somatic mutations, leading to the modulation of cryptic promoter activity, loss of promoter DNA methylation or alteration (including loss or gain) of key regulatory motifs (Figure 2A, [122]).
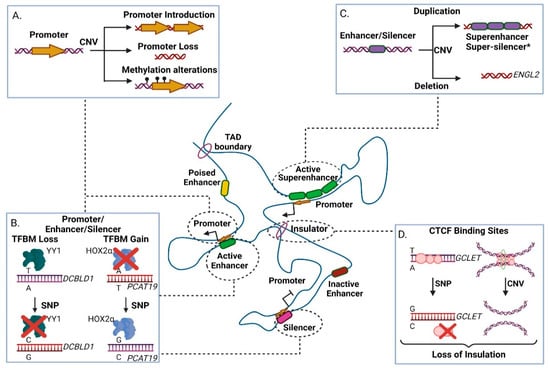
Figure 2.
The effect of genomic variants in non-transcribed gene regulatory elements within a Topologically Associated Domain (TAD). (A) Effects of Copy Number Variations (CNVs) in promoters. Duplication events may lead to ectopic promoter introduction, while deletion event may result in loss of crucial promoter elements. Depending on the occasion, duplication or deletion may result in DNA methylation alterations, modulating transcriptional activity. (B) Effect of Single Nucleotide Polymorphisms (SNPs) in promoter, enhancer and silencer elements. Presence of SNPs may lead to either increased or decreased affinity in transcription factor binding motifs, thus altering the element’s function. (C) Effects of CNVs at enhancer and silencer elements. Duplications may result in creation of super-enhancers or, hypothetically*, super-silencers. Deletions may lead to loss of crucial transcription factor binding motifs, thus impairing regulatory element function. (D) Presence of a risk SNP to a CTCF site may lead to loss of insulation due to CTCF site disruption. CNV occurrence may also lead to loss of insulation due to deletion of a CTCF site. TFBM-Transcription Factor Binding Motif. CTCF-CCCTC-binding factor. Created with BioRender.com, permission: 15 July 2021.
For example, rs11672691 (G/A) and rs887391 (T/C/A), two risk-associated SNPs for poor prognosis in prostate cancer, map to a genomic region with bifunctional role acting either as promoter or enhancer. Presence of the cancer-predisposing alleles facilitates promoter-to-enhancer switching, leading to reduced binding capacity of the transcription factors NKX3.1 and YY1 to the promoter of the short isoform of the PCAT19 (Prostate Cancer Associated Transcript 19) lncRNA transcript. This favors accumulation of the long PCAT19 isoform that interacts with HNRNPAB and promotes the expression of cell cycle genes that subsequently fuel tumor growth and metastasis [68]. Another example refers to the high-risk SNP rs17079281 (C/T) that resides within the promoter of the DCBLD1 gene and predisposes for lung cancer in Asian and European populations. The predisposing C allele of this SNP reduces the binding affinity of the YY1 transcription factor that normally represses transcription of the gene, ultimately leading to increased levels of the DCBLD1 oncogenic protein in the mutated tissues (Figure 2B) [69]. Another study in mice showed a gene directly affected by SNPs and CNAs, Plekha5, which normally acts as a suppressor of metastasis. In presence of SNPs or CNAs, Plekha5 is deregulated, leading to an increase in the metastatic rate of the cells [123]. The rs2267531 SNP lies within the promoter of Glypican-3 gene (in Xq26) and the CC/C genotype of which has been correlated with susceptibility and reduced overall survival of patients with hepatocellular carcinoma (HCC) [70]. In addition, the G allele of rs2280059 SNP, which lies within the promoter of HSPH1, is able to increase its expression levels, leading to enhanced resistance of the cancer cells to treatment in patients with advanced non-small-cell lung cancer [71]. Apart from motif changes, CNVs may also alter the methylation status of oncogenic promoters (e.g., through the demise of methylation sites) leading to increased proliferation advantages for the mutated cell subpopulations, evidence of which have been extensively found in lung adenocarcinoma [124]. Collectively, genetic variability in promoter regions often associates with altered gene expression that links to disease progression of various cancer types.
3.2. Genetic Variability in Enhancers
Most cancer enhancers show cell- and/or stage selectivity in their activation patterns [125,126,127], therefore their associated genetic variability is ideal for assessing personalized predisposition or therapy. Since enhancers (and super-enhancers) function through DNA binding motifs, their activity is vulnerable to variation that modulates the binding capacity of transcription factor proteins, thus altering transcription of the target gene [128]. Presence of CNA (and other architectural disarrangements) combined with loss of insulation events can lead to ectopic enhancer creation or activity resulting into metaplastic differentiation associated with malignancy (Figure 2C, [129,130,131]). For example, genome-wide CNA studies have correlated a deletion in an ovarian-specific enhancer with altered expression of EGLN2, an enzyme that mediates hydroxylation and subsequent degradation of the HIF1A protein (a master regulator of oxygen homeostasis) in normoxia (Figure 2C, [75]).
Parallel to CNAs, GWAS studies have also pinpointed the association between SNPs and enhancer activity in cancer. For example, the variant rs11672691 (G/A), which resides in an intronic enhancer at the lncRNA PCAT19 locus, correlates with prostate cancer predisposition and aggressiveness [132,133,134]. More specifically, the risk allele rs11672691-G enhances the binding activity of the novel transcription factor HOXA2, which in turn regulates expression of PCAT19 in prostate cancer through enhancer–promoter loop formation (Figure 2B) [135]. Single nucleotide editing in combination with ChIP-seq (Chromatin Immunoprecipitation followed by Sequencing) experiments revealed that binding of HOXA2 positively regulates not only PCAT19 but also its neighboring locus CEACAM21. Thus, the interplay of rs11672691 with the regulatory circuit of HOXA2, PCAT19 and CEACAM21 is linked to advanced cell growth and invasion with a significant clinical impact on prostate cancer disease aggressiveness and severity, highlight the role of enhancer mutations in the regulation of neighboring coding and non-coding targets in cancer tissues [68,135].
With regards to SNPs, rs67311347 (G > A) shows a positive correlation with cancer cell proliferation in patients with Renal Cell Carcinoma (RCC). The A allele creates a binding site for ZNF8 within an enhancer element regulating the tumor-suppressor lncRNA ENTPD3-AS1, leading to its increased expression. ENTPD3-AS1 interacts with miR-155-5p and activates the expression of HIF-1a in RCC [76]. The SNP rs4693608 lies within an enhancer regulating the expression of HPSE, by affecting the self-regulation of the oncogenic transcription factor in acute lymphoblastic leukemia (ALL), with the A allele carriers escaping the methylation of the enhancer [77].
An independent study revealed another layer of complexity for this enhancer-like regulatory region, which seems to have a bifunctional role. The presence of additional variants that also reside in the PCAT19 locus plays a crucial role in PCAT19 transcript isoform generation (PCAT19-short and PCAT19-long isoforms respectively) with the PCAT19-long elevated mRNA levels determining progression of prostate cancer. Specifically, the SNPs variants rs11672691 and rs887391 that reside in the promoter region of the PCAT19-short isoform can switch the regulatory identity of the element from promoter to enhancer. Presence of these two risk alleles disturbs binding capacity of the transcription factors NKX3.1 and YY1 to the promoter of the PCAT19-short isoform. At the same time the same risk SNPs reinforce enhancer activity of the bifunctional regulatory element leading to increased expression of PCAT19-long isoform through a promoter–enhancer interaction. Subsequently PCAT19-long isoform interacts with HNRNPAB and thus influences expression of cell cycle genes leading to acceleration of tumor growth and metastasis [68].
The expression of another prostate related lncRNA, PCAT1 (Prostate Cancer Associated Transcript 1), is also modulated at the transcriptional level by a cancer-associated SNP with pivotal function in prostate cancer. Initially, PCAT1 was reported to be implicated in early prostate cancer cell proliferation, yet recently it was shown to be involved also in castration-resistant, advanced prostate tumors [84,136]. PCAT1 expression is modulated by the risk SNP variant rs7463708 (T > G) located within an enhancer regulatory element that lies 78 kb away from the PCAT1 Transcriptional Start Site (TSS). PCAT1 promoter and its enhancer reside within a conserved TAD domain, which indicates the potential of chromatin loop extrusion between them. The T allele intensifies the binding affinity of the ONECUT and AR transcription factors, which in turn regulate PCAT1 transcription. Subsequently, the PCAT1 transcript interacts with the LSD1 and AR proteins facilitating their recruitment to enhancer regulatory elements of GNMT and DHCR24 that are androgen-late response genes that correlate with prostate cancer progression [72,137,138].
Apart from prostate, SNPs in enhancers also affect other forms of cancer. rs35252396 (AC > CG) refers to a two base pair substitution variant that is strongly associated with clear cell renal cell carcinoma. This particular variant resides in an enhancer element at 8q24.21 between the genomic loci of MYC and PVT1 and along with the SNP rs6983267, whose regulatory function is well characterized in colorectal and prostate carcinoma. rs35252396 affects chromatin accessibility in this area, increasing binding of hypoxia inducible factors in this enhancer element [73,74,139,140]. rs6983267 together with rs35252396, highlight the predisposing effect of neighboring yet separately segregating regulatory genetic lesions in carcinogenesis.
3.3. Genetic Variability in Silencer Elements
Mutations in distal silencer elements are less understood due to the biased focus on activating enhancers, even though the latter may also act as silencers and vice versa in different tissues and cell types [141]. Silencer elements, just like enhancers, contain transcription factor binding sites, that form chromatin loops with promoters (Figure 2B,C), preferably those with high levels of trimethylation of Lysine 27 in Histone 3 (H3K27me3) epigenetic marker [142]. Supposedly, the formation of a super-silencer is possible, but so far there are insufficient data that support their existence [143]. A putative silencer regulating ESR1 and RMND1 expression can be found in 6q25.2, and the SNP rs910416 contained within it shows allele specific binding of MYC. This disrupts the proper function of the silencer, leading to breast cancer development [144]. Another example that highlights the function of such repressive chromatin loops, refers to the regulation of Kit locus by GATA1, which has a repressive role in hematopoietic differentiation [145]. Other silencers are characterized by the presence of motifs of FRA1, USF1 and USF2, EBF1, BACH2, and the RFX family among others, which display repressing activities [146,147,148,149,150]. In contrast to the binding of activators in unmethylated or lowly methylated enhancer elements, a proportion of these suppressors can bind to methylated sequences as well, indicating that some silencers may show activity even in their DNA methylated form [151,152]. The SNP rs249473 and especially the risk allele A, which lies within a silencer of the AKT1 locus (encoding for the AKT protein, part of the PI3K/AKT/mTOR signaling pathway), abrogates its silencing activity by creating a binding site for YY1, which in turn activates AKT1 transcription and elevates the risk of endometrial cancer [78].
3.4. Genetic Variability in Insulator Elements
Insulators are DNA elements which are recognized by CTCF and facilitate creation of inter-domain boundaries, conferring separation of promoters and enhancers or insulation against the spread of heterochromatin regions [153,154]. Loss of insulator elements may occur due to the presence of SNPs that alter the CTCF binding site or the methylation status of the region [155]. Moreover, CNAs that promote genomic rearrangements of CTCF sites can lead to enhancer hijacking, that associates with increased levels of a putative oncogenes, such as MYCN that is one of the main drivers for neuroblastoma (Figure 2D) [80,156]. The SNP rs60507107 is correlated with increased risk of lung cancer, as the A allele reduces the binding affinity of CTCF at a CTCF binding site in the first intron of DAGLA (in 11q12.2), leading to its altered expression in lung cancer [157]. Collectively, these examples highlight the functional diversity through which genetic variability in regulatory elements predisposes for neoplastic development and progression.
Apart from coding genes, genetic aberrations may also disturb transcriptional regulation of lncRNAs at a chromatin architecture level. For instance, GCLET (Gastric Cancer Low-Expressed Transcript) is a novel lncRNA with a gastric cancer related variant rs3850997 T > G at 16p13 in the third intron of the GCLET genomic locus. Expression analysis, including eQTLs, revealed a strong association between high expression levels of GCLET and improved patient survival. Moreover, in vitro experiments showed that the rs3850997-T allele is bound by the CTCF transcription factor with higher affinity compared to G allele (Figure 2D). CTCF exerts an inhibitory function, so when bound to the relevant intronic region prevents chromatin loop formation between the intron/SNP variant and GCLET promoter region, ultimately precluding lncRNA transcription [79]. Furthermore, GCLET competes with miR-27a-3p to increase FOXP2 expression, therefore affecting lymph node invasion and metastasis. Inferentially, the T allele of the rs3850997 variant represses transcription of GCLET lncRNA and absence of the transcript contributes to gastric cancer progression with a significant impact on patient clinical prognosis [79,158,159,160].
4. Transcribed Non-Coding Variants
Apart from regulatory elements, cancer-related genetic variability is also embedded in transcribed, yet non-coding sequences. Transcribed non-coding Variants (referred to as TncVs thereof) exist both in coding and non-coding transcriptional units and fuel carcinogenesis through a distinct set of mechanisms compared to their counterparts in coding sequences. For example, TncVs can modulate the stability of the resulting transcript through abnormal splicing patterns, UTR variations that create or disrupt miRNA binding pockets, or through alterations in lncRNA secondary structure that influence interaction with regulatory partners (both protein and RNA molecules) [107,161]. The latter can lead to differential regulation of target gene expression, via loss of RNA-chromatin and/or RNA–protein complex formation, concurrently with disruption of TAD architecture [161]. Such cancer-related transcribed variability is not restricted to the DNA level, but also arises at the RNA level, giving rise to the very promising and largely unexplored field of epitranscriptomics which again may operate from within coding and non-coding transcripts in a similar manner to inherited mutations [162].
4.1. Non-Coding Variants Affecting miRNA Targeting and Biogenesis
Small RNA sequencing efforts have identified hundreds of miRNAs involved in cancer progression and tumorigenesis for a variety of cancer types and stages [163]. miRNA signatures with significant prognostic and diagnostic properties often reflect the tissue- and/or cancer-specific properties that characterize the expression of this class of non-coding transcripts [164]. In terms of function, miRNAs act on the basis of sequence complementarity with their cognate target-mRNA(s) [165,166,167]. Thus, any sequence variation, even in the form of single nucleotide polymorphisms that occurs within the seed sequence of their genomic loci, can alter targeting affinity [168]. Although GWAS approaches have revealed the importance of SNPs in oncogenic or tumor-suppressing miRNAs, functional characterization for the majority of such alterations awaits experimental validation [168,169,170].
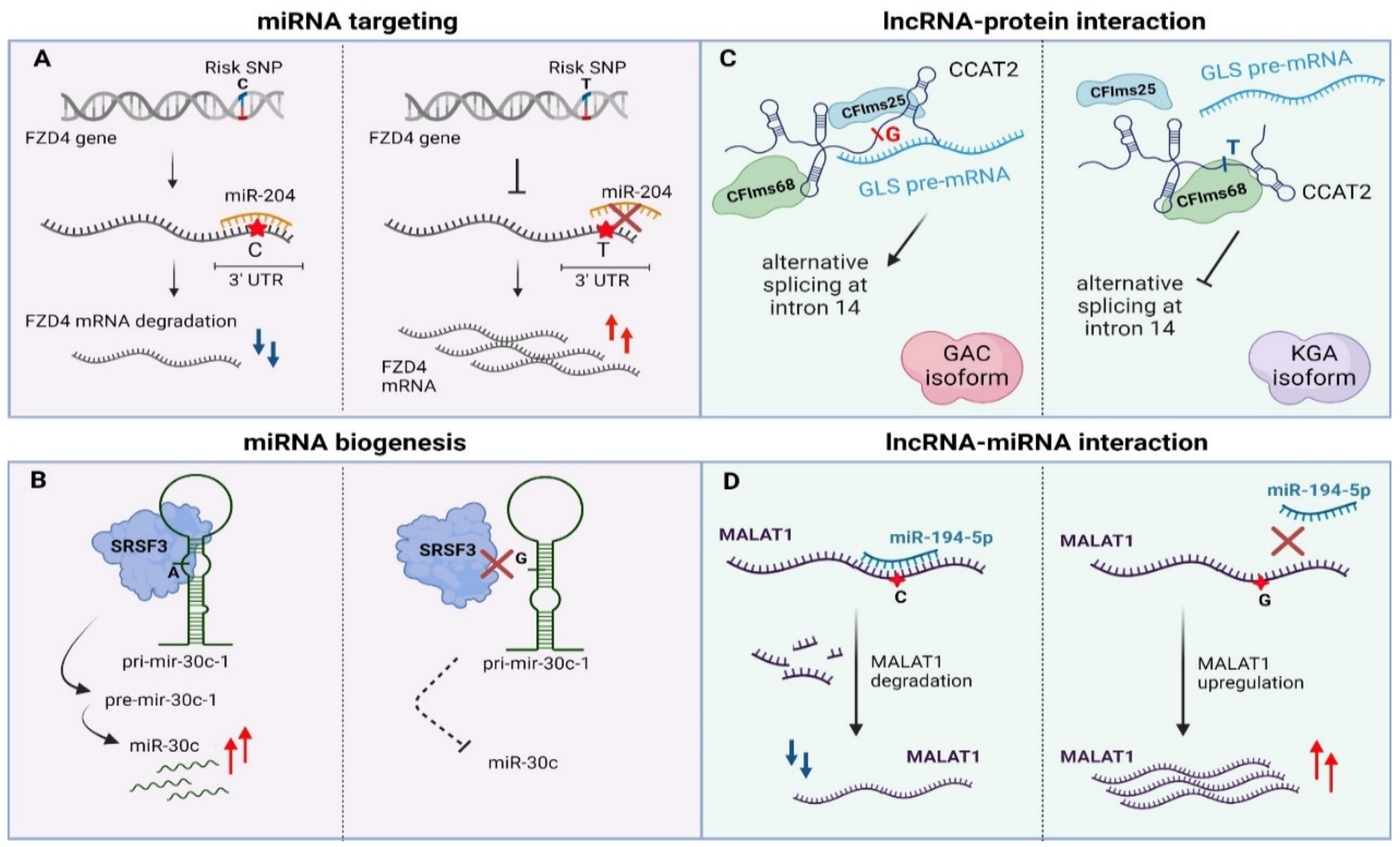
Apart from genetic lesions in miRNA transcripts, variability can also arise within miRNA binding sites in 3’UTRs of their target genes [171,172,173,174]. Such variability may ectopically create or disrupt a miRNA binding site in malignant or even pre-cancerous tissues. The miR-155-5p is highly expressed in melanoma patients and targets the 3′UTR region of TYRP1 (Tyrosinase Related Protein 1) mRNA in a SNP-dependent manner leading to decreased TYRP1 transcript levels. It has been shown that different combinations of AA/CC alleles of rs683/rs910 SNPs that lie in the 3′UTR region of TYRP1 mRNA affect the expression of TYRP1 at a post-transcriptional level while there is also a correlation with melanoma metastasis [81]. Another miR-SNP (rs713065, T to C change) in the 3′UTR region of FZD4, which is a consequential epidemiological biomarker for non-small-cell lung carcinoma (NSCLC), comprises a binding site for miR-204. The predisposing C allele of this SNP enhances binding of miR-204 compared to the wild type allele (T), leading to down-regulation of FZD4 through cleavage, uridylation and degradation of its mRNA. Subsequently, the miR-204-SNP mediated loss of FZD4 induces deregulation of key components of Wnt/Catenin signaling associated with impairment of colony formation and cell migration of NSCLC cancer cells (Figure 3A) [82].
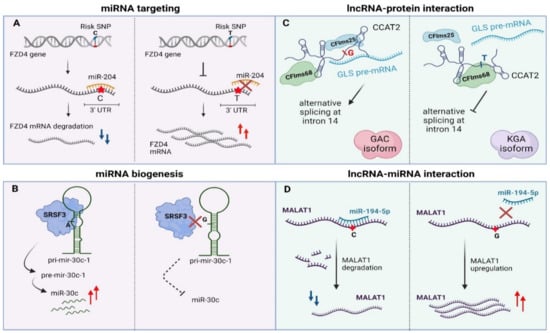
Figure 3.
Examples of genomic variants affecting non-coding RNAs function. (A) 3′UTR-occurring SNPs disturb miRNA targeting via sequence-based mismatches. (B) SNPs in pri-miRNA transcript can alter miRNA biogenesis. (C) SNPs can affect lncRNA–protein interactions via alterations in lncRNA secondary structure (D) SNPs can alter lncRNA stability via modulation of miRNA binding sites. UTR—Untranslated region, pri-miRNA—primary miRNA, pre-miRNA—precursor-miRNA, lncRNA—long non-coding RNA, miRNA—microRNA, GLS—glutaminase, GAC—glutaminase isoform C, KGA— kidney glutaminase isoform. Created with BioRender.com, permission: 15 July 2021.
An analogous example of TncV refers to SNP rs1071738 (G common allele, C minor allele in European individuals) at the miR-96/miR-182-binding site within the Palladin 3′-UTR with fundamental function in breast cancer metastasis. The ancestral C allele allows miRNA:mRNA binding while the alternate G allele disrupts it. miR-96 and miR-182 have anti-migration and anti-invasion roles in breast cancer cells that is associated with downregulation of Palladin, a phenotype which was confirmed by in vivo experiments. At the therapeutic level, in vivo delivery of miR-96 or miR-182 (fully complimentary with their binding site in Palladin-3′UTR) by using hydrogel-embedded gold nanoparticles with efficient release of miRNAs, led to a remarkable decrease of cancer cells’ metastatic capability [83]. Finally, the rs1048638 SNP that harbors within the 3′UTR of CA9 (Carbonic anhydrase IX) mRNA is strongly correlated with clinical features (overall survival, poor prognosis, recurrence) of HCC patients. The A allele of this SNP creates a binding site for miR-34a targeting that declines CA9 mRNA levels and affects cell proliferation and metastasis of HCC cells [175]. In conclusion, functional characterization of miRNA-associated TncVs in cancer progression can offer novel therapeutic opportunities at the genetic basis of cancer.
Parallel to miRNA binding sites, TncVs that reside in pri-miRNA sequences can affect cancer progression through defects in miRNA biogenesis [88,176]. A thoroughly characterized example is rs928508, referring to a G to A substitution that is present in the terminal loop of pri-mir-30c-1, perturbing its secondary RNA structure and subsequently leading to increased levels of mature miR-30c in breast cancer. Particularly, the G/A substitution, which lies in a CNNC motif, facilitates interaction of the pri-miRNA with SRSF3, a protein involved in alternative splicing and Drosha-mediated processing of pri-miRNA maturation [85,177,178]. Experimental validation with SHAPE (Selective 2’-hydroxyl acylation analyzed by primer extension, a technical approach for RNA structural analysis at single-nucleotide resolution) and toeprint assays (an assay using a fluorescent-labeled oligonucleotide to prime the reverse transcription step) proved that SRSF3 specifically recognizes the CNNC motif in a dose-dependent manner in MCF7 cancer cells [179,180]. Importantly, G/A variation promotes formation of a particular tertiary RNA structure of the pri-miRNA transcript, allowing stronger interaction with the SRSF3 protein and ultimately proper biogenesis of the miRNA (Figure 3B) [85]. Of note, the same variant has been previously linked to increased miR-30c expression in breast and gastric cancer patients [86,181]. Finally, miR-30c has also been shown to be a tumor prognostic biomarker for breast cancer, modulating chemoresistance through regulation of TWF1 and IL-11 [182].
In independent example of a mutation that affects miRNA biogenesis refers to rs7911488 (T > C), located in pre-miR-1307 and ultimately interfering with progression of colorectal cancer (CRC). Homozygous T alleles of this point mutation lead to elevated expression of mature miR-1307, which in turn binds to 3′UTR of the PRRX1 mRNA and diminishes its expression levels. Downregulation of PRRX1 enhanced proliferation and migration of CRC cells; however, the exact mechanism needs to be further clarified [87]. Finally, the A allele of rs11671784 SNP within the miR-27a is associated with high susceptibility to gastric cancer. Correlation studies indicated that this variant influence the maturation process of miR-27a leading to decreased expression levels of miR-27a in gastric cancer patients. The diminished levels of miR-27a activate the enhanced expression of HOXA (miR-27a-target gene) affecting tumor growth of gastric cancer cells [88]. In conclusion, genetic lesions that affect miRNA biogenesis/binding sites frequently associate with site-specific cancer progression.
4.2. Non-Coding Variants Affecting lncRNA Function
The last decade, lncRNAs gained particular interest in cancer biology given their cancer- and tissue-specific expression in various malignancies [161,183,184,185]. The intricate non-coding nature of their function relies on interactions with i) protein complexes (transcription factors, spliceosome, RNA binding proteins-RBPs, chromatin modifiers in the nucleus and RBPs, ribosomes and other proteins in the cytoplasm), ii) other non-coding transcripts (such as lncRNAs and miRNAs) or iii) DNA through triple helix formation [186,187]. As a result, genetic variability in lncRNA loci has also been associated with cancer predisposition, yet few cases have been experimentally validated, given the increased difficulty of functionally dissecting lncRNAs compared to miRNAs [188,189]. Among the most interesting examples of variations that occur in lncRNAs are SNPs that disorder lncRNA transcript functionality with a cancer-driving potential. Due to relaxed evolutionary constrains on primary sequence [190], lncRNA loci can easily accumulate genetic variability in cancer cells that can affect proper transcript folding, ultimately modulating lncRNA interactions with their protein partners or other regulatory molecules [191].
More specifically, lncRNAs may contribute to post-transcriptional regulation by affecting splicing [192], mRNA stability [193] or as precursors/regulators of miRNA biogenesis [194,195]. Presence of distinct sequence motifs in combination with a particular secondary structure facilitates binding of splicing factors and other RBPs enabling the lncRNA–protein functional interplay [161,196,197]. Cancer-risk variations that occur in these motifs may disturb this interaction, leading to deviant molecular signaling pathways and finally to malignant transformation [198]. An intriguing example is rs6983267 SNP (G/T) that resides in the lncRNA locus CCAT2 (Colon Cancer Associated Transcript 2) and is correlated with colon cancer metabolism (enhanced glutaminolysis) and cell proliferation. This transcribed SNP can recruit two subunits of the cleavage factor Im complex (CFIm, CFIms25 and CFIm68 subunits) in an allele-specific manner. CCAT2 transcripts containing the G-allele, allow binding of CFIms25 with higher specificity compared to T-allele transcripts, which in turn has stronger propinquity for the CFIm68 subunit. This dominant effect regulates glutaminase (GLS) pre-mRNA alternative splicing. The interaction between CCAT2 G-allele and CFIms25 directs binding of this RNA–protein complex to the poly(A) site in intron 14 of GLS pre-mRNA, inducing in this way splicing of glutaminase isoform C that associates with enhanced catalytic activity compared to the kidney glutaminase isoform. Biotinylated RNA pull-down experiments revealed that CCAT2 directly binds to GLS pre-mRNA, highlighting an example of RNA-RNA–protein complex which depends on the secondary structure of a scaffold-lncRNA that is mainly affected by the rs6983267 SNP (Figure 3C, [89]).
Another example of lncRNA–protein interactions that are altered by the presence of SNP variants is lncRNA NEXN-AS1 along with its associated SNP rs114020893 at 1p31.1. This variant is correlated with increased lung cancer susceptibility and is predicted to modulate the secondary structure of the NEXN-AS1 transcript [90,199]. These examples highlight the potential of TncVs that alter lncRNA–protein interactions in disease progression, yet detailed functional insights are still required for the majority of association studies that link SNP variation with lncRNA function in cancer [188].
Apart from affecting the interplay with protein partners, there are multiple levels of lncRNA-miRNA interactions that rely on mutations with a role in cancer [200,201]. This type of mechanism is sequence-dependent, which means that any alteration in the base sequence may influence base-to-base interplay. Similar to their role in mRNA:miRNA interactions, some TncVs can affect lncRNA transcript levels through differential miRNA binding. Such an example refers to MALAT, a thoroughly characterized lncRNA in many cancer types (e.g., oral squamous cell carcinoma, melanoma) with a predominant oncogenic activity, although a tumor-suppressive function has also been reported in breast cancer [202,203,204,205]. The first functionally characterized SNP of MALAT1 was rs664589, which is involved in colorectal cancer progression via its interaction with the miR-194-5p. MiR-194-5p targets MALAT1 for degradation in a rs664589 allele-dependent manner. Binding of miR-194-5p to the MALAT1 transcript with the rs664589-C genotype targets it for degradation in the nucleus, in contrast to the G allele that decreases overall binding affinity of the miRNA, leading to accumulation of MALAT1 and ultimately poor patient survival, increased distant metastasis and enhanced tumor growth (Figure 3D, [91]).
Another example of non-coding variant that affects miRNA-lncRNA interplay in cancer is CCSlnc362. CCSlnc362 (RP11-362K14.5) is a recently identified tumor-promoting lncRNA in colorectal cancer. Its expression correlates with the SNP variant rs1317082 (T > C), located at exon 1 of the CCSlnc362 locus. Functional experiments linked the oncogenic role of the CCSlnc362 with acceleration of the cell cycle parallel to apoptotic blockage. In vitro luciferase assays showed that miR-4658 binds to CCSlnc362 in an allele-specific manner. Binding affinity of miR-4658 is increased in the presence of homozygous C alleles in contrast to the T allele, highlighting an allele-dependent predilection of the miR-4658 seed [92]. Finally, a correlation study of rs11752942 (A > G) SNP located in LINC00951 (lincRNA-uc003opf.1) exon, conducted in 1493 Esophageal Squamous Cell carcinoma (ESCC) patients, revealed a distinct association of the G risk-allele with the reduced expression of LINC00951. The regulation of LINC00951 is miRNA-149 mediated and is involved in ESCC cell proliferation and tumor growth [93].
An independent study focused on the long intergenic noncoding RNA (lincRNA) LINC00673, which is correlated with an antitumor effect in pancreatic ductal adenocarcinoma. Rescue experiments divulged the significant role of this transcript in cell proliferation mechanism of pancreatic cancer cells while in vivo xenografts experiments showed its implication in pancreatic cancer tumor growth. LINC00673 promotes PTPN11 ubiquitination and degradation via mediation of an PRPF19–PTPN11 interaction, resulting into an elevated and STAT-dependent anti-tumor response. The function of LINC00673 was strongly linked to the germline variant rs11655237 (G > A transition), which creates a binding site for the miR-1231 preventing LINC00673 from exerting its regulatory role. Similar to the previous examples, miR-1231 acts with preference to the A allele of the rs11655237 variant, serving as a decoy for LINC00673 function [94]. Another study of lncRNA SNPs variants in a cohort of 505 nasopharyngeal carcinoma patients, uncovered variants associated with chemoradiotherapy sensitivity of patients. Specifically, MEG3 rs10132552 CC genotype was linked to elevated toxicity, LINC-PINT rs1059698 CC had a protective role against neutropenia and myelosuppression and pR-lncRNA-1 rs73594404 GA genotype patients had increased risk of toxic reactions. All the mentioned lncRNAs are involved in p53 signaling network, a fact that highlights their SNP potential for reducing treatment toxicity [206].
In the same context of ncRNA interactions, some polymorphisms indirectly modulate isoform selection [207]. An example of a miRNA:lncRNA interaction that relies on transcribed SNPs and associates with isoform stabilization of a Receptor tyrosine kinase (RTKs) target in cancer, is EGFR-AS1 (EGFR Antisense RNA 1). RTKs are of great importance in cancer progression with pivotal clinical and therapeutical applications [208,209,210,211,212,213,214]. rs10251977, which associates with the lncRNA EGFR-AS1, normally stabilizes isoform A of its RTK target EGFR in oral cancer patients. EGFR-AS1 was suggested to act as a scaffold for PTBP1 (member of the heterogeneous nuclear ribonucleoprotein family) to promote EGFR-A stabilization. The minor allele A of rs10251977 creates a binding site for miR-891b (which is downregulated in tumors), leading to degradation of EGFR-AS1 and thus is correlated with elevated levels of the alternative D isoform of the EGFR [95]. Although this study needs further experimental validation, it represents a notable example of a natural antisense transcript that regulates its mRNA target in cis through a genetic variant. This type of genetic variation that alters isoform selection of well-defined oncogenic drivers like EGFR may expand the prognostic toolbox of cancer or meliorate the personalized therapy for the patient. Genetic variation could also affect ectopic biogenesis of miRNAs from lncRNA loci cancer, however experimental validation of such cases in still pending.
5. Methodologies to Functionally Characterize Non-Coding Variants
There are many bioinformatic strategies and databases that take advantage of cancer genomic data to conduct significant correlations of cancer risk and predisposition (Table 2).

Table 2.
State-of-the art methodologies for functional genomic variant identification. Access: 15 July 2021.
However, validation and most importantly functional dissection of cancer-driver and passenger mutation requires innovative experimental approaches [215,216,217,218,219]. Alongside the advancement of genomic techniques and next generation sequencing technologies that improved our understanding regarding the function of the genome, came pioneer research strategies that identify, validate and finally characterize non-coding variability.
Discovery of functional variants in the non-transcribed portion of the genome is inextricably bound to experimental approaches designed to unveil novel regulatory sequences. A hallmark of a regulatory sequence is chromatin accessibility that subsequently allows functional activation of the region through binding of transcription factors [220]. Therefore, general approaches that scan the genome for open chromatin can serve as the first step towards the identification of regulatory variants, especially in cis regulatory elements [221,222,223]. When it comes to trans regulation (enhancers, silencers, insulators), chromatin status needs to be complemented with experimental assessment of chromatin architecture in the cancer genome in order to pinpoint the target(s) of the regulatory sequence [224]. Most importantly, the causative motif(s) within these cis or trans regulatory sequences needs identification and experimental validation prior to any connection with genetic lesions. Below some of these experimental approaches are presented based on the function and position of the non-coding regulatory sequence that hosts the causative variant with regards to it target(s).
5.1. Scanning for Regulatory Sequences Based on Open-Chromatin State
DNase was first used during the 1980s to map regulatory elements through the identification of global chromatin accessibility [225]. Following nuclei isolation, permeabilization of the nuclear membrane and DNaseI incubation, DNA elements are enriched by size selection either with gel extraction or by ultracentrifuge purification. The methodology can then be coupled with next generation sequencing. The result of this process is the detection of DNaseI Hypersensitivity Sites (DHS), which are sites located within open-chromatin regions with median length of 300 bp that are protected from degradation by DNaseI due to presence of transcription factors (TF) (Table 2) [226]. DNase-seq has the advantage of detecting open-chromatin without requiring prior knowledge of the sequence of the TF bound to the DHS. Additionally, it has higher sensitivity than other approaches (see below) at promoters. Its drawbacks are linked to the sequence specific function of DNaseI, thus DHS global identification is not bias-free. Moreover, the purification steps may lead to loss of DNA sample, lowering overall sensitivity of detection [227,228,229,230]. Although DNase-seq vastly enriches the sample in promoter regions, it demonstrates decreased representation of regulatory elements in a condensed chromatin state. Such an example applies to cases of SNPs within promoters of imprinted genes, where the methodology would show allelic biases towards the imprinted allele [228].
Variants detected within DHS are strong candidates for having a key role in carcinogenesis. Such variants are the SNPs rs62331150 (within active promoter region) and rs73838678 (within strong enhancer region), which are correlated with increased risk for breast cancer [231]. Complementary to SNPs, presence of CNVs in DHS may result in large scale chromatin accessibility changes. Such an example is deletion of the DHS chr8:579137-581436, which leads to increased expression levels through enhanced promoter accessibility of several tumorigenic protein-coding and non-coding genes [232].
An improvement of DNase-seq refers to single-cell DNase-seq (scDNase-seq). Application of scDNase-seq enables study of gene promoters and enhancers at a single-cell level with highly reproducible results. One SNP identified with this methodology is the chr18:52417839 (G > C), the frequency of which increases in patients with thyroid carcinoma, leading to decreased expression of TXNL1, due to disruption of a p53 binding motif within its promoter [233].
Formaldehyde-Assisted Isolation of Regulatory Elements sequencing (FAIRE-seq) can be used independently or in combination with other genomic approaches for the discovery of accessible chromatin, as it enriches for nucleosome-depleted DNA [234]. Following a cross-linking step, the non-crosslinked and therefore accessible DNA that may host interactions with TFs is isolated. Coupled to next generation sequencing, this method serves as a first, yet often crude, option for identifying accessible chromatin. The simplicity of the method together with the fact that it does not require prior treatment of the sample, highlight its applicability (Table 2). In comparison with DNase-seq, it shows reduced bias for cis regulatory regions and greater sensitivity in detecting intronic and intragenic regions. Its downsides associate with low signal-to-noise ratio, demanding high fixation efficiency, making data analysis more difficult, while it does not provide direct functional clues and therefore demands coupling to other techniques [234]. Micrococcal nuclease digestion with deep sequencing (MNase-seq) utilizes a non-specific endo-exonuclease micrococcal nuclease to detect chromatin regions bound by proteins [235,236]. MNase-seq requires low sequencing depths [237] but relies on non-specific digestion [238].
Proposed as an improved strategy for identifying candidate regulatory elements [239], the Assay for Transposase-Accessible Chromatin with high-throughput sequencing (ATAC-seq) is currently the most efficient method of this category. It utilizes the use of a hyperactive Tn5 transposase (normally catalyzes the movement of a transposon to accessible chromatin) that adds deep sequencing adaptors to accessible genomic regions. The superiority of this method is linked to its significantly increased efficiency, allowing for reduced cell numbers as input sample while the duration of the protocol is relatively short (Table 2). Additionally, it enables interrogation of both nucleosome and TF occupancy in regulatory elements. Like FAIRE-seq though, it does not provide direct evidence for the function of the regulatory element, thus needs to be coupled to other methodologies tailored for functional dissection [237,239]. So far, it has been successfully used along with eQTL analysis to detect variants correlated with altered chromatin accessibility for schizophrenia [240].
5.2. Scanning for Regulatory Variants in Trans Regulatory Elements
Massively Parallel Reporter assays (MPRAs) [241] and CRE-seq [242] have been extensively used for enhancer mapping, by creating a library in which the candidate enhancer sequences are inserted upstream of a reporter gene, regulating its expression.
Apart from insertion of the enhancer sequence in the plasmid, an extra step for barcode insertion in the 3′UTR is required, to pinpoint single enhancer mapping. This barcoded library is then transfected to cells that are harvested after 24 h, optionally followed by sorting and deep sequencing of the positive clones. As a result, the normalized count of the individual barcodes can be used for the estimation of regulatory activity of each enhancer in a particular cell line. This method allows high-throughput examination of enhancer activity, as well as multiple independent examinations of each enhancer, by use of different barcodes (Table 2). As for its drawbacks, it is an episomal assay, thus the enhancer activity is measured outside its native context, while insertion of the candidate sequences upstream of the TSS of the reporter gene may add false positive biases for enhancer activity in cases that an inserted sequence acts as a promoter instead of enhancer [243,244,245,246].
Self-transcribing active regulatory region sequencing (STARR-seq) is a high-throughput methodology to study enhancer activity [247]. With this approach, candidate enhancer sequences from various samples may be tested as input. These putative enhancer elements can be sonicated fragments previously enriched in functional elements by a FAIRE step, selected Bacterial Artificial Chromosomes (to study parts of larger genomes, including the human genome) or even synthesized oligonucleotides (which are used for studying the role of SNPs and SNPs in enhancer function). STARR-seq exploits the ability of the enhancer to act independently of its position, as the putative enhancer sequence is inserted in the 3′UTR of a reporter gene (e.g., luciferase, GFP) that lies within a plasmid used to transfect cells in culture (Table 2). Since the candidate sequences lie within the transcript area, barcoding is not required to perform this analysis thus making library construction easier [129]. A single day post transfection, positive cells are sorted and subjected deep sequencing, to identify which candidate sequences indeed act as enhancers. Enrichment of each candidate enhancer in the cDNA sample provides quantitative data for the activity of each enhancer [248]. This offers the ability to parallelly distinguish the activity of the same enhancer in presence of different variant alleles [249,250]. STARR-seq drawbacks relate again with the episomal nature of the assay, the fact that sequence insertion within the transcript may affect its stability and thus the result of the measurement and finally the fact that the assay does not allow multi-vectoral examination of the enhancer’s activity [251]. Applications of this methodology extend beyond studying cell-type specific enhancer activity. It can be applied for studies focusing on finding enhancers with a hormone or drug response potential or for comparative genomic approaches [13,252,253]. One advantage that distinguishes STARR-seq from similar approaches is the use of candidate silencer regions, in which the drop in the reporter gene expression levels can been effectively used for silencer element characterization [141].
5.3. Scanning for Regulatory Variants Based on Chromatin Interactions
Identification of TREs (enhancers, silencers and insulators) presents technical challenges that relate with the distance and orientation of the regulatory element compared to target CREs. Strategies such as Hi-C interrogate chromatin architecture through a combination of proximity ligation and deep sequencing [254]. However, such general approaches can be misleading since not all chromatin interactions are functionally relevant. Approaches that combine Chromosome Conformation Capture methodologies with a Chromatin Immunoprecipitation (ChIP) approach, such as Chromatin interaction analysis with paired-end tag sequencing (ChIA-PET), HiChIP and Proximity Ligation-Assisted ChIP-sequencing (PLAC-seq), isolate the fraction of chromatin interactions that are functionally relevant because they coincide with binding of transcription factors, CTCF or RNA-Pol-II [255,256].
The ChIA-PET strategy relies on crosslinking that fixes nuclear architecture, followed by chromatin extraction and sonication. Subsequently, an antibody is used to enrich for chromatin loops that contain the interacting protein of interest (e.g., RNA Pol-II). After enrichment, a linker ligation step is performed, in which the sample is separated in two aliquots and a different half-linker oligonucleotide is added in each sample fraction. Then both aliquots are mixed once again to perform the proximity ligation assay of the half-linkers that interact with each other. Ultimately DNA fragments are extracted after a de-crosslinking step, followed by protein digestion with MmeI to create paired-end tags with a tag-linker-tag order. Sequencing of the sample, most prominently by utilization of the Illumina platform, followed by a complex bioinformatic analysis can reveal the fraction of chromatin interactions that involve the protein of interest [257]. Using transcription factors as baits, ChIA-PET has the advantage of recovering global maps of precise interaction points on a genome wide scale, enabling the study of SNPs that lie within DNA regulatory domains, making it a great tool to study variants in promoters, enhancers, silencers and insulator elements at once (Table 2). Its pitfalls mainly associate with its complexity both in terms of sample preparation and bioinformatic analysis, together with its inefficiency that warrants extreme sequencing depths and therefore significantly elevated cost [258]. Moreover, it does not provide any information with regards to actual transcription, thus coupling its results to an RNA-targeting approach is required [259]. Nevertheless, ChIA-PET has been utilized for proving chromatin looping that is mediated by estrogen receptor alpha in hormonal cancer [260], or for characterization of CTCF-mediated loop formation [261]. Development of long-read ChIA-PET (250 bp tags) has improved mapping efficiency, enabling the study of SNPs and/or haplotype-specific interactions in various cancer types, most prominently breast cancer [262,263]. Such an example is rs16904316 that lies within the gene-body of the enhancer-like lincRNA CCDC26. Its promoter overlaps with a super-enhancer and shows correlation with onset of ALL in children. ChIA-PET analysis showed a cell-type specific long-range interaction between CCDC26 and MYC, indicating the regulatory role of lincRNA loci in MYC overexpression in ALL [264].
HiChIP is a protein-centric chromatin conformation capture method, which is based on the principles of in situ Hi-C and transposase-mediated on-bead library construction [265]. The protocol relies on cross-linking in vivo, followed by nuclei isolation and in situ Hi-C contact generation. Following establishment of long-range DNA contacts, nuclei lysis and sonication of the sample is performed prior to ChIP. Hi-C contacts carrying the target protein are then used for on-bead Tn5 library generation for paired-end sequencing. In comparison to ChIA-PET, it shows higher efficiency (10-fold increase in the yield of conformation informative reads) and lower false-positive ratio (Table 2). Furthermore, HiChIP requires up to 100-fold less starting cell amount than ChIA-PET, while also having a 2-day span workload [265]. A computational method to analyze data derived from HiChIP experiments is FitHiChIP, which can be applied to compute statistical significance estimates, lower the background, and overcome possible biases of the method [266]. HiChIP has been utilized to detect SNPs within active chromatin regions (carrying H3K27ac labeling) interacting with TNFAIP3 promoter. Some of the SNPs detected by this methodology showed allele-specific expression profiles (such as rs538522, rs559766217) and allele-specific TF binding profile (the rs643177 for TF Pou2f1) [267]. Another study utilized HiChIP to detect promoter–enhancer loops within risk-associated loci for endometrial cancer. The results of this analysis found four high-risk correlative variants. Among those variants were rs882380, which regulates the oncogene SNX11 and tumor suppressor HOXB2 and rs937213 that regulates the oncogene SRP14 (prognostic marker in renal cancer). Additionally, SNP rs7579014 that regulates the context-dependent tumor suppressor BCL11A, was proven to be a high-risk correlative variant in endometrial tumor sample. Finally, SNP rs9600103 was found to be part of a 23 bp anchor looped to the promoter of KLF5 [268].
PLAC-seq has a similar philosophy to ChIA-PET, differing in the proximity ligation step that occurs in nuclei, prior to lysis and sonication of the chromatin [269]. Thusly, the efficiency and accuracy in detecting long-range chromatin interactions via PLAC-seq is again vastly increased compared to ChIA-PET (Table 2). The cells required for a PLAC-seq protocol are up to 200-fold less than those required for ChIA-PET. Additionally, it produces less inter-chromosomal pairs and more intra-chromosomal pairs than ChIA-PET, covering more regulatory elements (supported also by DHSs) [269]. So far, PLAC-seq is used along with other methodologies such as ATAC-seq, to identify putative functional SNPs that show correlation with Alzheimer’s disease (AD). This approach pinpointed SNP rs10130373 as a single variant with an important functional role in AD, along with various other SNPs like rs181391313, which causes a KLF4 site disruption in a putative microglia-specific intronic regulatory element in STAB1 [270].
5.4. Scanning for Regulatory Variants Based on RNA-Chromatin Interactions
In the metagenomic era, regulation of the genome largely depends on direct or indirect interactions between the chromatin of regulatory elements and lncRNA transcripts. Such RNA-chromatin interactions are crucial for the recruitment of histone writers [26,271], interaction with transcription factors [272,273], formation of triple helixes or R-loops with DNA [274,275] and/or maintenance of chromatin loops [276]. Chromatin Isolation by RNA Purification sequencing (ChIRP-seq) is among the most utilized methodologies to study RNA-chromatin interactions [277]. This approach again relies on cross-linking for stabilization of RNA-chromatin interactions, followed by capturing of target transcript with multiple 20-mer antisense DNA probes that are biotinylated. After elution, the enriched DNA fragments are deep-sequenced, enabling discovery of putative regulatory RNA binding sites in the genome. Among the pitfalls of ChIRP-seq are low expression issues of the endogenous RNA target that can lead to an increased number of false-positive hits by the capturing probes (Table 2). ChIRP-seq has been successfully used to identify SNPs in various DNA regulatory elements, with an example referring to the detection of polymorphisms related with prostate cancer progression in lncRNA regulatory elements, such as rs72725879 and rs7463708 SNPs which lie within an enhancer element of PCAT1 [72].
Apart from ChIRP-seq, similar methodologies referring to Capture Hybridization Analysis of RNA Targets sequencing (CHART-seq), Mapping RNA-Genome Interactions (MARGI) and in situ MARGI (iMARGI), Global RNA Interactions with DNA by Deep Sequencing (GRID-seq), Chromatin Associated RNA sequencing (ChAR-seq) and RNA And DNA Interacting Complexes Ligated and sequenced (RADICL-seq) have also been used to clarify the role of SNPs in RNA-chromatin interactions [278]. CHART-seq incorporates an extra step of RNase H sensitivity assay to further improve specificity compared to ChIRP-seq, while also provides larger fragments for the analysis, albeit with lower sensitivity [279,280]. MARGI and its upgraded version iMARGI) have also been applied in similar approaches due to minimization of potential sequence biases given the increased read lengths that they offer [281]. These protocols are more straightforward but require millions of cells and comes with a lengthy workload. Additionally, difficulties in sensitivity and specificity evaluation do exist, without a clear statistical model being optimal for downstream bioinformatic processing [282].
GRID-seq has more specificity for binding to RNA or DNA, with low background noise along with an established computational pipeline for detecting background RNA-DNA interactions [283]. GRID-seq detects multiple RNA classes on chromatin, both acting in cis and in trans, while it also provides information on interactions in the 3D genome as it showcases formation of enhancer–promoter loops [283]. As for its limitations it requires extensive deep sequencing for creating the contact maps, otherwise RNAs with low abundance may not be detected, while it poses mapping issues in low complexity regions [284]. Alternative approaches include ChAR-seq that offers increased technical specificity and improved resolution in comparison to MARGI, while in comparison to GRID-seq, it has lower chance of false mapping [285]. Finally, RADICL-seq is the most recent method for studying genome wide RNA-chromatin interactions, allowing study of 3D nuclear structures. It offers four advantages in comparison to other methodologies that can be summarized to greater resolution, decreased background noise, reduced fraction of nascent RNA-chromatin interactions and improvement of fragment selection as well as downstream alignment steps. Overall, it offers an improved performance/cost ratio than GRID-seq [64,196].
5.5. Methodologies to Functionally Dissect Transcribed Non-Coding Variants
Given their transcribed nature and radically different mechanism of function, TncVs require a radically different set of experimental strategies compared to their non-transcribed regulatory counterparts. RNA Antisense Purification Sequencing (RAP-seq) is a methodology applied to the characterization of TncVs in the context of ncRNA secondary structure and interaction [286]. Based on the cross-linking of macromolecular complexes formed by ncRNAs alongside with the use of antisense biotinylated oligos, RAP-seq enables capture of ribonucleoprotein complexes and thus identifies proteins and/or RNA that interact with the target RNA on a genome-wide scale (Table 3).

Table 3.
Methodologies used to study the effect of variants in transcript-based regulatory networks. Access: 15 July 2021.
Its main disadvantage is background noise that is significantly alleviated through stringent hybridization and/or wash conditions [286,287]. RAP-seq has been effectively used along with other tools to identify the SNP rs199971565 as a novel indel biomarker for gastric cancer susceptibility, as it affects the secondary structure formation of miR-302c [288].
Another promising methodology for studying RNA–protein interactions is RNP network analysis by mutational profiling (RNP-MaP) [289]. Use of the cell-permeable reagent NHS-diazirine (SDA) allows rapid labeling of RNA molecules at in live cells. Activation of SDA by UV leads to formation of bonds between lysine residues in proteins with ribose base moieties at a 4–9 Å distance. After crosslinking lysis is performed, followed by protein digestion, leaving short peptide adducts. Utilization of the MaP reverse transcriptase, that reads through the adducts with relaxed fidelity, creates mutations at the RNA–protein interacting sites. RNP-MaP can be applied both for single-stranded and double stranded parts of the RNA molecule, showing a slight preference for single-stranded molecules (Table 3). Bond formation on the RNA molecule is independent of the nucleotide(s) found in the binding site, although it has higher reactivity with adenosine and uridine. RNP-MaP can be efficiently coupled with mass spectrometry approaches for identifying the interacting proteins. Thusly, RNA-MaP can be effectively used to identify the critical regions for protein interaction in ncRNAs and mRNA, and how these networks form and dissociate in different cell-types in presence of variants [289].
Another in vivo methodology for detecting RNA–protein interactions that relies on the Clustered Regularly Interspersed Palindromic Repeats (CRISPR) technology, is CRISPR-Assisted RNA–protein Interaction Detection (CARPID) [290]. CARPID utilizes a nuclease-activity-free form of the RNA-targeting VI-D CRISPR single effector dCasRx system, fused with the BASU biotin ligase. The dCasRx effector is capable of processing two sgRNAs simultaneously, thus increasing specificity. The BASU biotin ligase adds biotin groups to proteins interacting with the target lncRNAs, ultimately facilitating immunoprecipitation using streptavidin-bound beads and identification by mass spectrometry (Table 3). Coupled to RNA-seq, CARPID determine allelic expression and formation of RNA–protein interactions, based on the availability of SNPs and indels in the genetic background [290].
Post-Transcriptional Regulatory Element sequencing (PTRE-seq) is a high-throughput massively parallel methodology to study the effect of 3′ UTR sequence in post-transcriptional regulation via miRNA targeting [291]. It utilizes plasmid vectors that carry a reporter gene, with an insertion site for the candidate regulatory element (along with a unique barcode) in the 3′ end of the reporter. These vectors are then used for library construction of all candidates 3′ UTR regulatory elements followed by cell transfection. RNA-seq can be used to estimate barcode counts for each regulatory element (Table 3). The fact that PTRE-seq allows use of synthetic 3′ UTR sequences as inserts, may allow future applications aiming towards the characterization of TncVs in this regulatory context [291].
Parallel analysis of RNA-structure sequencing (PARS-seq) is a methodology that provides information about the secondary and tertiary structures of RNA molecules [292]. The principle of the methodology is based on the digestion of the RNA molecules with RNases that are specific for single-stranded and double-stranded RNA. The resulting fragments are reverse-transcribed to cDNA and deep-sequenced. This results in high-resolution sequences of the RNA, that can be used to deduce the RNA structure based on the comparison of the different digestion patterns by the various RNases (Table 3). PARS-seq has the advantage of providing RNA structural information that distinguishes between paired and unpaired bases. As for its disadvantages, use of RNases might not be specific, while digestion conditions need to be very well optimized, as the RNA can be over-digested. Finally, this application can only be performed in vitro [293]. Variants that may affect an RNA secondary structure might be detected by this approach, although there not any examples yet [294].
5.6. Validating Regulatory Variants with CRISPR-Based Approaches
Many applications of the CRISPR associated protein (Cas) system have been used to validate regulatory variants. Classical CRISPR-Cas9 editing can be deployed for altering the DNA of regulatory sequences in which a variant occurs (mostly tailored to SNPs but can be applied to CNAs in some cases) (Table 4). The editing strategy can rely either on correcting the cancer predisposing allele or creating it in an otherwise non-cancerous background. Mutated versions of Cas9 with nickase activity combined to the use of two distinct sgRNAs for a single target, which effectively reduces off-target effects are the preferred strategy for precise SNP editing [295,296].

Table 4.
CRISPR-Cas systems utilized in functional non-coding variant validation. Access: 15 July 2021.
CRISPR-activation (CRISPR-a) [297,298] or CRISPR-inhibition (CRISPR-i) [299], both of which rely on catalytically inactive forms of Cas9 that are coupled to transcriptional activators or inhibitors to endogenously modulate transcriptional activity, can precede SNP editing in order to precisely link the surrounding sequence of the variant with gene expression of specific targets [300,301]. Examples include a CRISPR-a based methodology that has been applied in the study of mutations in regulatory elements of KRAS in colorectal cancer [302].
A CRISPR-i approach has been used in the study of SNP rs11986220 (A > T), which resides in an enhancer that regulates expression of MYC and PVT1. This study correlated this SNP with increased DNA methylation levels at a nearby CTCF site that lies between the enhancer and the promoter of MYC and PVT1, leading to loss of insulation and formation of an enhancer–promoter loop which increases the expression level of MYC and PVT1 in prostate cancer (the same occurs also in presence of CNA that destroys the CTCF binding site) [303]. Application of the CRISPR armament can be expanded with the use of Cas9 homologs from species other than Streptococcus pyogenes, which recognize different PAM sequences allowing the study of loci that are not decorated with classical PAM sequences, or other Cas proteins like Cas12 and Cas13 that can be used for increased specificity and RNA targeting, respectively [304,305,306].
A CRISPR-deletion based approach was used to study the effect of two prostate cancer risk-associated SNPs, rs12144978 and rs4919742, involved in loop formation of two distinct prostate cancer risk-associated CTCF sites. The two CTCF sites regulate the expression of KCNN3 and KRT78, by insulating the promoters of each gene from active enhancer. Deletion of these CTCF sites leads to ~100-fold increase in the expression levels of each gene due to enhancer adoption [307]. Another example of a CRISPR-based approach for the functional characterization of an SNP focused on rs2431697. This SNP lies within a cell-type specific enhancer that forms a cognate enhancer–promoter loop with the promoter of miR-146a, a miRNA with significantly downregulated expression in patients with Systemic lupus erythematosus. Presence of the high-risk T allele lowers the binding affinity of NF-κB binding site, which then leads to the decreased expression levels of miR-146a (this result was also verified by FAIRE-seq and ATAC-seq) [300].
6. Utilizing Non-Coding Variants in Clinomics
Clinical interpretation of genomic variants in cancer relies on a multidimensional methodology, which incorporates an array of computational algorithms and bioinformatic approaches coupled to large scale genomic data of cancer patient cohorts from different types of cancer [308]. So far focus has been given on the identification of somatic mutations in coding sequences with cancer-driving potential together with their clinical pertinence, reflecting the wealth of exome sequencing data that is currently available at an improved depth/cost ratio compared to whole genome sequencing [309]. In addition, annotation of human protein coding genes is well defined and together with availability of amino acid sequences, allows precise evaluation of mutational events on protein function [310].
More recently, whole-genome sequencing data have shed light into the non-coding part of the genome and comprehensive studies focusing on non-coding variants [311]. A continuously growing list of genome-wide studies that focus on non-coding variants, reveal their crucial potential in cancer clinomics and diagnostics (Table 5, [312,313,314]). Robust statistical algorithms are now available to detect rare genomic variants with prognostic potential, a methodology which leads to the development of precision therapeutics [110]. Nonetheless, comprehensive studies focused on the clinical utilization of non-coding variants in cancer are limited. A recent pan-cancer analysis of solid tumors revealed 21.574 recurrent non-coding mutations in patient genomes, 580 of which were cancer related when correlated with TCGA clinical features [315]. This study emphasized on mutations that occur in TF binding sites through analysis of epigenomic and chromatin structural data, divulging TEAD1- CX3CR1- and NFYB-bound enhancer elements as the top-scored regulatory sites with high mutational significance in cancer.

Table 5.
Summary of non-coding variants associated with cancer clinomics.
Another recent study focused on the correlation of 3′UTR variants with clinicopathological features of breast cancer patients as well as with chemotherapy response. The investigated variants included the DPYD-associated rs291593-CC, which is correlated with increased risk of toxicity in cancer patients receiving 5-fluorouracil chemotherapy and the AKR1C3-associated rs3209896-AG, which is linked to elevated risk for breast cancer recurrence. Moreover, the progesterone receptor-associated rs1824125-GG was found to be associated with shortening of progression-free survival, while the ALDH5A1-associated rs1054899-AG/AA correlated with reduced chemotherapeutic response to 5-fluorouracil, doxorubicin and cyclophosphamide [316,320,321]. Survival analysis indicated shortening of survival time in patients carrying the rs7756222-CC and rs9487402-TG/GG variations in SLC22A16 gene [316,322]. These data demonstrate the potent existence of clinically associated variations in progesterone signaling pathways.
Ancestry across tissue and cancer types has been interconnected with the molecular and genetic background of African, European, South Asian and East Asian cancer patients [36]. Despite limitations in sample size (only 17% non-European samples), this approach revealed a significant correlation of ancestry with miRNA variations with greater differences in individual cancer types when compared to pan-cancer analysis. Approximately 80% of ancestry-associated miRs were located within host genes, indicating the need of considering (epi)genetic factors in the association of miRs with ancestry. This pilot study reflects the need of considering ancestry lineage in genomic variant research, especially for non-coding variants which may present a distinct alteration pattern among lineages.
A similar study of genetic variation in 1524 miRNA genes aimed to investigate the distribution of variation in diverse human populations including European, Asian and African populations (69 unrelated individuals in total among 14 global populations). Intriguingly, novel pre-miRNA hairpin mutations, located in highly conserved miRNA seed genomic loci with different frequencies among the individuals, were identified. Linked to cancer, the T allele of SNP rs12355840 in hsa-mir-202, previously reported to interfere in breast cancer mortality, had a significantly higher frequency in non-African populations (65%) when compared to African ones (26%) [323]. The same T-allele was also linked with a reduced risk of breast cancer mortality but was related to Hodgkin lymphoma [324,325,326].
A significant correlation was performed between 505 colorectal cancer (CRC) patients’ clinical profile and the SNP variant rs13230517 that lies in the promoter region of RP11-3N2.1 lncRNA. RP11-3N2.1 was shown to be downregulated in colon cancer biopsies and patients with rs13230517 GA/AA genotype had lower risk of developing colorectal cancer compared to AT/TT genotype [317]. In the context of the same cancer type, another study was conducted based on 900 CRC patient biopsies focusing on rs531564, C > G in the pri-miR-124. Although the exact molecular function of this polymorphism is currently unknown, a detrimental connection to clinicopathological features was achieved, revealing an association of rs531564 with lymph node metastasis and poor differentiation [317].
An independent association study of 359 HCC patient clinical characteristics with non-coding variants emphasizes in gene polymorphisms of H19 lncRNA. This study divulged a novel SNP variation (rs3741219) in the fifth exon of the transcript that is linked to elevated risk of developing HCC [318]. Additionally, a meta-analysis based on 11,821 HCC patients in total, shed light into miRNA SNPs serving as biomarkers for HCC. Specifically, the hsa-mir-146a residing rs2910164 (G to C variation) was linked to reduced HCC risk for the C allele while hsa-mir-34b/c rs4938723 (T to C variation) was associated with increased HCC risk. Furthermore, hsa-mir-196a-2 rs11614913 and hsa-mir-149 rs2292832 variations were all correlated with elevated HCC risk [319].
A pan-cancer study focused on enhancers of approximately 9000 samples in 33 different cancer types, highlighted the positive correlation of overall enhancer activation with aneuploidy in cancer. FANTOM Project (annotated enhancers based on epigenetic marks, TFs binding and open chromatin state) and TCGA data were utilized for this systematic analysis [327]. More specifically, tumor enhancers were divided into three subgroups based on their active state and then each group was interrelated with single CNAs and point mutations. Interestingly, highly active enhancers were more prone to structural mutations (compared to point mutations) due to their open chromatin state that increases the possibility of genomic rearrangements. Juxtaposed with eQTLs and Hi-C approaches, such data can pinpoint genes with clinical implications (oncogenes, tumor-suppressor genes, tumor biomarkers) that are regulated by these enhancers [125]. For example, enhancers associated with expression of genes with clinical implications include enhancer 9 for PD-L1 which is a target for immunotherapy for melanoma and lung cancer [125,328]. Collectively, these studies spotlight the importance of regulatory element variants in cancer progression and propose combinational clinomic approaches that can lead to targets with clinical significance.
Current steps towards personalized treatment include patient categorization based on cancer-driver mutations in protein coding genes, with novel therapy trials like NCI-MATCH and SHIVA highlighting the importance for precision oncology [329,330,331]. Although these kinds of approaches seem to have encouraging preliminary results, there are limitations that mainly underlie the specificity of drug combinations [332]. This specificity depends on classification of patients based on the genomic profile, which demands cancer- and tissue-specific polymorphisms that confront treatment side effects or medicine inefficiency [333]. The regulatory role of non-coding variants comes to fill the gap of these restrictions as they can specify the patient drug-response [334]. For example, in breast cancer, patients carrying LINK-A rs12095274 A allele are prone to develop resistance to AKT inhibitors when compared to patients with C allele [335]. Focusing on regulatory non-coding variants that affect TF binding sites such as NFKB and STAT3 can also provide novel therapeutic approaches [336]. It is imperative to continue unveiling the function of non-coding variants independently or alongside with their associated ncRNAs not only to ensure risk assignment for genetic predisposition to the disease but also to tailor existing and future therapies to individual cancer genomes.
7. Conclusions and Future Perspectives
Genome instability and mutation is an essential feature of carcinogenesis that was first added in the cancer hallmark framework in 2011 [337]. It is undoubtedly a main characteristic that underlies the rapid evolution of cancer genomes during progression of the disease. Characterization of genetic variants with cancer development association can provide information on population risk stratification and prioritize the subgroups within a population for monitoring and primary prevention [338]. Combined with additional molecular biomarkers, determination of variant’s involvement in a cancer-related phenotype can be used as a predictor for the patient’s clinical outcome [339].
Uncovering the mutational landscape in cancer remains a huge challenge in the field of genomics. Tumor heterogeneity among patients with different genetic background in combination with the low conservation of non-coding sequences perplexes the discovery of their functional role during disease progression [67,340]. Although studies have shown a correlation of non-coding variants with predisposition and clinical outcome of the disease, the underlying mechanism of many variants is still unclear [341]. Moreover, discrimination between cancer-driver and passenger mutations adds another level of complexity in dissecting the mutational non-coding landscape [342]. Functional analysis of GWAS variants remains challenging [343], but novel techniques like the ones presented in this review can functionally dissect the role of non-coding regulatory variants involved in disease mechanisms. Future experimental efforts should also focus on dissection the role of non-coding variants in the biogenesis of miRNAs from lncRNA loci. By evaluating an individual’s risk of cancer development, their application for personalized prevention and monitoring programs will be enabled [344]. Should a person succumb to the disease, clarification of their genetic profile will enable risk assessment as a basis for personalized treatment [345]. In the era of precision medicine, where mutation and variation information are a top priority, investigating the role of non-coding genetic variants in regulating cancer cell function is of outmost importance [346]. This can lead to the detection of suitable and non-invasive molecular biomarkers for disease predisposition [347].
When it comes to treatment approaches in cancer, existing studies that focus on protein-coding genes, provided promising results. Parallel to their coding counterparts, ncRNAs are rising as more accessible candidates for pharmaceutical intervention [348]. In comparison to pharmaceutical molecules and antibodies used in the clinical act, there are no reports stating that the cancer cells show resistance to non-coding RNA therapy, while chemical modification of the ncRNAs further improve the half-time of ncRNA drugs compared to the other approaches [349,350]. For example, administration of miRNA molecules (common targets of TncV) has already given hopeful results in preclinical trials, by utilization for pathological left ventricular hypertrophy in mice [351].
The field of RNA-based therapeutics has yet to overcome multiple obstacles, such as the delivery system, target specificity and immunogenicity [352]. One of the biggest concerns is the identification of the best (and most functional) target, as most ncRNAs are not extensively characterized. Utilization of a miRNA-based cancer treatment is not yet applied because a miRNA might have different targets in various cell types which are not characterized, thus leading to off-target effects. Identification of a ncRNA’s mechanism in cancer alongside with its associated genetic variation remains a priority, which can be tackled by utilizing novel, RNA-centric methods [352,353,354]. Therefore, focusing our efforts on understanding and uncovering the non-coding portion of cancer variability not only can accommodate early diagnosis of malignancies but can also lead to the development of personalized therapeutic strategies.
Author Contributions
Conceptualization, A.G., M.L. and R.B. writing—original draft preparation, M.L. and R.B.; writing—review and editing, A.G.; visualization, M.L. and R.B.; supervision, A.G.; project administration, A.G.; funding acquisition, A.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Fondation Santé (5935), ELIDEK POST-DOC SUPPORT (TINCeR 1407) and a Joint RT&D Greece—China research grant (“HEALTHAEGEAN”—MIS 5050733).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors would like to thank Pantelis Hatzis for his critical reading and suggestions on the manuscript. We regret for not including the excellent work of many more colleagues due to space limitations.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Della Chiara, G.; Gervasoni, F.; Fakiola, M.; Godano, C.; D’Oria, C.; Azzolin, L.; Bonnal, R.J.P.; Moreni, G.; Drufuca, L.; Rossetti, G.; et al. Epigenomic landscape of human colorectal cancer unveils an aberrant core of pan-cancer enhancers orchestrated by YAP/TAZ. Nat. Commun. 2021, 12, 2340. [Google Scholar] [CrossRef]
- Huang, H.; Hu, J.; Maryam, A.; Huang, Q.; Zhang, Y.; Ramakrishnan, S.; Li, J.; Ma, H.; Ma, V.W.S.; Cheuk, W.; et al. Defining super-enhancer landscape in triple-negative breast cancer by multiomic profiling. Nat. Commun. 2021, 12, 2242. [Google Scholar] [CrossRef]
- Xiong, L.; Wu, F.; Wu, Q.; Xu, L.; Cheung, O.K.; Kang, W.; Mok, M.T.; Szeto, L.L.M.; Lun, C.Y.; Lung, R.W.; et al. Aberrant enhancer hypomethylation contributes to hepatic carcinogenesis through global transcriptional reprogramming. Nat. Commun. 2019, 10, 335. [Google Scholar] [CrossRef]
- Sur, I.; Taipale, J. The role of enhancers in cancer. Nat. Rev. Cancer 2016, 16, 483–493. [Google Scholar] [CrossRef]
- See, Y.X.; Wang, B.Z.; Fullwood, M.J. Chromatin Interactions and Regulatory Elements in Cancer: From Bench to Bedside. Trends Genet. 2019, 35, 145–158. [Google Scholar] [CrossRef]
- Savarese, F.; Grosschedl, R. Blurring cis and trans in gene regulation. Cell 2006, 126, 248–250. [Google Scholar] [CrossRef]
- Andersson, R.; Sandelin, A. Determinants of enhancer and promoter activities of regulatory elements. Nat. Rev. Genet 2020, 21, 71–87. [Google Scholar] [CrossRef]
- Schoenfelder, S.; Fraser, P. Long-range enhancer-promoter contacts in gene expression control. Nat. Rev. Genet. 2019, 20, 437–455. [Google Scholar] [CrossRef]
- Schoenfelder, S.; Furlan-Magaril, M.; Mifsud, B.; Tavares-Cadete, F.; Sugar, R.; Javierre, B.M.; Nagano, T.; Katsman, Y.; Sakthidevi, M.; Wingett, S.W.; et al. The pluripotent regulatory circuitry connecting promoters to their long-range interacting elements. Genome Res. 2015, 25, 582–597. [Google Scholar] [CrossRef]
- Furlong, E.E.M.; Levine, M. Developmental enhancers and chromosome topology. Science 2018, 361, 1341–1345. [Google Scholar] [CrossRef]
- Maass, P.G.; Barutcu, A.R.; Rinn, J.L. Interchromosomal interactions: A genomic love story of kissing chromosomes. J. Cell Biol. 2019, 218, 27–38. [Google Scholar] [CrossRef]
- Gasperini, M.; Tome, J.M.; Shendure, J. Towards a comprehensive catalogue of validated and target-linked human enhancers. Nat. Rev. Genet. 2020, 21, 292–310. [Google Scholar] [CrossRef]
- Shlyueva, D.; Stampfel, G.; Stark, A. Transcriptional enhancers: From properties to genome-wide predictions. Nat. Rev. Genet. 2014, 15, 272–286. [Google Scholar] [CrossRef] [PubMed]
- Roadmap Epigenomics, C.; Kundaje, A.; Meuleman, W.; Ernst, J.; Bilenky, M.; Yen, A.; Heravi-Moussavi, A.; Kheradpour, P.; Zhang, Z.; Wang, J.; et al. Integrative analysis of 111 reference human epigenomes. Nature 2015, 518, 317–330. [Google Scholar] [CrossRef]
- Soutourina, J. Transcription regulation by the Mediator complex. Nat. Rev. Mol. Cell Biol. 2018, 19, 262–274. [Google Scholar] [CrossRef]
- Szabo, Q.; Bantignies, F.; Cavalli, G. Principles of genome folding into topologically associating domains. Sci. Adv. 2019, 5, eaaw1668. [Google Scholar] [CrossRef]
- Vietri Rudan, M.; Barrington, C.; Henderson, S.; Ernst, C.; Odom, D.T.; Tanay, A.; Hadjur, S. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 2015, 10, 1297–1309. [Google Scholar] [CrossRef]
- Nora, E.P.; Goloborodko, A.; Valton, A.L.; Gibcus, J.H.; Uebersohn, A.; Abdennur, N.; Dekker, J.; Mirny, L.A.; Bruneau, B.G. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell 2017, 169, 930.e922–944.e922. [Google Scholar] [CrossRef]
- Wutz, G.; Varnai, C.; Nagasaka, K.; Cisneros, D.A.; Stocsits, R.R.; Tang, W.; Schoenfelder, S.; Jessberger, G.; Muhar, M.; Hossain, M.J.; et al. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 2017, 36, 3573–3599. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.S.; Huntley, M.H.; Durand, N.C.; Stamenova, E.K.; Bochkov, I.D.; Robinson, J.T.; Sanborn, A.L.; Machol, I.; Omer, A.D.; Lander, E.S.; et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014, 159, 1665–1680. [Google Scholar] [CrossRef]
- Kim, S.; Yu, N.K.; Kaang, B.K. CTCF as a multifunctional protein in genome regulation and gene expression. Exp. Mol. Med. 2015, 47, e166. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Notani, D.; Rosenfeld, M.G. Enhancers as non-coding RNA transcription units: Recent insights and future perspectives. Nat. Rev. Genet. 2016, 17, 207–223. [Google Scholar] [CrossRef]
- Kim, T.K.; Hemberg, M.; Gray, J.M. Enhancer RNAs: A class of long noncoding RNAs synthesized at enhancers. Cold Spring Harb. Perspect. Biol. 2015, 7, a018622. [Google Scholar] [CrossRef] [PubMed]
- Jia, Q.; Chen, S.; Tan, Y.; Li, Y.; Tang, F. Oncogenic super-enhancer formation in tumorigenesis and its molecular mechanisms. Exp. Mol. Med. 2020, 52, 713–723. [Google Scholar] [CrossRef]
- Pott, S.; Lieb, J.D. What are super-enhancers? Nat. Genet. 2015, 47, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Sartorelli, V.; Lauberth, S.M. Enhancer RNAs are an important regulatory layer of the epigenome. Nat. Struct. Mol. Biol. 2020, 27, 521–528. [Google Scholar] [CrossRef]
- Zhang, Z.; Lee, J.H.; Ruan, H.; Ye, Y.; Krakowiak, J.; Hu, Q.; Xiang, Y.; Gong, J.; Zhou, B.; Wang, L.; et al. Transcriptional landscape and clinical utility of enhancer RNAs for eRNA-targeted therapy in cancer. Nat. Commun. 2019, 10, 4562. [Google Scholar] [CrossRef]
- Sakabe, N.J.; Savic, D.; Nobrega, M.A. Transcriptional enhancers in development and disease. Genome Biol. 2012, 13, 238. [Google Scholar] [CrossRef]
- Xia, J.H.; Wei, G.H. Enhancer Dysfunction in 3D Genome and Disease. Cells 2019, 8, 1281. [Google Scholar] [CrossRef]
- Chakravarty, D.; Solit, D.B. Clinical cancer genomic profiling. Nat. Rev. Genet. 2021. [Google Scholar] [CrossRef]
- Ramirez, M.; Rajaram, S.; Steininger, R.J.; Osipchuk, D.; Roth, M.A.; Morinishi, L.S.; Evans, L.; Ji, W.; Hsu, C.H.; Thurley, K.; et al. Diverse drug-resistance mechanisms can emerge from drug-tolerant cancer persister cells. Nat. Commun. 2016, 7, 10690. [Google Scholar] [CrossRef]
- Spielmann, M.; Lupianez, D.G.; Mundlos, S. Structural variation in the 3D genome. Nat. Rev. Genet. 2018, 19, 453–467. [Google Scholar] [CrossRef]
- Beckmann, J.S.; Estivill, X.; Antonarakis, S.E. Copy number variants and genetic traits: Closer to the resolution of phenotypic to genotypic variability. Nat. Rev. Genet. 2007, 8, 639–646. [Google Scholar] [CrossRef]
- Bu, D.; Yu, K.; Sun, S.; Xie, C.; Skogerbo, G.; Miao, R.; Xiao, H.; Liao, Q.; Luo, H.; Zhao, G.; et al. NONCODE v3.0: Integrative annotation of long noncoding RNAs. Nucleic Acids Res. 2012, 40, D210–D215. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research, N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008, 455, 1061–1068. [Google Scholar] [CrossRef]
- Carrot-Zhang, J.; Chambwe, N.; Damrauer, J.S.; Knijnenburg, T.A.; Robertson, A.G.; Yau, C.; Zhou, W.; Berger, A.C.; Huang, K.L.; Newberg, J.Y.; et al. Comprehensive Analysis of Genetic Ancestry and Its Molecular Correlates in Cancer. Cancer Cell 2020, 37, 639.e636–654.e636. [Google Scholar] [CrossRef]
- Hong, E.L.; Sloan, C.A.; Chan, E.T.; Davidson, J.M.; Malladi, V.S.; Strattan, J.S.; Hitz, B.C.; Gabdank, I.; Narayanan, A.K.; Ho, M.; et al. Principles of metadata organization at the ENCODE data coordination center. Database 2016, 2016. [Google Scholar] [CrossRef]
- Zhang, J.; Lee, D.; Dhiman, V.; Jiang, P.; Xu, J.; McGillivray, P.; Yang, H.; Liu, J.; Meyerson, W.; Clarke, D.; et al. An integrative ENCODE resource for cancer genomics. Nat. Commun. 2020, 11, 3696. [Google Scholar] [CrossRef]
- Liu, C.; Bai, B.; Skogerbo, G.; Cai, L.; Deng, W.; Zhang, Y.; Bu, D.; Zhao, Y.; Chen, R. NONCODE: An integrated knowledge database of non-coding RNAs. Nucleic Acids Res. 2005, 33, D112–D115. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research, N.; Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Karki, R.; Pandya, D.; Elston, R.C.; Ferlini, C. Defining “mutation” and “polymorphism” in the era of personal genomics. BMC Med. Genom. 2015, 8, 37. [Google Scholar] [CrossRef]
- Maston, G.A.; Evans, S.K.; Green, M.R. Transcriptional regulatory elements in the human genome. Annu Rev. Genom. Hum. Genet. 2006, 7, 29–59. [Google Scholar] [CrossRef]
- Kalender Atak, Z.; Imrichova, H.; Svetlichnyy, D.; Hulselmans, G.; Christiaens, V.; Reumers, J.; Ceulemans, H.; Aerts, S. Identification of cis-regulatory mutations generating de novo edges in personalized cancer gene regulatory networks. Genome Med. 2017, 9, 80. [Google Scholar] [CrossRef]
- Erson-Bensan, A.E. RNA-biology ruling cancer progression? Focus on 3’UTRs and splicing. Cancer Metastasis Rev. 2020, 39, 887–901. [Google Scholar] [CrossRef] [PubMed]
- Slack, F.J.; Chinnaiyan, A.M. The Role of Non-coding RNAs in Oncology. Cell 2019, 179, 1033–1055. [Google Scholar] [CrossRef]
- Anna, A.; Monika, G. Splicing mutations in human genetic disorders: Examples, detection, and confirmation. J. Appl. Genet. 2018, 59, 253–268. [Google Scholar] [CrossRef]
- Bashyam, M.D.; Animireddy, S.; Bala, P.; Naz, A.; George, S.A. The Yin and Yang of cancer genes. Gene 2019, 704, 121–133. [Google Scholar] [CrossRef]
- Baylin, S.B.; Jones, P.A. Epigenetic Determinants of Cancer. Cold Spring Harb. Perspect. Biol. 2016, 8. [Google Scholar] [CrossRef]
- Futreal, P.A.; Coin, L.; Marshall, M.; Down, T.; Hubbard, T.; Wooster, R.; Rahman, N.; Stratton, M.R. A census of human cancer genes. Nat. Rev. Cancer 2004, 4, 177–183. [Google Scholar] [CrossRef]
- Tam, V.; Patel, N.; Turcotte, M.; Bosse, Y.; Pare, G.; Meyre, D. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019, 20, 467–484. [Google Scholar] [CrossRef]
- Yong, S.Y.; Raben, T.G.; Lello, L.; Hsu, S.D.H. Genetic architecture of complex traits and disease risk predictors. Sci. Rep. 2020, 10, 12055. [Google Scholar] [CrossRef]
- Slatkin, M. Linkage disequilibrium--understanding the evolutionary past and mapping the medical future. Nat. Rev. Genet. 2008, 9, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Soong, D.; Stratford, J.; Avet-Loiseau, H.; Bahlis, N.; Davies, F.; Dispenzieri, A.; Sasser, A.K.; Schecter, J.M.; Qi, M.; Brown, C.; et al. CNV Radar: An improved method for somatic copy number alteration characterization in oncology. BMC Bioinform. 2020, 21, 98. [Google Scholar] [CrossRef]
- Li, Y.; Roberts, N.D.; Wala, J.A.; Shapira, O.; Schumacher, S.E.; Kumar, K.; Khurana, E.; Waszak, S.; Korbel, J.O.; Haber, J.E.; et al. Patterns of somatic structural variation in human cancer genomes. Nature 2020, 578, 112–121. [Google Scholar] [CrossRef]
- Kumaran, M.; Cass, C.E.; Graham, K.; Mackey, J.R.; Hubaux, R.; Lam, W.; Yasui, Y.; Damaraju, S. Germline copy number variations are associated with breast cancer risk and prognosis. Sci. Rep. 2017, 7, 14621. [Google Scholar] [CrossRef]
- Granados-Soler, J.L.; Bornemann-Kolatzki, K.; Beck, J.; Brenig, B.; Schutz, E.; Betz, D.; Junginger, J.; Hewicker-Trautwein, M.; Murua Escobar, H.; Nolte, I. Analysis of Copy-Number Variations and Feline Mammary Carcinoma Survival. Sci. Rep. 2020, 10, 1003. [Google Scholar] [CrossRef] [PubMed]
- Camps, J.; Grade, M.; Nguyen, Q.T.; Hormann, P.; Becker, S.; Hummon, A.B.; Rodriguez, V.; Chandrasekharappa, S.; Chen, Y.; Difilippantonio, M.J.; et al. Chromosomal breakpoints in primary colon cancer cluster at sites of structural variants in the genome. Cancer Res. 2008, 68, 1284–1295. [Google Scholar] [CrossRef] [PubMed]
- Lahortiga, I.; De Keersmaecker, K.; Van Vlierberghe, P.; Graux, C.; Cauwelier, B.; Lambert, F.; Mentens, N.; Beverloo, H.B.; Pieters, R.; Speleman, F.; et al. Duplication of the MYB oncogene in T cell acute lymphoblastic leukemia. Nat. Genet. 2007, 39, 593–595. [Google Scholar] [CrossRef]
- Basecke, J.; Whelan, J.T.; Griesinger, F.; Bertrand, F.E. The MLL partial tandem duplication in acute myeloid leukaemia. Br. J. Haematol. 2006, 135, 438–449. [Google Scholar] [CrossRef]
- Kumaran, M.; Krishnan, P.; Cass, C.E.; Hubaux, R.; Lam, W.; Yasui, Y.; Damaraju, S. Breast cancer associated germline structural variants harboring small noncoding RNAs impact post-transcriptional gene regulation. Sci Rep. 2018, 8, 7529. [Google Scholar] [CrossRef]
- Li, Y.R.; Glessner, J.T.; Coe, B.P.; Li, J.; Mohebnasab, M.; Chang, X.; Connolly, J.; Kao, C.; Wei, Z.; Bradfield, J.; et al. Rare copy number variants in over 100,000 European ancestry subjects reveal multiple disease associations. Nat. Commun. 2020, 11, 255. [Google Scholar] [CrossRef] [PubMed]
- Hennessey, R.C.; Brown, K.M. Cancer regulatory variation. Curr Opin Genet. Dev. 2021, 66, 41–49. [Google Scholar] [CrossRef]
- Spielmann, M.; Klopocki, E. CNVs of noncoding cis-regulatory elements in human disease. Curr. Opin. Genet. Dev. 2013, 23, 249–256. [Google Scholar] [CrossRef]
- Bonetti, A.; Agostini, F.; Suzuki, A.M.; Hashimoto, K.; Pascarella, G.; Gimenez, J.; Roos, L.; Nash, A.J.; Ghilotti, M.; Cameron, C.J.F.; et al. RADICL-seq identifies general and cell type-specific principles of genome-wide RNA-chromatin interactions. Nat. Commun. 2020, 11, 1018. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Deng, L.; Xia, Y.; Wei, X.; Cao, Y.; Guo, R.; Zhang, R.; Guo, J.; Liang, D.; Wu, L. A clear bias in parental origin of de novo pathogenic CNVs related to intellectual disability, developmental delay and multiple congenital anomalies. Sci. Rep. 2017, 7, 44446. [Google Scholar] [CrossRef]
- Wang, B.; Ji, T.; Zhou, X.; Wang, J.; Wang, X.; Wang, J.; Zhu, D.; Zhang, X.; Sham, P.C.; Zhang, X.; et al. CNV analysis in Chinese children of mental retardation highlights a sex differentiation in parental contribution to de novo and inherited mutational burdens. Sci. Rep. 2016, 6, 25954. [Google Scholar] [CrossRef]
- Rheinbay, E.; Nielsen, M.M.; Abascal, F.; Wala, J.A.; Shapira, O.; Tiao, G.; Hornshoj, H.; Hess, J.M.; Juul, R.I.; Lin, Z.; et al. Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature 2020, 578, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Hua, J.T.; Ahmed, M.; Guo, H.; Zhang, Y.; Chen, S.; Soares, F.; Lu, J.; Zhou, S.; Wang, M.; Li, H.; et al. Risk SNP-Mediated Promoter-Enhancer Switching Drives Prostate Cancer through lncRNA PCAT19. Cell 2018, 174, 564.e518–575.e518. [Google Scholar] [CrossRef]
- Wang, Y.; Ma, R.; Liu, B.; Kong, J.; Lin, H.; Yu, X.; Wang, R.; Li, L.; Gao, M.; Zhou, B.; et al. SNP rs17079281 decreases lung cancer risk through creating an YY1-binding site to suppress DCBLD1 expression. Oncogene 2020, 39, 4092–4102. [Google Scholar] [CrossRef]
- Motawi, T.M.K.; Sadik, N.A.H.; Sabry, D.; Shahin, N.N.; Fahim, S.A. rs2267531, a promoter SNP within glypican-3 gene in the X chromosome, is associated with hepatocellular carcinoma in Egyptians. Sci. Rep. 2019, 9, 6868. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, N.; Zhang, L.; Song, Y.; Liu, J.; Yu, J.; Yang, M. Oncogene HSPH1 modulated by the rs2280059 genetic variant diminishes EGFR-TKIs efficiency in advanced lung adenocarcinoma. Carcinogenesis 2020, 41, 1195–1202. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ahmed, M.; Zhang, F.; Yao, C.Q.; Li, S.; Liang, Y.; Hua, J.; Soares, F.; Sun, Y.; Langstein, J.; et al. Modulation of long noncoding RNAs by risk SNPs underlying genetic predispositions to prostate cancer. Nat. Genet. 2016, 48, 1142–1150. [Google Scholar] [CrossRef]
- Grampp, S.; Platt, J.L.; Lauer, V.; Salama, R.; Kranz, F.; Neumann, V.K.; Wach, S.; Stohr, C.; Hartmann, A.; Eckardt, K.U.; et al. Genetic variation at the 8q24.21 renal cancer susceptibility locus affects HIF binding to a MYC enhancer. Nat. Commun. 2016, 7, 13183. [Google Scholar] [CrossRef]
- Tuupanen, S.; Turunen, M.; Lehtonen, R.; Hallikas, O.; Vanharanta, S.; Kivioja, T.; Bjorklund, M.; Wei, G.; Yan, J.; Niittymaki, I.; et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat. Genet. 2009, 41, 885–890. [Google Scholar] [CrossRef]
- Walker, L.C.; BCFR; Marquart, L.; Pearson, J.F.; Wiggins, G.A.R.; O’Mara, T.A.; Parsons, M.T.; Barrowdale, D.; McGuffog, L.; Dennis, J.; et al. Evaluation of copy-number variants as modifiers of breast and ovarian cancer risk for BRCA1 pathogenic variant carriers. Eur. J. Hum. Genet. 2017, 25, 432–438. [Google Scholar] [CrossRef]
- Wang, J.; Zou, Y.; Du, B.; Li, W.; Yu, G.; Li, L.; Zhou, L.; Gu, X.; Song, S.; Liu, Y.; et al. SNP-mediated lncRNA-ENTPD3-AS1 upregulation suppresses renal cell carcinoma via miR-155/HIF-1alpha signaling. Cell Death Dis. 2021, 12, 672. [Google Scholar] [CrossRef] [PubMed]
- Ostrovsky, O.; Grushchenko-Polaq, A.H.; Beider, K.; Mayorov, M.; Canaani, J.; Shimoni, A.; Vlodavsky, I.; Nagler, A. Identification of strong intron enhancer in the heparanase gene: Effect of functional rs4693608 variant on HPSE enhancer activity in hematological and solid malignancies. Oncogenesis 2018, 7, 51. [Google Scholar] [CrossRef]
- Painter, J.N.; Kaufmann, S.; O’Mara, T.A.; Hillman, K.M.; Sivakumaran, H.; Darabi, H.; Cheng, T.H.T.; Pearson, J.; Kazakoff, S.; Waddell, N.; et al. A Common Variant at the 14q32 Endometrial Cancer Risk Locus Activates AKT1 through YY1 Binding. Am. J. Hum. Genet. 2016, 98, 1159–1169. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Zheng, R.; Ma, G.; Chu, H.; Lu, J.; Li, S.; Xin, J.; Tong, N.; Zhang, G.; Wang, W.; et al. Remote modulation of lncRNA GCLET by risk variant at 16p13 underlying genetic susceptibility to gastric cancer. Sci. Adv. 2020, 6, eaay5525. [Google Scholar] [CrossRef]
- Helmsauer, K.; Valieva, M.E.; Ali, S.; Chamorro Gonzalez, R.; Schopflin, R.; Roefzaad, C.; Bei, Y.; Dorado Garcia, H.; Rodriguez-Fos, E.; Puiggros, M.; et al. Enhancer hijacking determines extrachromosomal circular MYCN amplicon architecture in neuroblastoma. Nat. Commun. 2020, 11, 5823. [Google Scholar] [CrossRef]
- El Hajj, P.; Gilot, D.; Migault, M.; Theunis, A.; van Kempen, L.C.; Sales, F.; Fayyad-Kazan, H.; Badran, B.; Larsimont, D.; Awada, A.; et al. SNPs at miR-155 binding sites of TYRP1 explain discrepancy between mRNA and protein and refine TYRP1 prognostic value in melanoma. Br. J. Cancer 2015, 113, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Zandi, R.; Shao, R.; Gu, J.; Ye, Y.; Wang, J.; Zhao, Y.; Pertsemlidis, A.; Wistuba, I.I.; Wu, X.; et al. A miR-SNP biomarker linked to an increased lung cancer survival by miRNA-mediated down-regulation of FZD4 expression and Wnt signaling. Sci. Rep. 2017, 7, 9029. [Google Scholar] [CrossRef] [PubMed]
- Gilam, A.; Conde, J.; Weissglas-Volkov, D.; Oliva, N.; Friedman, E.; Artzi, N.; Shomron, N. Local microRNA delivery targets Palladin and prevents metastatic breast cancer. Nat. Commun. 2016, 7, 12868. [Google Scholar] [CrossRef]
- Prensner, J.R.; Iyer, M.K.; Balbin, O.A.; Dhanasekaran, S.M.; Cao, Q.; Brenner, J.C.; Laxman, B.; Asangani, I.A.; Grasso, C.S.; Kominsky, H.D.; et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat. Biotechnol. 2011, 29, 742–749. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, N.; Cordiner, R.A.; Young, R.S.; Hug, N.; Macias, S.; Caceres, J.F. Genetic variation and RNA structure regulate microRNA biogenesis. Nat. Commun. 2017, 8, 15114. [Google Scholar] [CrossRef] [PubMed]
- Mu, Y.P.; Su, X.L. Polymorphism in pre-miR-30c contributes to gastric cancer risk in a Chinese population. Med. Oncol. 2012, 29, 1723–1732. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Liu, X.; Meng, F.; Zhang, Y.; Wang, M.; Chen, Y.; Guo, X.; Chen, W.; Wang, W. The rs7911488-T allele promotes the growth and metastasis of colorectal cancer through modulating miR-1307/PRRX1. Cell Death Dis. 2020, 11, 651. [Google Scholar] [CrossRef]
- Yang, Q.; Jie, Z.; Ye, S.; Li, Z.; Han, Z.; Wu, J.; Yang, C.; Jiang, Y. Genetic variations in miR-27a gene decrease mature miR-27a level and reduce gastric cancer susceptibility. Oncogene 2014, 33, 193–202. [Google Scholar] [CrossRef]
- Redis, R.S.; Vela, L.E.; Lu, W.; Ferreira de Oliveira, J.; Ivan, C.; Rodriguez-Aguayo, C.; Adamoski, D.; Pasculli, B.; Taguchi, A.; Chen, Y.; et al. Allele-Specific Reprogramming of Cancer Metabolism by the Long Non-coding RNA CCAT2. Mol. Cell 2016, 61, 520–534. [Google Scholar] [CrossRef]
- Yuan, H.; Liu, H.; Liu, Z.; Owzar, K.; Han, Y.; Su, L.; Wei, Y.; Hung, R.J.; McLaughlin, J.; Brhane, Y.; et al. A Novel Genetic Variant in Long Non-coding RNA Gene NEXN-AS1 is Associated with Risk of Lung Cancer. Sci. Rep. 2016, 6, 34234. [Google Scholar] [CrossRef]
- Wu, S.; Sun, H.; Wang, Y.; Yang, X.; Meng, Q.; Yang, H.; Zhu, H.; Tang, W.; Li, X.; Aschner, M.; et al. MALAT1 rs664589 Polymorphism Inhibits Binding to miR-194-5p, Contributing to Colorectal Cancer Risk, Growth, and Metastasis. Cancer Res. 2019, 79, 5432–5441. [Google Scholar] [CrossRef]
- Shen, C.; Yan, T.; Wang, Z.; Su, H.C.; Zhu, X.; Tian, X.; Fang, J.Y.; Chen, H.; Hong, J. Variant of SNP rs1317082 at CCSlnc362 (RP11-362K14.5) creates a binding site for miR-4658 and diminishes the susceptibility to CRC. Cell Death Dis. 2018, 9, 1177. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zheng, J.; Deng, J.; Hu, M.; You, Y.; Li, N.; Li, W.; Lu, J.; Zhou, Y. A genetic polymorphism in lincRNA-uc003opf.1 is associated with susceptibility to esophageal squamous cell carcinoma in Chinese populations. Carcinogenesis 2013, 34, 2908–2917. [Google Scholar] [CrossRef]
- Zheng, J.; Huang, X.; Tan, W.; Yu, D.; Du, Z.; Chang, J.; Wei, L.; Han, Y.; Wang, C.; Che, X.; et al. Pancreatic cancer risk variant in LINC00673 creates a miR-1231 binding site and interferes with PTPN11 degradation. Nat. Genet. 2016, 48, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Dhamodharan, S.; Rose, M.M.; Chakkarappan, S.R.; Umadharshini, K.V.; Arulmurugan, R.; Subbiah, S.; Inoue, I.; Munirajan, A.K. Genetic variant rs10251977 (G > A) in EGFR-AS1 modulates the expression of EGFR isoforms A and D. Sci. Rep. 2021, 11, 8808. [Google Scholar] [CrossRef]
- Lee, D.S.M.; Park, J.; Kromer, A.; Baras, A.; Rader, D.J.; Ritchie, M.D.; Ghanem, L.R.; Barash, Y. Disrupting upstream translation in mRNAs is associated with human disease. Nat. Commun. 2021, 12, 1515. [Google Scholar] [CrossRef] [PubMed]
- Steri, M.; Idda, M.L.; Whalen, M.B.; Orru, V. Genetic variants in mRNA untranslated regions. Wiley Interdiscip. Rev. RNA 2018, 9, e1474. [Google Scholar] [CrossRef] [PubMed]
- Powers, M.P. The ever-changing world of gene fusions in cancer: A secondary gene fusion and progression. Oncogene 2019, 38, 7197–7199. [Google Scholar] [CrossRef]
- Rashkin, S.R.; Graff, R.E.; Kachuri, L.; Thai, K.K.; Alexeeff, S.E.; Blatchins, M.A.; Cavazos, T.B.; Corley, D.A.; Emami, N.C.; Hoffman, J.D.; et al. Pan-cancer study detects genetic risk variants and shared genetic basis in two large cohorts. Nat. Commun. 2020, 11, 4423. [Google Scholar] [CrossRef] [PubMed]
- Giral, H.; Landmesser, U.; Kratzer, A. Into the Wild: GWAS Exploration of Non-coding RNAs. Front. Cardiovasc. Med. 2018, 5, 181. [Google Scholar] [CrossRef]
- Hrdlickova, B.; de Almeida, R.C.; Borek, Z.; Withoff, S. Genetic variation in the non-coding genome: Involvement of micro-RNAs and long non-coding RNAs in disease. Biochim. Biophys. Acta 2014, 1842, 1910–1922. [Google Scholar] [CrossRef] [PubMed]
- Consortium, G.T. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 2015, 348, 648–660. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Wen, J.; Huang, Z.; Chen, X.P.; Zhang, B.X.; Chu, L. Small Nucleolar RNAs: Insight into Their Function in Cancer. Front. Oncol. 2019, 9, 587. [Google Scholar] [CrossRef]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 2019, 20, 89–108. [Google Scholar] [CrossRef] [PubMed]
- Morris, K.V.; Mattick, J.S. The rise of regulatory RNA. Nat. Rev. Genet. 2014, 15, 423–437. [Google Scholar] [CrossRef] [PubMed]
- Khurana, E.; Fu, Y.; Chakravarty, D.; Demichelis, F.; Rubin, M.A.; Gerstein, M. Role of non-coding sequence variants in cancer. Nat. Rev. Genet. 2016, 17, 93–108. [Google Scholar] [CrossRef]
- Zhu, H.; Uuskula-Reimand, L.; Isaev, K.; Wadi, L.; Alizada, A.; Shuai, S.; Huang, V.; Aduluso-Nwaobasi, D.; Paczkowska, M.; Abd-Rabbo, D.; et al. Candidate Cancer Driver Mutations in Distal Regulatory Elements and Long-Range Chromatin Interaction Networks. Mol. Cell 2020, 77, 1307.e1310–1321.e1310. [Google Scholar] [CrossRef]
- Cheng, Y.; Quinn, J.F.; Weiss, L.A. An eQTL mapping approach reveals that rare variants in the SEMA5A regulatory network impact autism risk. Hum. Mol. Genet. 2013, 22, 2960–2972. [Google Scholar] [CrossRef]
- Li, X.; Kim, Y.; Tsang, E.K.; Davis, J.R.; Damani, F.N.; Chiang, C.; Hess, G.T.; Zappala, Z.; Strober, B.J.; Scott, A.J.; et al. The impact of rare variation on gene expression across tissues. Nature 2017, 550, 239–243. [Google Scholar] [CrossRef]
- Huyghe, J.R.; Bien, S.A.; Harrison, T.A.; Kang, H.M.; Chen, S.; Schmit, S.L.; Conti, D.V.; Qu, C.; Jeon, J.; Edlund, C.K.; et al. Discovery of common and rare genetic risk variants for colorectal cancer. Nat. Genet. 2019, 51, 76–87. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Luo, Y.; Sun, Y.; Guo, W.; Zhao, X.; Xi, Y.; Ma, Y.; Shao, M.; Tan, W.; Gao, G.; et al. Genomic and transcriptomic alterations associated with drug vulnerabilities and prognosis in adenocarcinoma at the gastroesophageal junction. Nat. Commun. 2020, 11, 6091. [Google Scholar] [CrossRef] [PubMed]
- Bi, H.; Tian, T.; Zhu, L.; Zhou, H.; Hu, H.; Liu, Y.; Li, X.; Hu, F.; Zhao, Y.; Wang, G. Copy number variation of E3 ubiquitin ligase genes in peripheral blood leukocyte and colorectal cancer. Sci. Rep. 2016, 6, 29869. [Google Scholar] [CrossRef][Green Version]
- Mamlouk, S.; Childs, L.H.; Aust, D.; Heim, D.; Melching, F.; Oliveira, C.; Wolf, T.; Durek, P.; Schumacher, D.; Blaker, H.; et al. DNA copy number changes define spatial patterns of heterogeneity in colorectal cancer. Nat. Commun. 2017, 8, 14093. [Google Scholar] [CrossRef] [PubMed]
- Fagny, M.; Platig, J.; Kuijjer, M.L.; Lin, X.; Quackenbush, J. Nongenic cancer-risk SNPs affect oncogenes, tumour-suppressor genes, and immune function. Br. J. Cancer 2020, 122, 569–577. [Google Scholar] [CrossRef]
- Liu, X.Q.; Liu, X.S.; Rong, J.Y.; Gao, F.; Wu, Y.D.; Deng, C.H.; Jiang, H.Y.; Li, X.F.; Chen, Y.Q.; Zhao, Z.G.; et al. Precise diagnosis of three top cancers using dbGaP data. Sci. Rep. 2021, 11, 823. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Song, J.; Lu, Y.; Fall, K.; Chen, X.; Fang, F.; Landen, M.; Hultman, C.M.; Czene, K.; Sullivan, P.; et al. A shared genetic contribution to breast cancer and schizophrenia. Nat. Commun. 2020, 11, 4637. [Google Scholar] [CrossRef]
- Zhong, Q.; Lu, M.; Yuan, W.; Cui, Y.; Ouyang, H.; Fan, Y.; Wang, Z.; Wu, C.; Qiao, J.; Hang, J. Eight-lncRNA signature of cervical cancer were identified by integrating DNA methylation, copy number variation and transcriptome data. J. Transl. Med. 2021, 19, 58. [Google Scholar] [CrossRef]
- Diederichs, S.; Bartsch, L.; Berkmann, J.C.; Frose, K.; Heitmann, J.; Hoppe, C.; Iggena, D.; Jazmati, D.; Karschnia, P.; Linsenmeier, M.; et al. The dark matter of the cancer genome: Aberrations in regulatory elements, untranslated regions, splice sites, non-coding RNA and synonymous mutations. EMBO Mol. Med. 2016, 8, 442–457. [Google Scholar] [CrossRef] [PubMed]
- Poulos, R.C.; Sloane, M.A.; Hesson, L.B.; Wong, J.W. The search for cis-regulatory driver mutations in cancer genomes. Oncotarget 2015, 6, 32509–32525. [Google Scholar] [CrossRef] [PubMed]
- Haberle, V.; Stark, A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell Biol. 2018, 19, 621–637. [Google Scholar] [CrossRef] [PubMed]
- Muratani, M.; Deng, N.; Ooi, W.F.; Lin, S.J.; Xing, M.; Xu, C.; Qamra, A.; Tay, S.T.; Malik, S.; Wu, J.; et al. Nanoscale chromatin profiling of gastric adenocarcinoma reveals cancer-associated cryptic promoters and somatically acquired regulatory elements. Nat. Commun. 2014, 5, 4361. [Google Scholar] [CrossRef]
- Liu, J.; Adhav, R.; Miao, K.; Su, S.M.; Mo, L.; Chan, U.I.; Zhang, X.; Xu, J.; Li, J.; Shu, X.; et al. Characterization of BRCA1-deficient premalignant tissues and cancers identifies Plekha5 as a tumor metastasis suppressor. Nat. Commun. 2020, 11, 4875. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Estecio, M.R.; Chen, R.; Reuben, A.; Wang, L.; Fujimoto, J.; Carrot-Zhang, J.; McGranahan, N.; Ying, L.; Fukuoka, J.; et al. Evolution of DNA methylome from precancerous lesions to invasive lung adenocarcinomas. Nat. Commun. 2021, 12, 687. [Google Scholar] [CrossRef]
- Chen, H.; Li, C.; Peng, X.; Zhou, Z.; Weinstein, J.N.; Cancer Genome Atlas Research, N.; Liang, H. A Pan-Cancer Analysis of Enhancer Expression in Nearly 9000 Patient Samples. Cell 2018, 173, 386.e312–399.e312. [Google Scholar] [CrossRef]
- Lidschreiber, K.; Jung, L.A.; von der Emde, H.; Dave, K.; Taipale, J.; Cramer, P.; Lidschreiber, M. Transcriptionally active enhancers in human cancer cells. Mol. Syst. Biol. 2021, 17, e9873. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Liao, Z.; Gritsch, D.; Hadzhiev, Y.; Bai, Y.; Locascio, J.J.; Guennewig, B.; Liu, G.; Blauwendraat, C.; Wang, T.; et al. Enhancers active in dopamine neurons are a primary link between genetic variation and neuropsychiatric disease. Nat. Neurosci. 2018, 21, 1482–1492. [Google Scholar] [CrossRef] [PubMed]
- Garieri, M.; Delaneau, O.; Santoni, F.; Fish, R.J.; Mull, D.; Carninci, P.; Dermitzakis, E.T.; Antonarakis, S.E.; Fort, A. The effect of genetic variation on promoter usage and enhancer activity. Nat. Commun. 2017, 8, 1358. [Google Scholar] [CrossRef]
- Elkon, R.; Agami, R. Characterization of noncoding regulatory DNA in the human genome. Nat. Biotechnol. 2017, 35, 732–746. [Google Scholar] [CrossRef]
- Thandapani, P. Super-enhancers in cancer. Pharmacol. Ther. 2019, 199, 129–138. [Google Scholar] [CrossRef]
- Li, Y.; He, Y.; Peng, J.; Su, Z.; Li, Z.; Zhang, B.; Ma, J.; Zhuo, M.; Zou, D.; Liu, X.; et al. Mutant Kras co-opts a proto-oncogenic enhancer network in inflammation-induced metaplastic progenitor cells to initiate pancreatic cancer. Nature Cancer 2021, 2, 49–65. [Google Scholar] [CrossRef]
- Chen, R.; Ren, S.; Sun, Y. Genome-wide association studies on prostate cancer: The end or the beginning? Protein Cell 2013, 4, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Amin Al Olama, A.; Kote-Jarai, Z.; Schumacher, F.R.; Wiklund, F.; Berndt, S.I.; Benlloch, S.; Giles, G.G.; Severi, G.; Neal, D.E.; Hamdy, F.C.; et al. A meta-analysis of genome-wide association studies to identify prostate cancer susceptibility loci associated with aggressive and non-aggressive disease. Hum. Mol. Genet. 2013, 22, 408–415. [Google Scholar] [CrossRef]
- Shui, I.M.; Lindstrom, S.; Kibel, A.S.; Berndt, S.I.; Campa, D.; Gerke, T.; Penney, K.L.; Albanes, D.; Berg, C.; Bueno-de-Mesquita, H.B.; et al. Prostate cancer (PCa) risk variants and risk of fatal PCa in the National Cancer Institute Breast and Prostate Cancer Cohort Consortium. Eur. Urol. 2014, 65, 1069–1075. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Xia, J.H.; Sipeky, C.; Dong, X.M.; Zhang, Q.; Yang, Y.; Zhang, P.; Cruz, S.P.; Zhang, K.; Zhu, J.; et al. Biology and Clinical Implications of the 19q13 Aggressive Prostate Cancer Susceptibility Locus. Cell 2018, 174, 576.e518–589.e518. [Google Scholar] [CrossRef]
- Shang, Z.; Yu, J.; Sun, L.; Tian, J.; Zhu, S.; Zhang, B.; Dong, Q.; Jiang, N.; Flores-Morales, A.; Chang, C.; et al. LncRNA PCAT1 activates AKT and NF-kappaB signaling in castration-resistant prostate cancer by regulating the PHLPP/FKBP51/IKKalpha complex. Nucleic Acids Res. 2019, 47, 4211–4225. [Google Scholar] [CrossRef]
- Song, Y.H.; Shiota, M.; Kuroiwa, K.; Naito, S.; Oda, Y. The important role of glycine N-methyltransferase in the carcinogenesis and progression of prostate cancer. Mod. Pathol 2011, 24, 1272–1280. [Google Scholar] [CrossRef]
- Bonaccorsi, L.; Luciani, P.; Nesi, G.; Mannucci, E.; Deledda, C.; Dichiara, F.; Paglierani, M.; Rosati, F.; Masieri, L.; Serni, S.; et al. Androgen receptor regulation of the seladin-1/DHCR24 gene: Altered expression in prostate cancer. Lab. Investig. 2008, 88, 1049–1056. [Google Scholar] [CrossRef]
- Wasserman, N.F.; Aneas, I.; Nobrega, M.A. An 8q24 gene desert variant associated with prostate cancer risk confers differential in vivo activity to a MYC enhancer. Genome Res. 2010, 20, 1191–1197. [Google Scholar] [CrossRef]
- Wright, J.B.; Brown, S.J.; Cole, M.D. Upregulation of c-MYC in cis through a large chromatin loop linked to a cancer risk-associated single-nucleotide polymorphism in colorectal cancer cells. Mol. Cell Biol. 2010, 30, 1411–1420. [Google Scholar] [CrossRef]
- Doni Jayavelu, N.; Jajodia, A.; Mishra, A.; Hawkins, R.D. Candidate silencer elements for the human and mouse genomes. Nat. Commun. 2020, 11, 1061. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Zhang, Y.; Loh, Y.P.; Tng, J.Q.; Lim, M.C.; Cao, Z.; Raju, A.; Lieberman Aiden, E.; Li, S.; Manikandan, L.; et al. H3K27me3-rich genomic regions can function as silencers to repress gene expression via chromatin interactions. Nat. Commun. 2021, 12, 719. [Google Scholar] [CrossRef] [PubMed]
- Pang, B.; Snyder, M.P. Systematic identification of silencers in human cells. Nat. Genet. 2020, 52, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Dunning, A.M.; Michailidou, K.; Kuchenbaecker, K.B.; Thompson, D.; French, J.D.; Beesley, J.; Healey, C.S.; Kar, S.; Pooley, K.A.; Lopez-Knowles, E.; et al. Breast cancer risk variants at 6q25 display different phenotype associations and regulate ESR1, RMND1 and CCDC170. Nat. Genet. 2016, 48, 374–386. [Google Scholar] [CrossRef]
- Jing, H.; Vakoc, C.R.; Ying, L.; Mandat, S.; Wang, H.; Zheng, X.; Blobel, G.A. Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol. Cell 2008, 29, 232–242. [Google Scholar] [CrossRef]
- Grotsch, B.; Brachs, S.; Lang, C.; Luther, J.; Derer, A.; Schlotzer-Schrehardt, U.; Bozec, A.; Fillatreau, S.; Berberich, I.; Hobeika, E.; et al. The AP-1 transcription factor Fra1 inhibits follicular B cell differentiation into plasma cells. J. Exp. Med. 2014, 211, 2199–2212. [Google Scholar] [CrossRef]
- Hadjiagapiou, C.; Borthakur, A.; Dahdal, R.Y.; Gill, R.K.; Malakooti, J.; Ramaswamy, K.; Dudeja, P.K. Role of USF1 and USF2 as potential repressor proteins for human intestinal monocarboxylate transporter 1 promoter. Am. J. Physiol. Gastrointest. Liver Physiol. 2005, 288, G1118–G1126. [Google Scholar] [CrossRef]
- Nakayama, A.; Murakami, H.; Maeyama, N.; Yamashiro, N.; Sakakibara, A.; Mori, N.; Takahashi, M. Role for RFX transcription factors in non-neuronal cell-specific inactivation of the microtubule-associated protein MAP1A promoter. J. Biol. Chem 2003, 278, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Timblin, G.A.; Schlissel, M.S. Ebf1 and c-Myb repress rag transcription downstream of Stat5 during early B cell development. J. Immunol. 2013, 191, 4676–4687. [Google Scholar] [CrossRef] [PubMed]
- Tsukumo, S.; Unno, M.; Muto, A.; Takeuchi, A.; Kometani, K.; Kurosaki, T.; Igarashi, K.; Saito, T. Bach2 maintains T cells in a naive state by suppressing effector memory-related genes. Proc. Natl. Acad. Sci. USA 2013, 110, 10735–10740. [Google Scholar] [CrossRef]
- Jolma, A.; Yan, J.; Whitington, T.; Toivonen, J.; Nitta, K.R.; Rastas, P.; Morgunova, E.; Enge, M.; Taipale, M.; Wei, G.; et al. DNA-binding specificities of human transcription factors. Cell 2013, 152, 327–339. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Morgunova, E.; Jolma, A.; Kaasinen, E.; Sahu, B.; Khund-Sayeed, S.; Das, P.K.; Kivioja, T.; Dave, K.; Zhong, F.; et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 2017, 356. [Google Scholar] [CrossRef]
- Gaszner, M.; Felsenfeld, G. Insulators: Exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 2006, 7, 703–713. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Maurano, M.T.; Wang, H.; Qi, H.; Song, C.Z.; Navas, P.A.; Emery, D.W.; Stamatoyannopoulos, J.A.; Stamatoyannopoulos, G. Genomic discovery of potent chromatin insulators for human gene therapy. Nat. Biotechnol. 2015, 33, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Lou, D.; Wang, Z. Crosstalk of Genetic Variants, Allele-Specific DNA Methylation, and Environmental Factors for Complex Disease Risk. Front. Genet. 2018, 9, 695. [Google Scholar] [CrossRef] [PubMed]
- Deng, N.; Zhou, H.; Fan, H.; Yuan, Y. Single nucleotide polymorphisms and cancer susceptibility. Oncotarget 2017, 8, 110635–110649. [Google Scholar] [CrossRef]
- Dai, J.; Zhu, M.; Wang, C.; Shen, W.; Zhou, W.; Sun, J.; Liu, J.; Jin, G.; Ma, H.; Hu, Z.; et al. Systematical analyses of variants in CTCF-binding sites identified a novel lung cancer susceptibility locus among Chinese population. Sci. Rep. 2015, 5, 7833. [Google Scholar] [CrossRef]
- Wang, J.; Guan, X.; Zhang, Y.; Ge, S.; Zhang, L.; Li, H.; Wang, X.; Liu, R.; Ning, T.; Deng, T.; et al. Exosomal miR-27a Derived from Gastric Cancer Cells Regulates the Transformation of Fibroblasts into Cancer-Associated Fibroblasts. Cell Physiol. Biochem. 2018, 49, 869–883. [Google Scholar] [CrossRef]
- Zhou, L.; Liang, X.; Zhang, L.; Yang, L.; Nagao, N.; Wu, H.; Liu, C.; Lin, S.; Cai, G.; Liu, J. MiR-27a-3p functions as an oncogene in gastric cancer by targeting BTG2. Oncotarget 2016, 7, 51943–51954. [Google Scholar] [CrossRef]
- Kim, J.H.; Hwang, J.; Jung, J.H.; Lee, H.J.; Lee, D.Y.; Kim, S.H. Molecular networks of FOXP family: Dual biologic functions, interplay with other molecules and clinical implications in cancer progression. Mol. Cancer 2019, 18, 180. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, I.; Kouzarides, T. Role of RNA modifications in cancer. Nat. Rev. Cancer 2020, 20, 303–322. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef]
- Qi, Y.; Lai, Y.L.; Shen, P.C.; Chen, F.H.; Lin, L.J.; Wu, H.H.; Peng, P.H.; Hsu, K.W.; Cheng, W.C. Identification and validation of a miRNA-based prognostic signature for cervical cancer through an integrated bioinformatics approach. Sci. Rep. 2020, 10, 22270. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Helwak, A.; Kudla, G.; Dudnakova, T.; Tollervey, D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 2013, 153, 654–665. [Google Scholar] [CrossRef] [PubMed]
- Ryan, B.M.; Robles, A.I.; Harris, C.C. Genetic variation in microRNA networks: The implications for cancer research. Nat. Rev. Cancer 2010, 10, 389–402. [Google Scholar] [CrossRef] [PubMed]
- Dhawan, A.; Scott, J.G.; Harris, A.L.; Buffa, F.M. Pan-cancer characterisation of microRNA across cancer hallmarks reveals microRNA-mediated downregulation of tumour suppressors. Nat. Commun. 2018, 9, 5228. [Google Scholar] [CrossRef]
- Lin, W.C.; Shih, P.H.; Wang, W.; Wu, C.H.; Hsia, S.M.; Wang, H.J.; Hwang, P.A.; Wang, C.Y.; Chen, S.H.; Kuo, Y.T. Inhibitory effects of high stability fucoxanthin on palmitic acid-induced lipid accumulation in human adipose-derived stem cells through modulation of long non-coding RNA. Food Funct. 2015, 6, 2215–2223. [Google Scholar] [CrossRef]
- Lee, A.R.; Park, J.; Jung, K.J.; Jee, S.H.; Kim-Yoon, S. Genetic variation rs7930 in the miR-4273-5p target site is associated with a risk of colorectal cancer. Onco. Targets Ther. 2016, 9, 6885–6895. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, M.; Du, M.; Ma, L.; Chu, H.; Lv, Q.; Ye, D.; Guo, J.; Gu, C.; Xia, G.; Zhu, Y.; et al. A functional variant in TP63 at 3q28 associated with bladder cancer risk by creating an miR-140-5p binding site. Int. J. Cancer 2016, 139, 65–74. [Google Scholar] [CrossRef]
- Wynendaele, J.; Bohnke, A.; Leucci, E.; Nielsen, S.J.; Lambertz, I.; Hammer, S.; Sbrzesny, N.; Kubitza, D.; Wolf, A.; Gradhand, E.; et al. An illegitimate microRNA target site within the 3’ UTR of MDM4 affects ovarian cancer progression and chemosensitivity. Cancer Res. 2010, 70, 9641–9649. [Google Scholar] [CrossRef]
- Jacinta-Fernandes, A.; Xavier, J.M.; Magno, R.; Lage, J.G.; Maia, A.T. Allele-specific miRNA-binding analysis identifies candidate target genes for breast cancer risk. NPJ Genom. Med. 2020, 5, 4. [Google Scholar] [CrossRef]
- Hua, K.T.; Liu, Y.F.; Hsu, C.L.; Cheng, T.Y.; Yang, C.Y.; Chang, J.S.; Lee, W.J.; Hsiao, M.; Juan, H.F.; Chien, M.H.; et al. 3’UTR polymorphisms of carbonic anhydrase IX determine the miR-34a targeting efficiency and prognosis of hepatocellular carcinoma. Sci. Rep. 2017, 7, 4466. [Google Scholar] [CrossRef]
- Hogg, D.R.; Harries, L.W. Human genetic variation and its effect on miRNA biogenesis, activity and function. Biochem. Soc. Trans. 2014, 42, 1184–1189. [Google Scholar] [CrossRef]
- Song, X.; Wan, X.; Huang, T.; Zeng, C.; Sastry, N.; Wu, B.; James, C.D.; Horbinski, C.; Nakano, I.; Zhang, W.; et al. SRSF3-Regulated RNA Alternative Splicing Promotes Glioblastoma Tumorigenicity by Affecting Multiple Cellular Processes. Cancer Res. 2019, 79, 5288–5301. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.; Triboulet, R.; Mohseni, M.; Schlegelmilch, K.; Shrestha, K.; Camargo, F.D.; Gregory, R.I. Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell 2014, 156, 893–906. [Google Scholar] [CrossRef]
- Gould, P.S.; Bird, H.; Easton, A.J. Translation toeprinting assays using fluorescently labeled primers and capillary electrophoresis. Biotechniques 2005, 38, 397–400. [Google Scholar] [CrossRef]
- Wilkinson, K.A.; Merino, E.J.; Weeks, K.M. Selective 2’-hydroxyl acylation analyzed by primer extension (SHAPE): Quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006, 1, 1610–1616. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Ambrosone, C.B.; Zhao, H. Novel genetic variants in microRNA genes and familial breast cancer. Int. J. Cancer 2009, 124, 1178–1182. [Google Scholar] [CrossRef] [PubMed]
- Bockhorn, J.; Dalton, R.; Nwachukwu, C.; Huang, S.; Prat, A.; Yee, K.; Chang, Y.F.; Huo, D.; Wen, Y.; Swanson, K.E.; et al. MicroRNA-30c inhibits human breast tumour chemotherapy resistance by regulating TWF1 and IL-11. Nat. Commun. 2013, 4, 1393. [Google Scholar] [CrossRef] [PubMed]
- Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef] [PubMed]
- Yousefi, H.; Maheronnaghsh, M.; Molaei, F.; Mashouri, L.; Reza Aref, A.; Momeny, M.; Alahari, S.K. Long noncoding RNAs and exosomal lncRNAs: Classification, and mechanisms in breast cancer metastasis and drug resistance. Oncogene 2020, 39, 953–974. [Google Scholar] [CrossRef] [PubMed]
- Chiu, H.S.; Somvanshi, S.; Patel, E.; Chen, T.W.; Singh, V.P.; Zorman, B.; Patil, S.L.; Pan, Y.; Chatterjee, S.S.; Cancer Genome Atlas Research, N.; et al. Pan-Cancer Analysis of lncRNA Regulation Supports Their Targeting of Cancer Genes in Each Tumor Context. Cell Rep. 2018, 23, 297.e212–312.e212. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Z.; Liu, H.; Chen, S.R. Mechanisms of Long Non-Coding RNAs in Cancers and Their Dynamic Regulations. Cancers 2020, 12, 1245. [Google Scholar] [CrossRef]
- Begolli, R.; Sideris, N.; Giakountis, A. LncRNAs as Chromatin Regulators in Cancer: From Molecular Function to Clinical Potential. Cancers 2019, 11, 1524. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C.; Kanhere, A. The Missing Link Between Cancer-Associated Variants and LncRNAs. Trends Genet. 2021, 37, 410–413. [Google Scholar] [CrossRef]
- Weinhold, N.; Jacobsen, A.; Schultz, N.; Sander, C.; Lee, W. Genome-wide analysis of noncoding regulatory mutations in cancer. Nat. Genet. 2014, 46, 1160–1165. [Google Scholar] [CrossRef]
- Johnsson, P.; Lipovich, L.; Grander, D.; Morris, K.V. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim. Biophys. Acta 2014, 1840, 1063–1071. [Google Scholar] [CrossRef]
- Kapusta, A.; Feschotte, C. Volatile evolution of long noncoding RNA repertoires: Mechanisms and biological implications. Trends Genet. 2014, 30, 439–452. [Google Scholar] [CrossRef]
- Jonas, K.; Calin, G.A.; Pichler, M. RNA-Binding Proteins as Important Regulators of Long Non-Coding RNAs in Cancer. Int. J. Mol. Sci. 2020, 21, 2969. [Google Scholar] [CrossRef]
- Pisignano, G.; Ladomery, M. Post-Transcriptional Regulation through Long Non-Coding RNAs (lncRNAs). Noncoding RNA 2021, 7, 29. [Google Scholar] [CrossRef]
- Liz, J.; Portela, A.; Soler, M.; Gomez, A.; Ling, H.; Michlewski, G.; Calin, G.A.; Guil, S.; Esteller, M. Regulation of pri-miRNA processing by a long noncoding RNA transcribed from an ultraconserved region. Mol. Cell 2014, 55, 138–147. [Google Scholar] [CrossRef]
- Dhir, A.; Dhir, S.; Proudfoot, N.J.; Jopling, C.L. Microprocessor mediates transcriptional termination of long noncoding RNA transcripts hosting microRNAs. Nat. Struct. Mol. Biol. 2015, 22, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Gil, N.; Ulitsky, I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat. Rev. Genet. 2020, 21, 102–117. [Google Scholar] [CrossRef] [PubMed]
- Zampetaki, A.; Albrecht, A.; Steinhofel, K. Long Non-coding RNA Structure and Function: Is There a Link? Front. Physiol. 2018, 9, 1201. [Google Scholar] [CrossRef] [PubMed]
- Aznaourova, M.; Schmerer, N.; Schmeck, B.; Schulte, L.N. Disease-Causing Mutations and Rearrangements in Long Non-coding RNA Gene Loci. Front. Genet. 2020, 11, 527484. [Google Scholar] [CrossRef]
- Gao, P.; Wei, G.H. Genomic Insight into the Role of lncRNA in Cancer Susceptibility. Int. J. Mol. Sci. 2017, 18, 1239. [Google Scholar] [CrossRef]
- Tang, X.; Feng, D.; Li, M.; Zhou, J.; Li, X.; Zhao, D.; Hao, B.; Li, D.; Ding, K. Transcriptomic Analysis of mRNA-lncRNA-miRNA Interactions in Hepatocellular Carcinoma. Sci. Rep. 2019, 9, 16096. [Google Scholar] [CrossRef]
- Xia, T.; Liao, Q.; Jiang, X.; Shao, Y.; Xiao, B.; Xi, Y.; Guo, J. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci. Rep. 2014, 4, 6088. [Google Scholar] [CrossRef]
- Zhou, X.; Liu, S.; Cai, G.; Kong, L.; Zhang, T.; Ren, Y.; Wu, Y.; Mei, M.; Zhang, L.; Wang, X. Long Non Coding RNA MALAT1 Promotes Tumor Growth and Metastasis by inducing Epithelial-Mesenchymal Transition in Oral Squamous Cell Carcinoma. Sci. Rep. 2015, 5, 15972. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Li, X.; Qiao, L.; Liu, W.; Xu, C.; Wang, X. MALAT1 regulates miR-34a expression in melanoma cells. Cell Death Dis. 2019, 10, 389. [Google Scholar] [CrossRef]
- Han, Y.; Wu, Z.; Wu, T.; Huang, Y.; Cheng, Z.; Li, X.; Sun, T.; Xie, X.; Zhou, Y.; Du, Z. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by downregulation of MMP2 and inactivation of ERK/MAPK signaling. Cell Death Dis. 2016, 7, e2123. [Google Scholar] [CrossRef]
- Kim, J.; Piao, H.L.; Kim, B.J.; Yao, F.; Han, Z.; Wang, Y.; Xiao, Z.; Siverly, A.N.; Lawhon, S.E.; Ton, B.N.; et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat. Genet. 2018, 50, 1705–1715. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Guo, Z.; Zhao, Y.; Jin, Y.; An, L.; Wu, B.; Liu, Z.; Chen, X.; Chen, X.; Zhou, H.; et al. Genetic polymorphisms of lncRNA-p53 regulatory network genes are associated with concurrent chemoradiotherapy toxicities and efficacy in nasopharyngeal carcinoma patients. Sci. Rep. 2017, 7, 8320. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Liu, C.; Li, H.; Zhang, L.; Luo, G.; Liang, S.; Lu, M. Research progress on the interactions between long non-coding RNAs and microRNAs in human cancer. Oncol. Lett. 2020, 19, 595–605. [Google Scholar] [CrossRef]
- Jin, N.; Bi, A.; Lan, X.; Xu, J.; Wang, X.; Liu, Y.; Wang, T.; Tang, S.; Zeng, H.; Chen, Z.; et al. Identification of metabolic vulnerabilities of receptor tyrosine kinases-driven cancer. Nat. Commun. 2019, 10, 2701. [Google Scholar] [CrossRef]
- Lo, R.S. Receptor tyrosine kinases in cancer escape from BRAF inhibitors. Cell Res. 2012, 22, 945–947. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Du, Z.; Lovly, C.M. Mechanisms of receptor tyrosine kinase activation in cancer. Mol. Cancer 2018, 17, 58. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Brown, B.P.; Kim, S.; Ferguson, D.; Pavlick, D.C.; Jayakumaran, G.; Benayed, R.; Gallant, J.N.; Zhang, Y.K.; Yan, Y.; et al. Structure-function analysis of oncogenic EGFR Kinase Domain Duplication reveals insights into activation and a potential approach for therapeutic targeting. Nat. Commun. 2021, 12, 1382. [Google Scholar] [CrossRef] [PubMed]
- Banys-Paluchowski, M.; Witzel, I.; Riethdorf, S.; Rack, B.; Janni, W.; Fasching, P.A.; Solomayer, E.F.; Aktas, B.; Kasimir-Bauer, S.; Pantel, K.; et al. Evaluation of serum epidermal growth factor receptor (EGFR) in correlation to circulating tumor cells in patients with metastatic breast cancer. Sci. Rep. 2017, 7, 17307. [Google Scholar] [CrossRef]
- Nakagawa, T.; Takeuchi, S.; Yamada, T.; Nanjo, S.; Ishikawa, D.; Sano, T.; Kita, K.; Nakamura, T.; Matsumoto, K.; Suda, K.; et al. Combined therapy with mutant-selective EGFR inhibitor and Met kinase inhibitor for overcoming erlotinib resistance in EGFR-mutant lung cancer. Mol. Cancer Ther. 2012, 11, 2149–2157. [Google Scholar] [CrossRef] [PubMed]
- Fernandes Neto, J.M.; Nadal, E.; Bosdriesz, E.; Ooft, S.N.; Farre, L.; McLean, C.; Klarenbeek, S.; Jurgens, A.; Hagen, H.; Wang, L.; et al. Multiple low dose therapy as an effective strategy to treat EGFR inhibitor-resistant NSCLC tumours. Nat. Commun. 2020, 11, 3157. [Google Scholar] [CrossRef]
- Gyorffy, B.; Pongor, L.; Bottai, G.; Li, X.; Budczies, J.; Szabo, A.; Hatzis, C.; Pusztai, L.; Santarpia, L. An integrative bioinformatics approach reveals coding and non-coding gene variants associated with gene expression profiles and outcome in breast cancer molecular subtypes. Br. J. Cancer 2018, 118, 1107–1114. [Google Scholar] [CrossRef] [PubMed]
- Soltis, A.R.; Dalgard, C.L.; Pollard, H.B.; Wilkerson, M.D. MutEnricher: A flexible toolset for somatic mutation enrichment analysis of tumor whole genomes. BMC Bioinform. 2020, 21, 338. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.R.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; et al. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018, 46, D1062–D1067. [Google Scholar] [CrossRef]
- Firth, H.V.; Richards, S.M.; Bevan, A.P.; Clayton, S.; Corpas, M.; Rajan, D.; Van Vooren, S.; Moreau, Y.; Pettett, R.M.; Carter, N.P. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 2009, 84, 524–533. [Google Scholar] [CrossRef]
- Fulco, C.P.; Nasser, J.; Jones, T.R.; Munson, G.; Bergman, D.T.; Subramanian, V.; Grossman, S.R.; Anyoha, R.; Doughty, B.R.; Patwardhan, T.A.; et al. Activity-by-contact model of enhancer-promoter regulation from thousands of CRISPR perturbations. Nat. Genet. 2019, 51, 1664–1669. [Google Scholar] [CrossRef]
- Sheffield, N.C.; Furey, T.S. Identifying and characterizing regulatory sequences in the human genome with chromatin accessibility assays. Genes 2012, 3, 651–670. [Google Scholar] [CrossRef] [PubMed]
- Fadason, T.; Schierding, W.; Lumley, T.; O’Sullivan, J.M. Chromatin interactions and expression quantitative trait loci reveal genetic drivers of multimorbidities. Nat. Commun. 2018, 9, 5198. [Google Scholar] [CrossRef] [PubMed]
- Keele, G.R.; Quach, B.C.; Israel, J.W.; Chappell, G.A.; Lewis, L.; Safi, A.; Simon, J.M.; Cotney, P.; Crawford, G.E.; Valdar, W.; et al. Integrative QTL analysis of gene expression and chromatin accessibility identifies multi-tissue patterns of genetic regulation. PLoS Genet. 2020, 16, e1008537. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Rendon, A.; Wernisch, L. Transcription factor and chromatin features predict genes associated with eQTLs. Nucleic Acids Res. 2013, 41, 1450–1463. [Google Scholar] [CrossRef]
- Kim, K.; Jang, I.; Kim, M.; Choi, J.; Kim, M.S.; Lee, B.; Jung, I. 3DIV update for 2021: A comprehensive resource of 3D genome and 3D cancer genome. Nucleic Acids Res. 2021, 49, D38–D46. [Google Scholar] [CrossRef] [PubMed]
- Wu, C. The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature 1980, 286, 854–860. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Chen, D.; Chen, Y. Advances of DNase-seq for mapping active gene regulatory elements across the genome in animals. Gene 2018, 667, 83–94. [Google Scholar] [CrossRef]
- Kumar, V.; Muratani, M.; Rayan, N.A.; Kraus, P.; Lufkin, T.; Ng, H.H.; Prabhakar, S. Uniform, optimal signal processing of mapped deep-sequencing data. Nat. Biotechnol. 2013, 31, 615–622. [Google Scholar] [CrossRef]
- Lu, F.; Liu, Y.; Inoue, A.; Suzuki, T.; Zhao, K.; Zhang, Y. Establishing Chromatin Regulatory Landscape during Mouse Preimplantation Development. Cell 2016, 165, 1375–1388. [Google Scholar] [CrossRef]
- Yan, H.; Tian, S.; Slager, S.L.; Sun, Z.; Ordog, T. Genome-Wide Epigenetic Studies in Human Disease: A Primer on -Omic Technologies. Am. J. Epidemiol. 2016, 183, 96–109. [Google Scholar] [CrossRef]
- Zentner, G.E.; Henikoff, S. Surveying the epigenomic landscape, one base at a time. Genome Biol. 2012, 13, 250. [Google Scholar] [CrossRef]
- Guo, F.; Li, L.; Li, J.; Wu, X.; Hu, B.; Zhu, P.; Wen, L.; Tang, F. Single-cell multi-omics sequencing of mouse early embryos and embryonic stem cells. Cell Res. 2017, 27, 967–988. [Google Scholar] [CrossRef] [PubMed]
- D’antonio, M.; Weghorn, D.; D’antonio-Chronowska, A.; Coulet, F.; Olson, K.M.; DeBoever, C.; Drees, F.; Arias, A.; Alakus, H.; Richardson, A.L.; et al. Identifying DNase I hypersensitive sites as driver distal regulatory elements in breast cancer. Nat. Commun. 2017, 8, 436. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Tang, Q.; Wan, M.; Cui, K.; Zhang, Y.; Ren, G.; Ni, B.; Sklar, J.; Przytycka, T.M.; Childs, R.; et al. Genome-wide detection of DNase I hypersensitive sites in single cells and FFPE tissue samples. Nature 2015, 528, 142–146. [Google Scholar] [CrossRef]
- Simon, J.M.; Giresi, P.G.; Davis, I.J.; Lieb, J.D. Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat. Protoc. 2012, 7, 256–267. [Google Scholar] [CrossRef]
- Schones, D.E.; Cui, K.; Cuddapah, S.; Roh, T.Y.; Barski, A.; Wang, Z.; Wei, G.; Zhao, K. Dynamic regulation of nucleosome positioning in the human genome. Cell 2008, 132, 887–898. [Google Scholar] [CrossRef] [PubMed]
- Klein, D.C.; Hainer, S.J. Genomic methods in profiling DNA accessibility and factor localization. Chromosome Res. 2020, 28, 69–85. [Google Scholar] [CrossRef] [PubMed]
- Buenrostro, J.D.; Giresi, P.G.; Zaba, L.C.; Chang, H.Y.; Greenleaf, W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 2013, 10, 1213–1218. [Google Scholar] [CrossRef]
- Cui, K.; Zhao, K. Genome-wide approaches to determining nucleosome occupancy in metazoans using MNase-Seq. Methods Mol. Biol. 2012, 833, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Shashikant, T.; Ettensohn, C.A. Genome-wide analysis of chromatin accessibility using ATAC-seq. Methods Cell Biol. 2019, 151, 219–235. [Google Scholar] [CrossRef]
- Bryois, J.; Garrett, M.E.; Song, L.; Safi, A.; Giusti-Rodriguez, P.; Johnson, G.D.; Shieh, A.W.; Buil, A.; Fullard, J.F.; Roussos, P.; et al. Evaluation of chromatin accessibility in prefrontal cortex of individuals with schizophrenia. Nat. Commun. 2018, 9, 3121. [Google Scholar] [CrossRef]
- Kheradpour, P.; Ernst, J.; Melnikov, A.; Rogov, P.; Wang, L.; Zhang, X.; Alston, J.; Mikkelsen, T.S.; Kellis, M. Systematic dissection of regulatory motifs in 2000 predicted human enhancers using a massively parallel reporter assay. Genome Res. 2013, 23, 800–811. [Google Scholar] [CrossRef]
- Kwasnieski, J.C.; Mogno, I.; Myers, C.A.; Corbo, J.C.; Cohen, B.A. Complex effects of nucleotide variants in a mammalian cis-regulatory element. Proc. Natl. Acad. Sci. USA 2012, 109, 19498–19503. [Google Scholar] [CrossRef] [PubMed]
- Birnbaum, R.Y.; Patwardhan, R.P.; Kim, M.J.; Findlay, G.M.; Martin, B.; Zhao, J.; Bell, R.J.; Smith, R.P.; Ku, A.A.; Shendure, J.; et al. Systematic dissection of coding exons at single nucleotide resolution supports an additional role in cell-specific transcriptional regulation. PLoS Genet. 2014, 10, e1004592. [Google Scholar] [CrossRef] [PubMed]
- Melnikov, A.; Murugan, A.; Zhang, X.; Tesileanu, T.; Wang, L.; Rogov, P.; Feizi, S.; Gnirke, A.; Callan, C.G., Jr.; Kinney, J.B.; et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat. Biotechnol. 2012, 30, 271–277. [Google Scholar] [CrossRef]
- Patwardhan, R.P.; Hiatt, J.B.; Witten, D.M.; Kim, M.J.; Smith, R.P.; May, D.; Lee, C.; Andrie, J.M.; Lee, S.I.; Cooper, G.M.; et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nat. Biotechnol. 2012, 30, 265–270. [Google Scholar] [CrossRef]
- White, M.A.; Myers, C.A.; Corbo, J.C.; Cohen, B.A. Massively parallel in vivo enhancer assay reveals that highly local features determine the cis-regulatory function of ChIP-seq peaks. Proc. Natl. Acad. Sci. USA 2013, 110, 11952–11957. [Google Scholar] [CrossRef]
- Arnold, C.D.; Gerlach, D.; Stelzer, C.; Boryn, L.M.; Rath, M.; Stark, A. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science 2013, 339, 1074–1077. [Google Scholar] [CrossRef]
- Muerdter, F.; Boryn, L.M.; Arnold, C.D. STARR-seq - principles and applications. Genomics 2015, 106, 145–150. [Google Scholar] [CrossRef]
- Klein, J.C.; Keith, A.; Rice, S.J.; Shepherd, C.; Agarwal, V.; Loughlin, J.; Shendure, J. Functional testing of thousands of osteoarthritis-associated variants for regulatory activity. Nat. Commun. 2019, 10, 2434. [Google Scholar] [CrossRef]
- Zhang, P.; Xia, J.H.; Zhu, J.; Gao, P.; Tian, Y.J.; Du, M.; Guo, Y.C.; Suleman, S.; Zhang, Q.; Kohli, M.; et al. High-throughput screening of prostate cancer risk loci by single nucleotide polymorphisms sequencing. Nat. Commun. 2018, 9, 2022. [Google Scholar] [CrossRef] [PubMed]
- Vanhille, L.; Griffon, A.; Maqbool, M.A.; Zacarias-Cabeza, J.; Dao, L.T.; Fernandez, N.; Ballester, B.; Andrau, J.C.; Spicuglia, S. High-throughput and quantitative assessment of enhancer activity in mammals by CapStarr-seq. Nat. Commun. 2015, 6, 6905. [Google Scholar] [CrossRef]
- Arnold, C.D.; Gerlach, D.; Spies, D.; Matts, J.A.; Sytnikova, Y.A.; Pagani, M.; Lau, N.C.; Stark, A. Quantitative genome-wide enhancer activity maps for five Drosophila species show functional enhancer conservation and turnover during cis-regulatory evolution. Nat. Genet. 2014, 46, 685–692. [Google Scholar] [CrossRef] [PubMed]
- Johnson, G.D.; Barrera, A.; McDowell, I.C.; D’Ippolito, A.M.; Majoros, W.H.; Vockley, C.M.; Wang, X.; Allen, A.S.; Reddy, T.E. Human genome-wide measurement of drug-responsive regulatory activity. Nat. Commun. 2018, 9, 5317. [Google Scholar] [CrossRef]
- Gong, H.; Yang, Y.; Zhang, S.; Li, M.; Zhang, X. Application of Hi-C and other omics data analysis in human cancer and cell differentiation research. Comput. Struct. Biotechnol. J. 2021, 19, 2070–2083. [Google Scholar] [CrossRef]
- Fullwood, M.J.; Ruan, Y. ChIP-based methods for the identification of long-range chromatin interactions. J. Cell Biochem. 2009, 107, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhao, K. The Toolbox for Untangling Chromosome Architecture in Immune Cells. Front. Immunol. 2021, 12, 670884. [Google Scholar] [CrossRef]
- Li, G.; Cai, L.; Chang, H.; Hong, P.; Zhou, Q.; Kulakova, E.V.; Kolchanov, N.A.; Ruan, Y. Chromatin Interaction Analysis with Paired-End Tag (ChIA-PET) sequencing technology and application. BMC Genom. 2014, 15 (Suppl. S12), S11. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Luo, O.J.; Wang, P.; Zheng, M.; Wang, D.; Piecuch, E.; Zhu, J.J.; Tian, S.Z.; Tang, Z.; Li, G.; et al. Long-read ChIA-PET for base-pair-resolution mapping of haplotype-specific chromatin interactions. Nat. Protoc. 2017, 12, 899–915. [Google Scholar] [CrossRef]
- Al Bkhetan, Z.; Plewczynski, D. Three-dimensional Epigenome Statistical Model: Genome-wide Chromatin Looping Prediction. Sci. Rep. 2018, 8, 5217. [Google Scholar] [CrossRef] [PubMed]
- Fullwood, M.J.; Liu, M.H.; Pan, Y.F.; Liu, J.; Xu, H.; Mohamed, Y.B.; Orlov, Y.L.; Velkov, S.; Ho, A.; Mei, P.H.; et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature 2009, 462, 58–64. [Google Scholar] [CrossRef]
- Xi, W.; Beer, M.A. Loop competition and extrusion model predicts CTCF interaction specificity. Nat. Commun. 2021, 12, 1046. [Google Scholar] [CrossRef] [PubMed]
- Couch, F.J.; Kuchenbaecker, K.B.; Michailidou, K.; Mendoza-Fandino, G.A.; Nord, S.; Lilyquist, J.; Olswold, C.; Hallberg, E.; Agata, S.; Ahsan, H.; et al. Identification of four novel susceptibility loci for oestrogen receptor negative breast cancer. Nat. Commun. 2016, 7, 11375. [Google Scholar] [CrossRef] [PubMed]
- Milne, R.L.; Kuchenbaecker, K.B.; Michailidou, K.; Beesley, J.; Kar, S.; Lindstrom, S.; Hui, S.; Lemacon, A.; Soucy, P.; Dennis, J.; et al. Identification of ten variants associated with risk of estrogen-receptor-negative breast cancer. Nat. Genet. 2017, 49, 1767–1778. [Google Scholar] [CrossRef]
- Cai, L.; Chang, H.; Fang, Y.; Li, G. A Comprehensive Characterization of the Function of LincRNAs in Transcriptional Regulation Through Long-Range Chromatin Interactions. Sci. Rep. 2016, 6, 36572. [Google Scholar] [CrossRef] [PubMed]
- Mumbach, M.R.; Rubin, A.J.; Flynn, R.A.; Dai, C.; Khavari, P.A.; Greenleaf, W.J.; Chang, H.Y. HiChIP: Efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods 2016, 13, 919–922. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Chandra, V.; Vijayanand, P.; Ay, F. Identification of significant chromatin contacts from HiChIP data by FitHiChIP. Nat. Commun. 2019, 10, 4221. [Google Scholar] [CrossRef]
- Ray, J.P.; de Boer, C.G.; Fulco, C.P.; Lareau, C.A.; Kanai, M.; Ulirsch, J.C.; Tewhey, R.; Ludwig, L.S.; Reilly, S.K.; Bergman, D.T.; et al. Prioritizing disease and trait causal variants at the TNFAIP3 locus using functional and genomic features. Nat. Commun. 2020, 11, 1237. [Google Scholar] [CrossRef]
- O’Mara, T.A.; Spurdle, A.B.; Glubb, D.M.; Endometrial Cancer Association, C. Analysis of Promoter-Associated Chromatin Interactions Reveals Biologically Relevant Candidate Target Genes at Endometrial Cancer Risk Loci. Cancers 2019, 11, 1440. [Google Scholar] [CrossRef]
- Fang, R.; Yu, M.; Li, G.; Chee, S.; Liu, T.; Schmitt, A.D.; Ren, B. Mapping of long-range chromatin interactions by proximity ligation-assisted ChIP-seq. Cell Res. 2016, 26, 1345–1348. [Google Scholar] [CrossRef]
- Corces, M.R.; Shcherbina, A.; Kundu, S.; Gloudemans, M.J.; Fresard, L.; Granja, J.M.; Louie, B.H.; Eulalio, T.; Shams, S.; Bagdatli, S.T.; et al. Single-cell epigenomic analyses implicate candidate causal variants at inherited risk loci for Alzheimer’s and Parkinson’s diseases. Nat. Genet. 2020, 52, 1158–1168. [Google Scholar] [CrossRef]
- Deng, X.; Kong, F.; Li, S.; Jiang, H.; Dong, L.; Xu, X.; Zhang, X.; Yuan, H.; Xu, Y.; Chu, Y.; et al. A KLF4/PiHL/EZH2/HMGA2 regulatory axis and its function in promoting oxaliplatin-resistance of colorectal cancer. Cell Death Dis. 2021, 12, 485. [Google Scholar] [CrossRef]
- Feng, Y.C.; Liu, X.Y.; Teng, L.; Ji, Q.; Wu, Y.; Li, J.M.; Gao, W.; Zhang, Y.Y.; La, T.; Tabatabaee, H.; et al. c-Myc inactivation of p53 through the pan-cancer lncRNA MILIP drives cancer pathogenesis. Nat. Commun. 2020, 11, 4980. [Google Scholar] [CrossRef]
- Xu, L.; Huan, L.; Guo, T.; Wu, Y.; Liu, Y.; Wang, Q.; Huang, S.; Xu, Y.; Liang, L.; He, X. LncRNA SNHG11 facilitates tumor metastasis by interacting with and stabilizing HIF-1alpha. Oncogene 2020, 39, 7005–7018. [Google Scholar] [CrossRef] [PubMed]
- Mondal, T.; Subhash, S.; Vaid, R.; Enroth, S.; Uday, S.; Reinius, B.; Mitra, S.; Mohammed, A.; James, A.R.; Hoberg, E.; et al. MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat. Commun. 2015, 6, 7743. [Google Scholar] [CrossRef] [PubMed]
- Feretzaki, M.; Pospisilova, M.; Valador Fernandes, R.; Lunardi, T.; Krejci, L.; Lingner, J. RAD51-dependent recruitment of TERRA lncRNA to telomeres through R-loops. Nature 2020, 587, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Laffleur, B.; Lim, J.; Zhang, W.; Chen, Y.; Pefanis, E.; Bizarro, J.; Batista, C.R.; Wu, L.; Economides, A.N.; Wang, J.; et al. Noncoding RNA processing by DIS3 regulates chromosomal architecture and somatic hypermutation in B cells. Nat. Genet. 2021, 53, 230–242. [Google Scholar] [CrossRef]
- Chu, C.; Qu, K.; Zhong, F.L.; Artandi, S.E.; Chang, H.Y. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 2011, 44, 667–678. [Google Scholar] [CrossRef]
- Nguyen, T.C.; Zaleta-Rivera, K.; Huang, X.; Dai, X.; Zhong, S. RNA, Action through Interactions. Trends Genet. 2018, 34, 867–882. [Google Scholar] [CrossRef]
- Kato, M.; Carninci, P. Genome-Wide Technologies to Study RNA-Chromatin Interactions. Noncoding RNA 2020, 6, 20. [Google Scholar] [CrossRef]
- Machyna, M.; Simon, M.D. Catching RNAs on chromatin using hybridization capture methods. Brief. Funct. Genom. 2018, 17, 96–103. [Google Scholar] [CrossRef]
- Sridhar, B.; Rivas-Astroza, M.; Nguyen, T.C.; Chen, W.; Yan, Z.; Cao, X.; Hebert, L.; Zhong, S. Systematic Mapping of RNA-Chromatin Interactions In Vivo. Curr. Biol. 2017, 27, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Yan, Z.; Nguyen, T.C.; Bouman Chen, Z.; Chien, S.; Zhong, S. Mapping RNA-chromatin interactions by sequencing with iMARGI. Nat. Protoc. 2019, 14, 3243–3272. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhou, B.; Chen, L.; Gou, L.T.; Li, H.; Fu, X.D. GRID-seq reveals the global RNA-chromatin interactome. Nat. Biotechnol. 2017, 35, 940–950. [Google Scholar] [CrossRef]
- Zhou, B.; Li, X.; Luo, D.; Lim, D.H.; Zhou, Y.; Fu, X.D. GRID-seq for comprehensive analysis of global RNA-chromatin interactions. Nat. Protoc. 2019, 14, 2036–2068. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.C.; Jukam, D.; Teran, N.A.; Risca, V.I.; Smith, O.K.; Johnson, W.L.; Skotheim, J.M.; Greenleaf, W.J.; Straight, A.F. Chromatin-associated RNA sequencing (ChAR-seq) maps genome-wide RNA-to-DNA contacts. eLife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Engreitz, J.; Lander, E.S.; Guttman, M. RNA antisense purification (RAP) for mapping RNA interactions with chromatin. Methods Mol. Biol. 2015, 1262, 183–197. [Google Scholar] [CrossRef] [PubMed]
- D’Antonio, M.; D’Onorio De Meo, P.; Pallocca, M.; Picardi, E.; D’Erchia, A.M.; Calogero, R.A.; Castrignano, T.; Pesole, G. RAP: RNA-Seq Analysis Pipeline, a new cloud-based NGS web application. BMC Genom. 2015, 16, S3. [Google Scholar] [CrossRef]
- Grillone, K.; Riillo, C.; Scionti, F.; Rocca, R.; Tradigo, G.; Guzzi, P.H.; Alcaro, S.; Di Martino, M.T.; Tagliaferri, P.; Tassone, P. Non-coding RNAs in cancer: Platforms and strategies for investigating the genomic “dark matter”. J. Exp. Clin. Cancer Res. 2020, 39, 117. [Google Scholar] [CrossRef]
- Weidmann, C.A.; Mustoe, A.M.; Jariwala, P.B.; Calabrese, J.M.; Weeks, K.M. Analysis of RNA-protein networks with RNP-MaP defines functional hubs on RNA. Nat. Biotechnol. 2021, 39, 347–356. [Google Scholar] [CrossRef]
- Yi, W.; Li, J.; Zhu, X.; Wang, X.; Fan, L.; Sun, W.; Liao, L.; Zhang, J.; Li, X.; Ye, J.; et al. CRISPR-assisted detection of RNA-protein interactions in living cells. Nat. Methods 2020, 17, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Cottrell, K.A.; Chaudhari, H.G.; Cohen, B.A.; Djuranovic, S. PTRE-seq reveals mechanism and interactions of RNA binding proteins and miRNAs. Nat. Commun. 2018, 9, 301. [Google Scholar] [CrossRef]
- Wan, Y.; Qu, K.; Ouyang, Z.; Chang, H.Y. Genome-wide mapping of RNA structure using nuclease digestion and high-throughput sequencing. Nat. Protoc. 2013, 8, 849–869. [Google Scholar] [CrossRef]
- Kertesz, M.; Wan, Y.; Mazor, E.; Rinn, J.L.; Nutter, R.C.; Chang, H.Y.; Segal, E. Genome-wide measurement of RNA secondary structure in yeast. Nature 2010, 467, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Solomon, O.; Di Segni, A.; Cesarkas, K.; Porath, H.T.; Marcu-Malina, V.; Mizrahi, O.; Stern-Ginossar, N.; Kol, N.; Farage-Barhom, S.; Glick-Saar, E.; et al. RNA editing by ADAR1 leads to context-dependent transcriptome-wide changes in RNA secondary structure. Nat. Commun. 2017, 8, 1440. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A.; Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef] [PubMed]
- Sander, J.D.; Joung, J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014, 32, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Bortesi, L.; Fischer, R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 2015, 33, 41–52. [Google Scholar] [CrossRef]
- Wang, H.; La Russa, M.; Qi, L.S. CRISPR/Cas9 in Genome Editing and Beyond. Annu. Rev. Biochem. 2016, 85, 227–264. [Google Scholar] [CrossRef]
- Larson, M.H.; Gilbert, L.A.; Wang, X.; Lim, W.A.; Weissman, J.S.; Qi, L.S. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 2013, 8, 2180–2196. [Google Scholar] [CrossRef]
- Hou, G.; Harley, I.T.W.; Lu, X.; Zhou, T.; Xu, N.; Yao, C.; Qin, Y.; Ouyang, Y.; Ma, J.; Zhu, X.; et al. SLE non-coding genetic risk variant determines the epigenetic dysfunction of an immune cell specific enhancer that controls disease-critical microRNA expression. Nat. Commun. 2021, 12, 135. [Google Scholar] [CrossRef]
- Nakamura, M.; Gao, Y.; Dominguez, A.A.; Qi, L.S. CRISPR technologies for precise epigenome editing. Nat. Cell Biol. 2021, 23, 11–22. [Google Scholar] [CrossRef]
- Palin, K.; Pitkanen, E.; Turunen, M.; Sahu, B.; Pihlajamaa, P.; Kivioja, T.; Kaasinen, E.; Valimaki, N.; Hanninen, U.A.; Cajuso, T.; et al. Contribution of allelic imbalance to colorectal cancer. Nat. Commun. 2018, 9, 3664. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.; Soares, F.; Xia, J.H.; Yang, Y.; Li, J.; Guo, H.; Su, P.; Tian, Y.; Lee, H.J.; Wang, M.; et al. CRISPRi screens reveal a DNA methylation-mediated 3D genome dependent causal mechanism in prostate cancer. Nat. Commun. 2021, 12, 1781. [Google Scholar] [CrossRef]
- Nichols, C.A.; Gibson, W.J.; Brown, M.S.; Kosmicki, J.A.; Busanovich, J.P.; Wei, H.; Urbanski, L.M.; Curimjee, N.; Berger, A.C.; Gao, G.F.; et al. Loss of heterozygosity of essential genes represents a widespread class of potential cancer vulnerabilities. Nat. Commun. 2020, 11, 2517. [Google Scholar] [CrossRef] [PubMed]
- Pickar-Oliver, A.; Gersbach, C.A. The next generation of CRISPR-Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 2019, 20, 490–507. [Google Scholar] [CrossRef]
- Smargon, A.A.; Shi, Y.J.; Yeo, G.W. RNA-targeting CRISPR systems from metagenomic discovery to transcriptomic engineering. Nat. Cell Biol. 2020, 22, 143–150. [Google Scholar] [CrossRef]
- Guo, Y.; Perez, A.A.; Hazelett, D.J.; Coetzee, G.A.; Rhie, S.K.; Farnham, P.J. CRISPR-mediated deletion of prostate cancer risk-associated CTCF loop anchors identifies repressive chromatin loops. Genome Biol. 2018, 19, 160. [Google Scholar] [CrossRef]
- Hasin, Y.; Seldin, M.; Lusis, A. Multi-omics approaches to disease. Genome Biol. 2017, 18, 83. [Google Scholar] [CrossRef] [PubMed]
- Sims, D.; Sudbery, I.; Ilott, N.E.; Heger, A.; Ponting, C.P. Sequencing depth and coverage: Key considerations in genomic analyses. Nat. Rev. Genet. 2014, 15, 121–132. [Google Scholar] [CrossRef]
- Wagner, A.H.; Walsh, B.; Mayfield, G.; Tamborero, D.; Sonkin, D.; Krysiak, K.; Deu-Pons, J.; Duren, R.P.; Gao, J.; McMurry, J.; et al. A harmonized meta-knowledgebase of clinical interpretations of somatic genomic variants in cancer. Nat. Genet. 2020, 52, 448–457. [Google Scholar] [CrossRef]
- Piraino, S.W.; Furney, S.J. Beyond the exome: The role of non-coding somatic mutations in cancer. Ann. Oncol. 2016, 27, 240–248. [Google Scholar] [CrossRef]
- Liu, Y.; Li, C.; Shen, S.; Chen, X.; Szlachta, K.; Edmonson, M.N.; Shao, Y.; Ma, X.; Hyle, J.; Wright, S.; et al. Discovery of regulatory noncoding variants in individual cancer genomes by using cis-X. Nat. Genet. 2020, 52, 811–818. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Drubay, D.; Michiels, S.; Gautheret, D. Mining the coding and non-coding genome for cancer drivers. Cancer Lett. 2015, 369, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Wu, L.X.; Tan, L.; Shang, F.F.; Zhou, H.H. Significance of Single-Nucleotide Variants in Long Intergenic Non-protein Coding RNAs. Front. Cell Dev. Biol. 2020, 8, 347. [Google Scholar] [CrossRef]
- Kikutake, C.; Yoshihara, M.; Suyama, M. Pan-cancer analysis of non-coding recurrent mutations and their possible involvement in cancer pathogenesis. NAR Cancer 2021, 3. [Google Scholar] [CrossRef]
- Pamula-Pilat, J.; Tecza, K.; Kalinowska-Herok, M.; Grzybowska, E. Genetic 3’UTR variations and clinical factors significantly contribute to survival prediction and clinical response in breast cancer patients. Sci. Rep. 2020, 10, 5736. [Google Scholar] [CrossRef] [PubMed]
- Ye, D.; Hu, Y.; Jing, F.; Li, Y.; Gu, S.; Jiang, X.; Mao, Y.; Li, Q.; Jin, M.; Chen, K. A novel SNP in promoter region of RP11-3N2.1 is associated with reduced risk of colorectal cancer. J. Hum. Genet. 2018, 63, 47–54. [Google Scholar] [CrossRef]
- Wu, E.R.; Chou, Y.E.; Liu, Y.F.; Hsueh, K.C.; Lee, H.L.; Yang, S.F.; Su, S.C. Association of lncRNA H19 Gene Polymorphisms with the Occurrence of Hepatocellular Carcinoma. Genes 2019, 10, 506. [Google Scholar] [CrossRef]
- Wang, B.G.; Jiang, L.Y.; Xu, Q. Comprehensive assessment for miRNA polymorphisms in hepatocellular cancer risk: A systematic review and meta-analysis. BioSci. Rep. 2018, 38. [Google Scholar] [CrossRef]
- Endo, S.; Oguri, H.; Segawa, J.; Kawai, M.; Hu, D.; Xia, S.; Okada, T.; Irie, K.; Fujii, S.; Gouda, H.; et al. Development of Novel AKR1C3 Inhibitors as New Potential Treatment for Castration-Resistant Prostate Cancer. J. Med. Chem. 2020, 63, 10396–10411. [Google Scholar] [CrossRef] [PubMed]
- Kaur, H.; Mao, S.; Li, Q.; Sameni, M.; Krawetz, S.A.; Sloane, B.F.; Mattingly, R.R. RNA-Seq of human breast ductal carcinoma in situ models reveals aldehyde dehydrogenase isoform 5A1 as a novel potential target. PLoS ONE 2012, 7, e50249. [Google Scholar] [CrossRef]
- Bray, J.; Sludden, J.; Griffin, M.J.; Cole, M.; Verrill, M.; Jamieson, D.; Boddy, A.V. Influence of pharmacogenetics on response and toxicity in breast cancer patients treated with doxorubicin and cyclophosphamide. Br. J. Cancer 2010, 102, 1003–1009. [Google Scholar] [CrossRef] [PubMed]
- Rawlings-Goss, R.A.; Campbell, M.C.; Tishkoff, S.A. Global population-specific variation in miRNA associated with cancer risk and clinical biomarkers. BMC Med. Genom. 2014, 7, 53. [Google Scholar] [CrossRef]
- Hoffman, A.E.; Liu, R.; Fu, A.; Zheng, T.; Slack, F.; Zhu, Y. Targetome profiling, pathway analysis and genetic association study implicate miR-202 in lymphomagenesis. Cancer Epidemiol. Biomark. Prev. 2013, 22, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Pipan, V.; Zorc, M.; Kunej, T. MicroRNA Polymorphisms in Cancer: A Literature Analysis. Cancers 2015, 7, 1806–1814. [Google Scholar] [CrossRef] [PubMed]
- Qian, F.; Feng, Y.; Zheng, Y.; Ogundiran, T.O.; Ojengbede, O.; Zheng, W.; Blot, W.; Ambrosone, C.B.; John, E.M.; Bernstein, L.; et al. Genetic variants in microRNA and microRNA biogenesis pathway genes and breast cancer risk among women of African ancestry. Hum. Genet. 2016, 135, 1145–1159. [Google Scholar] [CrossRef] [PubMed]
- Andersson, R.; Gebhard, C.; Miguel-Escalada, I.; Hoof, I.; Bornholdt, J.; Boyd, M.; Chen, Y.; Zhao, X.; Schmidl, C.; Suzuki, T.; et al. An atlas of active enhancers across human cell types and tissues. Nature 2014, 507, 455–461. [Google Scholar] [CrossRef]
- Topalian, S.L.; Taube, J.M.; Anders, R.A.; Pardoll, D.M. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat. Rev. Cancer 2016, 16, 275–287. [Google Scholar] [CrossRef]
- Flaherty, K.T.; Gray, R.J.; Chen, A.P.; Li, S.; McShane, L.M.; Patton, D.; Hamilton, S.R.; Williams, P.M.; Iafrate, A.J.; Sklar, J.; et al. Molecular Landscape and Actionable Alterations in a Genomically Guided Cancer Clinical Trial: National Cancer Institute Molecular Analysis for Therapy Choice (NCI-MATCH). J. Clin. Oncol. 2020, 38, 3883–3894. [Google Scholar] [CrossRef] [PubMed]
- Belin, L.; Kamal, M.; Mauborgne, C.; Plancher, C.; Mulot, F.; Delord, J.P.; Goncalves, A.; Gavoille, C.; Dubot, C.; Isambert, N.; et al. Randomized phase II trial comparing molecularly targeted therapy based on tumor molecular profiling versus conventional therapy in patients with refractory cancer: Cross-over analysis from the SHIVA trial. Ann. Oncol 2017, 28, 590–596. [Google Scholar] [CrossRef]
- Gambardella, V.; Tarazona, N.; Cejalvo, J.M.; Lombardi, P.; Huerta, M.; Rosello, S.; Fleitas, T.; Roda, D.; Cervantes, A. Personalized Medicine: Recent Progress in Cancer Therapy. Cancers 2020, 12, 1009. [Google Scholar] [CrossRef] [PubMed]
- Vasconcellos, V.F.; Colli, L.M.; Awada, A.; de Castro Junior, G. Precision oncology: As much expectations as limitations. Ecancermedicalscience 2018, 12, ed86. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cowie, P.; Hay, E.A.; MacKenzie, A. The noncoding human genome and the future of personalised medicine. Expert Rev. Mol. Med. 2015, 17, e4. [Google Scholar] [CrossRef]
- Zhang, Z.; Gu, M.; Gu, Z.; Lou, Y.R. Role of Long Non-Coding RNA Polymorphisms in Cancer Chemotherapeutic Response. J. Pers. Med. 2021, 11, 513. [Google Scholar] [CrossRef]
- Lin, A.; Hu, Q.; Li, C.; Xing, Z.; Ma, G.; Wang, C.; Li, J.; Ye, Y.; Yao, J.; Liang, K.; et al. The LINK-A lncRNA interacts with PtdIns(3,4,5)P3 to hyperactivate AKT and confer resistance to AKT inhibitors. Nat. Cell Biol. 2017, 19, 238–251. [Google Scholar] [CrossRef]
- Meddens, C.A.; van der List, A.C.J.; Nieuwenhuis, E.E.S.; Mokry, M. Non-coding DNA in IBD: From sequence variation in DNA regulatory elements to novel therapeutic potential. Gut 2019, 68, 928–941. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Wang, J.; Liu, Q.; Yuan, S.; Xie, W.; Liu, Y.; Xiang, Y.; Wu, N.; Wu, L.; Ma, X.; Cai, T.; et al. Genetic predisposition to lung cancer: Comprehensive literature integration, meta-analysis, and multiple evidence assessment of candidate-gene association studies. Sci. Rep. 2017, 7, 8371. [Google Scholar] [CrossRef]
- Yan, T.; Shen, C.; Jiang, P.; Yu, C.; Guo, F.; Tian, X.; Zhu, X.; Lu, S.; Han, B.; Zhong, M.; et al. Risk SNP-induced lncRNA-SLCC1 drives colorectal cancer through activating glycolysis signaling. Signal Transduct. Target. Ther. 2021, 6, 70. [Google Scholar] [CrossRef]
- Dentro, S.C.; Leshchiner, I.; Haase, K.; Tarabichi, M.; Wintersinger, J.; Deshwar, A.G.; Yu, K.; Rubanova, Y.; Macintyre, G.; Demeulemeester, J.; et al. Characterizing genetic intra-tumor heterogeneity across 2,658 human cancer genomes. Cell 2021, 184, 2239.e2239–2254.e2239. [Google Scholar] [CrossRef]
- Lu, Y.; Kweon, S.S.; Tanikawa, C.; Jia, W.H.; Xiang, Y.B.; Cai, Q.; Zeng, C.; Schmit, S.L.; Shin, A.; Matsuo, K.; et al. Large-Scale Genome-Wide Association Study of East Asians Identifies Loci Associated with Risk for Colorectal Cancer. Gastroenterology 2019, 156, 1455–1466. [Google Scholar] [CrossRef]
- Pon, J.R.; Marra, M.A. Driver and passenger mutations in cancer. Annu. Rev. Pathol. 2015, 10, 25–50. [Google Scholar] [CrossRef]
- Lin, Y.; Nakatochi, M.; Hosono, Y.; Ito, H.; Kamatani, Y.; Inoko, A.; Sakamoto, H.; Kinoshita, F.; Kobayashi, Y.; Ishii, H.; et al. Genome-wide association meta-analysis identifies GP2 gene risk variants for pancreatic cancer. Nat. Commun. 2020, 11, 3175. [Google Scholar] [CrossRef]
- Pashayan, N.; Antoniou, A.C.; Ivanus, U.; Esserman, L.J.; Easton, D.F.; French, D.; Sroczynski, G.; Hall, P.; Cuzick, J.; Evans, D.G.; et al. Personalized early detection and prevention of breast cancer: ENVISION consensus statement. Nat. Rev. Clin. Oncol. 2020, 17, 687–705. [Google Scholar] [CrossRef] [PubMed]
- Erichsen, H.C.; Chanock, S.J. SNPs in cancer research and treatment. Br. J. Cancer 2004, 90, 747–751. [Google Scholar] [CrossRef]
- Fu, X.; Shi, Y.; Qi, T.; Qiu, S.; Huang, Y.; Zhao, X.; Sun, Q.; Lin, G. Precise design strategies of nanomedicine for improving cancer therapeutic efficacy using subcellular targeting. Signal Transduct. Target. Ther. 2020, 5, 262. [Google Scholar] [CrossRef] [PubMed]
- Muthuirulan, P.; Zhao, D.; Young, M.; Richard, D.; Liu, Z.; Emami, A.; Portilla, G.; Hosseinzadeh, S.; Cao, J.; Maridas, D.; et al. Joint disease-specificity at the regulatory base-pair level. Nat. Commun. 2021, 12, 4161. [Google Scholar] [CrossRef]
- Russ, A.P.; Lampel, S. The druggable genome: An update. Drug Discov. Today 2005, 10, 1607–1610. [Google Scholar] [CrossRef]
- Geary, R.S.; Norris, D.; Yu, R.; Bennett, C.F. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug Deliv. Rev. 2015, 87, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Rogge, M. Targeting RNA: A Transformative Therapeutic Strategy. Clin. Transl. Sci. 2019, 12, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Kopechek, J.A.; McTiernan, C.F.; Chen, X.; Zhu, J.; Mburu, M.; Feroze, R.; Whitehurst, D.A.; Lavery, L.; Cyriac, J.; Villanueva, F.S. Ultrasound and Microbubble-targeted Delivery of a microRNA Inhibitor to the Heart Suppresses Cardiac Hypertrophy and Preserves Cardiac Function. Theranostics 2019, 9, 7088–7098. [Google Scholar] [CrossRef] [PubMed]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics—Challenges and potential solutions. Nat. Rev. Drug Discov. 2021. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wang, J.; He, Z.; Chen, P.; Jiang, X.; Chen, Y.; Liu, X.; Jiang, J. LncRNA CERS6-AS1 promotes proliferation and metastasis through the upregulation of YWHAG and activation of ERK signaling in pancreatic cancer. Cell Death Dis. 2021, 12, 648. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Wang, R.; Shen, D.; Cheng, S.; Wang, H.; Lu, Z.; Zheng, Q.; Wang, L.; Xia, L.; Li, G. Role of noncoding RNA in drug resistance of prostate cancer. Cell Death Dis. 2021, 12, 590. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).