Genome-Wide Screening and Characterization of Non-Coding RNAs in Coffea canephora
Abstract
1. Introduction
2. Results
2.1. Non-Coding RNA Loci Overview
2.2. Small Nuclear RNAs
2.3. Small Nucleolar RNAs
2.4. Transfer RNAs
2.5. Ribosomal RNAs
2.6. MicroRNAs Prediction
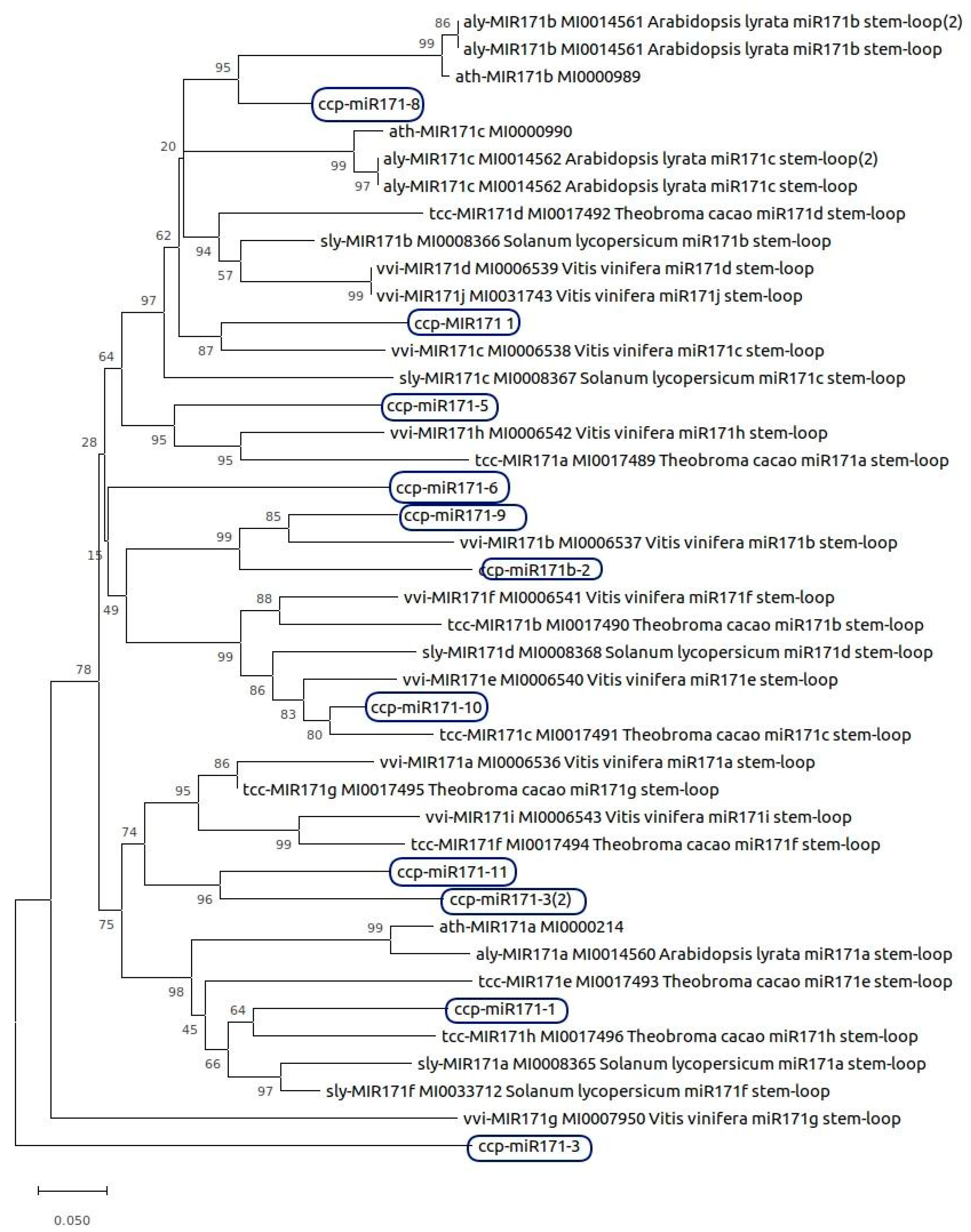
2.7. MicroRNAs Curation
2.8. Phylogenetic Analysis of MicroRNAs
2.9. MicroRNA Targets Prediction
2.10. MicroRNA Expression under Drought Stress
2.11. Long Non-Coding RNAs
3. Discussion
4. Materials and Methods
4.1. Sequence Datasets
4.2. Similarity Search
4.3. Structural Search
4.4. Small Nuclear RNAs Filtering
4.5. Small Nucleolar RNAs
4.6. Transfer RNAs
4.7. Ribosomal RNAs
4.8. MicroRNAs
4.9. Long Non-Coding RNAs
4.10. Transcriptional Evidence Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ariel, F.; Romero-Barrios, N.; Jégu, T.; Benhamed, M.; Crespi, M. Battles and hijacks: Noncoding transcription in plants. Trends Plant Sci. 2015, 20, 362–371. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Chu, C. MicroRNAs in crop improvement: Fine-tuners for complex traits. Nat. Plants 2017, 3, 17077. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Mewalal, R.; Hu, R.; Tuskan, G.A.; Yang, X. New technologies accelerate the exploration of non-coding RNAs in horticultural plants. Hortic. Res. 2017, 4, 17031. [Google Scholar] [CrossRef]
- D’Ario, M.; Griffiths-Jones, S.; Kim, M. Small RNAs: Big Impact on Plant Development. Trends Plant Sci. 2017, 22, 1056–1068. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W.; Bartel, D.P. Computational Identification of Plant MicroRNAs and Their Targets, Including a Stress-Induced miRNA. Mol. Cell 2004, 14, 787–799. [Google Scholar] [CrossRef] [PubMed]
- Chekanova, J.A. Long non-coding RNAs and their functions in plants. Curr. Opin. Plant Boil. 2015, 27, 207–216. [Google Scholar] [CrossRef]
- Diler, E.; Unver, T.; Karakülah, G. Differential Expression of Hyperhydricity Responsive Peach MicroRNAs. J. Integr. Bioinform. 2016, 13, 57–69. [Google Scholar] [CrossRef]
- Rai, M.I.; Alam, M.; Lightfoot, D.; Gurha, P.; Afzal, A.J. Classification and experimental identification of plant long non-coding RNAs. Genomics 2019, 111, 997–1005. [Google Scholar] [CrossRef]
- Zhang, B.; Pan, X.; Cannon, C.H.; Cobb, G.P.; Anderson, T.A. Conservation and divergence of plant microRNA genes. Plant J. 2006, 46, 243–259. [Google Scholar] [CrossRef]
- Nithin, C.; Thomas, A.; Basak, J.; Bahadur, R.P. Genome-wide identification of miRNAs and lncRNAs in Cajanus cajan. BMC Genom. 2017, 18, 878. [Google Scholar] [CrossRef]
- Lin, Z.; Li, Q.; Yin, Q.; Wang, J.; Zhang, B.; Gan, S.; Wu, A.M. Identification of novel miRNAs and their target genes in Eucalyptus grandis. Tree Genet. Genomes 2018, 14, 60. [Google Scholar] [CrossRef]
- Taylor, R.; Tarver, J.E.; Foroozani, A.; Donoghue, P.C.J. MicroRNA annotation of plant genomes—Do it right or not at all. BioEssays 2017, 39, 1600113. [Google Scholar] [CrossRef] [PubMed]
- Ambros, V.; Bartel, B.; Bartel, D.P.; Burge, C.B.; Carrington, J.C.; Chen, X.; Dreyfuss, G.; Eddy, S.R.; Griffiths-Jones, S.; Marshall, M.; et al. A uniform system for microRNA annotation. RNA 2003, 9, 277–279. [Google Scholar] [CrossRef] [PubMed]
- Meyers, B.C.; Axtell, M.J.; Bartel, B.; Bartel, D.P.; Baulcombe, D.C.; Bowman, J.L.; Cao, X.; Carrington, J.C.; Chen, X.; Green, P.J.; et al. Criteria for Annotation of Plant MicroRNAs. Plant Cell 2008, 20, 3186–3190. [Google Scholar] [CrossRef]
- Axtell, M.J.; Meyers, B.C. Revisiting Criteria for Plant MicroRNA Annotation in the Era of Big Data. Plant Cell 2018, 30, 272–284. [Google Scholar] [CrossRef]
- ICO. International Coffee Organization Statistics. Available online: http://www.ico.org (accessed on 20 July 2019).
- Dereeper, A.; Bocs, S.; Rouard, M.; Guignon, V.; Ravel, S.; Tranchant-Dubreuil, C.; Poncet, V.; Garsmeur, O.; Lashermes, P.; Droc, G. The coffee genome hub: A resource for coffee genomes. Nucleic Acids Res. 2015, 43, D1028–D1035. [Google Scholar] [CrossRef]
- Denoeud, F.; Carretero-Paulet, L.; Dereeper, A.; Droc, G.; Guyot, R.; Pietrella, M.; Zheng, C.; Alberti, A.; Anthony, F.; Aprea, G.; et al. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 2014, 345, 1181–1184. [Google Scholar] [CrossRef]
- Bibi, F.; Barozai, M.Y.K.; Din, M. Bioinformatics profiling and characterization of potentialmicroRNAs and their targets in the genus Coffea. Turk. J. Agric. For. 2017, 41, 191–200. [Google Scholar] [CrossRef]
- Nellikunnumal, S.M.; Chandrashekar, A. Computational Identification of Conserved microRNA and their Targets in Coffea canephora by EST Analysis. Dyn. Biochem. Process Biotechnol. Mol. Biol. 2012, 6, 70–76. [Google Scholar]
- Chaves, S.S.; Fernandes-Brum, C.N.; Silva, G.F.F.; Ferrara-Barbosa, B.C.; Paiva, L.V.; Nogueira, F.T.S.; Cardoso, T.C.S.; Amaral, L.R.; Gomes, M.D.S.; Chalfun-Junior, A. New Insights on Coffea miRNAs: Features and Evolutionary Conservation. Appl. Biochem. Biotechnol. 2015, 177, 879–908. [Google Scholar] [CrossRef]
- Fernandes-Brum, C.N.; Rezende, P.M.; Ribeiro, T.H.C.; De Oliveira, R.R.; Cardoso, T.C.D.S.; Amaral, L.R.D.; Gomes, M.D.S.; Chalfun-Junior, A. A genome-wide analysis of the RNA-guided silencing pathway in coffee reveals insights into its regulatory mechanisms. PLoS ONE 2017, 12, e0176333. [Google Scholar] [CrossRef] [PubMed]
- Loss-Morais, G.; Ferreira, D.C.; Margis, R.; Alves-Ferreira, M.; Correa, R.L. Identification of novel and conserved microRNAs in Coffea canephora and Coffea arabica. Genet. Mol. Boil. 2014, 37, 671–682. [Google Scholar] [CrossRef]
- Guedes, F.; Nobres, P.; Ferreira, D.C.R.; Menezes-Silva, P.E.; Ribeiro-Alves, M.; Correa, R.L.; DaMatta, F.M.; Alves-Ferreira, M. Transcriptional memory contributes to drought tolerance in coffee (Coffea canephora) plants. Environ. Exp. Bot. 2018, 147, 220–233. [Google Scholar] [CrossRef]
- Oróstica, K.Y.; Verdugo, R.A. chromPlot: Visualization of genomic data in chromosomal context. Bioinformatics 2016, 32, 2366–2368. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, M. Transcriptional regulation of snRNAs and its significance for plant development. J. Plant Res. 2016, 130, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.S.; Marquez, Y.; Kalyna, M.; Barta, A. Complexity of the alternative splicing landscape in plants. Plant Cell 2013, 25, 3657–3683. [Google Scholar] [CrossRef]
- Dohm, J.C.; Minoche, A.E.; Holtgräwe, D.; Capella-Gutierrez, S.; Zakrzewski, F.; Tafer, H.; Rupp, O.; Sörensen, T.R.; Stracke, R.; Reinhardt, R.; et al. The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature 2013, 505, 546–549. [Google Scholar] [CrossRef]
- Bhattacharya, D.P.; Canzler, S.; Kehr, S.; Hertel, J.; Grosse, I.; Stadler, P.F. Phylogenetic distribution of plant snoRNA families. BMC Genom. 2016, 17, 969. [Google Scholar] [CrossRef]
- Brown, J.W.; Clark, G.P.; Leader, D.J.; Simpson, C.G.; Lowe, T.M. Multiple snoRNA gene clusters from Arabidopsis. RNA 2001, 7, 1817–1832. [Google Scholar] [CrossRef]
- Chen, C.L.; Liang, D.; Zhou, H.; Zhuo, M.; Chen, Y.; Qu, L.H. The high diversity of snoRNAs in plants: Identification and comparative study of 120 snoRNA genes from Oryza sativa. Nucleic Acids Res. 2003, 31, 2601–2613. [Google Scholar] [CrossRef]
- Mohanta, T.K.; Bae, H. Analyses of Genomic tRNA Reveal Presence of Novel tRNAs in Oryza sativa. Front. Genet. 2017, 8, 90. [Google Scholar] [CrossRef]
- Michaud, M.; Cognat, V.; Duchêne, A.-M.; Maréchal-Drouard, L. A global picture of tRNA genes in plant genomes. Plant J. 2011, 66, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Santesmasses, D.; Mariotti, M.; Guigo, R. Computational identification of the selenocysteine tRNA (tRNASec) in genomes. PLoS Comput. Boil. 2017, 13, e1005383. [Google Scholar] [CrossRef] [PubMed]
- Lobanov, A.V.; Hatfield, D.L.; Gladyshev, V.N. Eukaryotic selenoproteins and selenoproteomes. Biochim. Biophys. Acta (BBA) 2009, 1790, 1424–1428. [Google Scholar] [CrossRef]
- Cech, T.R.; Steitz, J.A. The Noncoding RNA Revolution—Trashing Old Rules to Forge New Ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef] [PubMed]
- Iorizzo, M.; Ellison, S.; Senalik, D.; Zeng, P.; Satapoomin, P.; Huang, J.; Bowman, M.J.; Iovene, M.; Sanseverino, W.; Cavagnaro, P.; et al. A high-quality carrot genome assembly provides new insights into carotenoid accumulation and asterid genome evolution. Nat. Genet. 2016, 48, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Vlasova, A.; Capella-Gutierrez, S.; Rendón-Anaya, M.; Hernández-Oñate, M.A.; Minoche, A.E.; Erb, I.; Câmara, F.; Barja, P.P.; Corvelo, A.; Sanseverino, W.; et al. Genome and transcriptome analysis of the Mesoamerican common bean and the role of gene duplications in establishing tissue and temporal specialization of genes. Genome Boil. 2016, 17, 32. [Google Scholar] [CrossRef] [PubMed]
- Xia, E.H.; Zhang, H.B.; Sheng, J.; Li, K.; Zhang, Q.J.; Kim, C.; Zhang, Y.; Liu, Y.; Zhu, T.; Li, W.; et al. The Tea Tree Genome Provides Insights into Tea Flavor and Independent Evolution of Caffeine Biosynthesis. Mol. Plant 2017, 10, 866–877. [Google Scholar] [CrossRef]
- Leclercq, M.; Diallo, A.B.; Blanchette, M. Computational prediction of the localization of microRNAs within their pre-miRNA. Nucleic Acids Res. 2013, 41, 7200–7211. [Google Scholar] [CrossRef]
- Cuperus, J.T.; Fahlgren, N.; Carrington, J.C. Evolution and Functional Diversification of miRNA Genes. Plant Cell 2011, 23, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Datta, S.K.; Datta, K. miRNA regulation of nutrient homeostasis in plants. Front. Plant Sci. 2015, 6, 232. [Google Scholar] [CrossRef] [PubMed]
- Santos, T.B.; Soares, J.D.M.; Lima, J.E.; Silva, J.C.; Ivamoto-Suzuki, S.T.; Baba, V.Y.; Souza, S.G.H.; Lorenzetti, A.P.R.; Paschoal, A.R.; Meda, A.R.; et al. An integrated analysis of mRNA and sRNA transcriptional profiles in Coffea arabica L. roots: Insights on nitrogen starvation responses. Funct. Integr. Genom. 2018, 19, 151–169. [Google Scholar] [CrossRef]
- Pant, B.D.; Buhtz, A.; Kehr, J.; Scheible, W. MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 2008, 53, 731–738. [Google Scholar] [CrossRef] [PubMed]
- Rogans, S.J.; Rey, C. Unveiling the Micronome of Cassava (Manihot esculenta Crantz). PLoS ONE 2016, 11, e0147251. [Google Scholar] [CrossRef]
- Yang, T.; Wang, Y.; Teotia, S.; Zhang, Z.; Tang, G. The Making of Leaves: How Small RNA Networks Modulate Leaf Development. Front. Plant Sci. 2018, 9, 824. [Google Scholar] [CrossRef]
- Bel, M.V.; Diels, T.; Vancaester, E.; Kreft, L.; Botzki, A.; Van De Peer, Y.; Coppens, F.; Vandepoele, K. PLAZA 4.0: An integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res. 2018, 46, D1190–D1196. [Google Scholar] [CrossRef]
- Nejat, N.; Mantri, N. Emerging roles of long non-coding RNAs in plant response to biotic and abiotic stresses. Crit. Rev. Biotechnol. 2018, 38, 93–105. [Google Scholar] [CrossRef]
- He, J.; Xu, M.; Willmann, M.R.; McCormick, K.; Hu, T.; Yang, L.; Starker, C.G.; Voytas, D.F.; Meyers, B.C.; Poethig, R.S. Threshold-dependent repression of SPL gene expression by miR156/miR157 controls vegetative phase change in Arabidopsis thaliana. PLoS Genet. 2018, 14, e1007337. [Google Scholar] [CrossRef]
- Xue, M.; Yi, H.; Wang, H. Identification of miRNAs involved in SO2 preservation in Vitis vinifera L. by deep sequencing. Environ. Exp. Bot. 2018, 153, 218–228. [Google Scholar] [CrossRef]
- Lin, Y.; Lai, Z.; Tian, Q.; Lin, L.; Lai, R.; Yang, M.; Zhang, D.; Chen, Y.; Zhang, Z. Endogenous target mimics down-regulate miR160 mediation of ARF10, -16, and -17 cleavage during somatic embryogenesis in Dimocarpus longan Lour. Front. Plant Sci. 2015, 6, 956. [Google Scholar] [CrossRef] [PubMed]
- Hewezi, T.; Piya, S.; Qi, M.; Balasubramaniam, M.; Rice, J.H.; Baum, T.J. Arabidopsis miR827 mediates post-transcriptional gene silencing of its ubiquitin E3 ligase target gene in the syncytium of the cyst nematode Heterodera schachtii to enhance susceptibility. Plant J. 2016, 88, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Kaur, P.; Shukla, N.; Joshi, G.; Vijayakumar, C.; Jagannath, A.; Agarwal, M.; Goel, S.; Kumar, A. Genome-wide identification and characterization of miRNAome from tomato (Solanum lycopersicum) roots and root-knot nematode (Meloidogyne incognita) during susceptible interaction. PLoS ONE 2017, 12, e0175178. [Google Scholar] [CrossRef]
- Yates, A.; Achuthan, P.; Akanni, W.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; Bhai, J.; et al. Ensembl 2020. Nucleic Acids Res. 2020, 48, D682–D688. [Google Scholar] [CrossRef]
- Boratyn, G.M.; Camacho, C.; Cooper, P.S.; Coulouris, G.; Fong, A.; Ma, N.; Madden, T.L.; Matten, W.T.; McGinnis, S.D.; Merezhuk, Y.; et al. BLAST: A more efficient report with usability improvements. Nucleic Acids Res. 2013, 41, W29–W33. [Google Scholar] [CrossRef] [PubMed]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef]
- Kalvari, I.; Nawrocki, E.P.; Argasinska, J.; Quinones-Olvera, N.; Finn, R.D.; Bateman, A.; Petrov, A.I. Non-Coding RNA Analysis Using the Rfam Database. Curr. Protoc. Bioinform. 2018, 62, e51. [Google Scholar] [CrossRef]
- Oliveira, J.V.A.; Costa, F.; Backofen, R.; Stadler, P.F.; Walter, M.E.M.T.; Hertel, J. SnoReport 2.0: New features and a refined Support Vector Machine to improve snoRNA identification. BMC Bioinform. 2018, 17 (Suppl. 18), 464–486. [Google Scholar] [CrossRef]
- Kalinina, N.O.; Makarova, S.; Makhotenko, A.; Love, A.J.; Taliansky, M.E. The Multiple Functions of the Nucleolus in Plant Development, Disease and Stress Responses. Front. Plant Sci. 2018, 9, 132. [Google Scholar] [CrossRef]
- Liu, T.T.; Zhu, D.; Chen, W.; Deng, W.; He, H.; He, G.; Bai, B.; Qi, Y.; Chen, R.; Deng, X.W. A Global Identification and Analysis of Small Nucleolar RNAs and Possible Intermediate-Sized Non-Coding RNAs in Oryza sativa. Mol. Plant 2013, 6, 830–846. [Google Scholar] [CrossRef]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Stærfeldt, H.H.; Rognes, T.; Ussery, D.W.; Staerfeldt, H.H. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Boil. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef]
- Mitchell, A.; Attwood, T.K.; Babbitt, P.C.; Blum, M.; Bork, P.; Bridge, A.; Brown, S.D.; Chang, H.Y.; El-Gebali, S.; Fraser, M.I.; et al. InterPro in 2019: Improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2018, 47, D351–D360. [Google Scholar] [CrossRef]
- Mondego, J.M.C.; Vidal, R.O.; Carazzolle, M.F.; Tokuda, E.K.; Parizzi, L.P.; Costa, G.G.L.; Pereira, L.F.P.; Andrade, A.C.; Colombo, C.A.; Vieira, L.G.E.; et al. An EST-based analysis identifies new genes and reveals distinctive gene expression features of Coffea arabica and Coffea canephora. BMC Plant Boil. 2011, 11, 30. [Google Scholar] [CrossRef]
- Negri, T.D.C.; Alves, W.A.L.; Bugatti, P.H.; Saito, P.T.; Domingues, D.S.; Paschoal, A.R. Pattern recognition analysis on long noncoding RNAs: A tool for prediction in plants. Brief. Bioinform. 2019, 20, 682–689. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Kang, Y.J.; Yang, D.C.; Kong, L.; Hou, M.; Meng, Y.Q.; Wei, L.; Gao, G. CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017, 45, W12–W16. [Google Scholar] [CrossRef]
- Suzek, B.E.; Wang, Y.; Huang, H.; McGarvey, P.B.; Wu, C.H.; UniProt Consortium. The UniProt Consortium UniRef clusters: A comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 2015, 31, 926–932. [Google Scholar] [CrossRef]
- Quinlan, A.R. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr. Protoc. Bioinform. 2014, 47, 11.12.1–11.12.34. [Google Scholar] [CrossRef] [PubMed]
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Research 2018, 7, 1338. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Jiménez-Jacinto, V.; Sanchez-Flores, A.; Vega-Alvarado, L. Integrative Differential Expression Analysis for Multiple EXperiments (IDEAMEX): A Web Server Tool for Integrated RNA-Seq Data Analysis. Front. Genet. 2019, 10, 279. [Google Scholar] [CrossRef]
- Costa-Silva, J.; Domingues, D.S.; Lopes, F.M. RNA-Seq differential expression analysis: An extended review and a software tool. PLoS ONE 2017, 12, e0190152. [Google Scholar] [CrossRef]
- Mishra, A.; Bohra, A. Non-coding RNAs and plant male sterility: Current knowledge and future prospects. Plant Cell Rep. 2018, 37, 177–191. [Google Scholar] [CrossRef]
- Shin, S.Y.; Shin, C. Regulatory non-coding RNAs in plants: Potential gene resources for the improvement of agricultural traits. Plant Biotechnol. Rep. 2016, 10, 35–47. [Google Scholar] [CrossRef]
- Kim, N.H.; Xi, Y.; Sung, S. Modular function of long noncoding RNA, COLDAIR, in the vernalization response. PLoS Genet. 2017, 13, e1006939. [Google Scholar] [CrossRef]
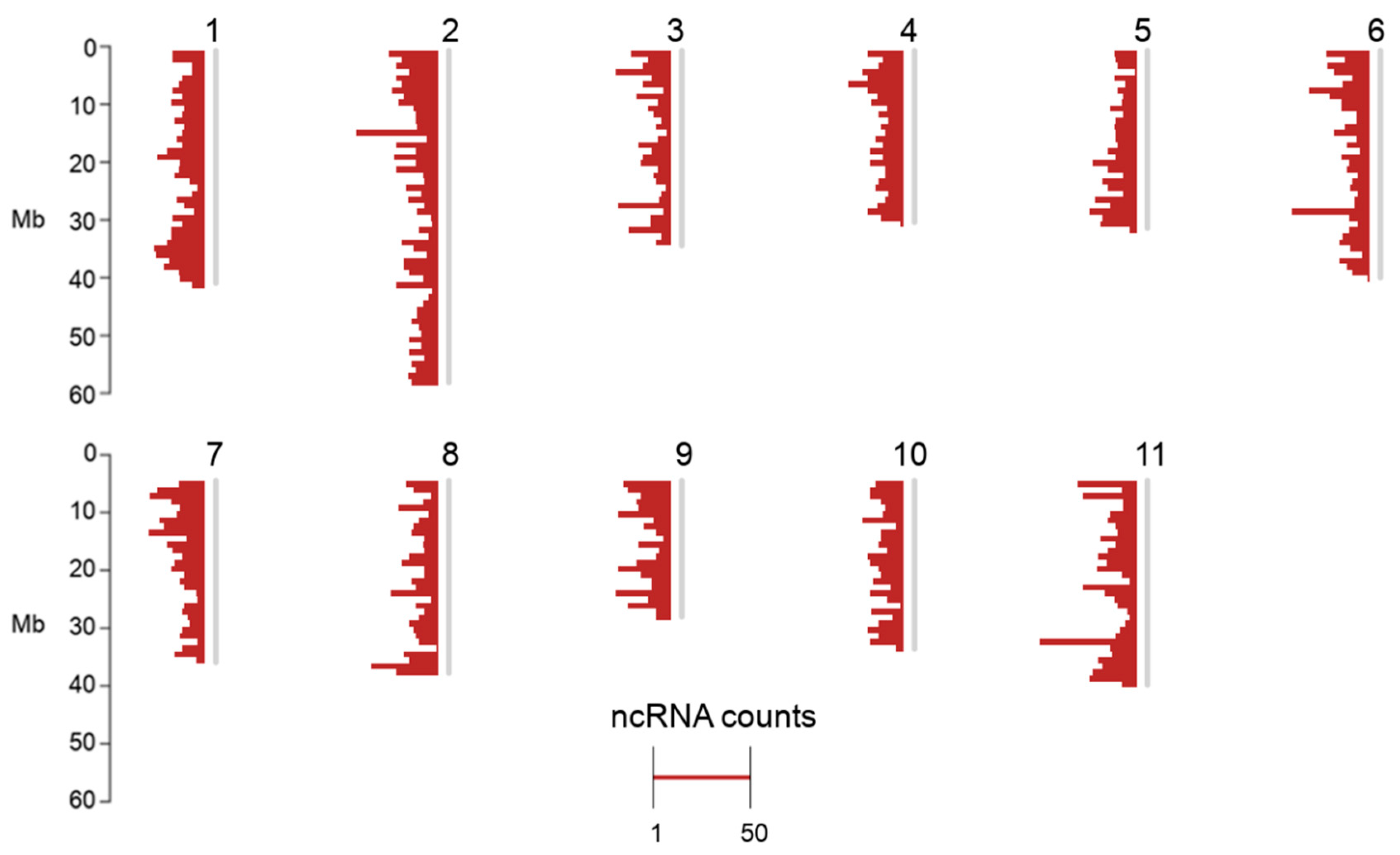
| NcRNA Class (Genome Size) | C. canephora (~569 Mb) | A. thaliana (~135 Mb) | C. sinensis (~3.2 Gb) | B. vulgaris (~567 Mb) | P. vulgaris (~550 Mb) |
|---|---|---|---|---|---|
| tRNA | 602 | 689 | 700 | 1043 | 712 |
| rRNA | 72 | 15 | 2860 | 231 | 145 |
| miRNA | 88 | 325 | 233 | 258 | 309 |
| snRNA | 115 | 82 | 223 | 121 | 165 |
| snoRNA | 1031 | 287 | 454 | 218 | 356 |
| lncRNA | 5064 | 3559 | 42,951 | 2546 | 1033 |
| Total | 6976 | 4957 | 47,421 | 4417 | 2720 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lemos, S.M.C.; Fonçatti, L.F.C.; Guyot, R.; Paschoal, A.R.; Domingues, D.S. Genome-Wide Screening and Characterization of Non-Coding RNAs in Coffea canephora. Non-Coding RNA 2020, 6, 39. https://doi.org/10.3390/ncrna6030039
Lemos SMC, Fonçatti LFC, Guyot R, Paschoal AR, Domingues DS. Genome-Wide Screening and Characterization of Non-Coding RNAs in Coffea canephora. Non-Coding RNA. 2020; 6(3):39. https://doi.org/10.3390/ncrna6030039
Chicago/Turabian StyleLemos, Samara M. C., Luiz F. C. Fonçatti, Romain Guyot, Alexandre R. Paschoal, and Douglas S. Domingues. 2020. "Genome-Wide Screening and Characterization of Non-Coding RNAs in Coffea canephora" Non-Coding RNA 6, no. 3: 39. https://doi.org/10.3390/ncrna6030039
APA StyleLemos, S. M. C., Fonçatti, L. F. C., Guyot, R., Paschoal, A. R., & Domingues, D. S. (2020). Genome-Wide Screening and Characterization of Non-Coding RNAs in Coffea canephora. Non-Coding RNA, 6(3), 39. https://doi.org/10.3390/ncrna6030039






