Non-Coding RNAs: Strategy for Viruses’ Offensive
Abstract
1. Introduction
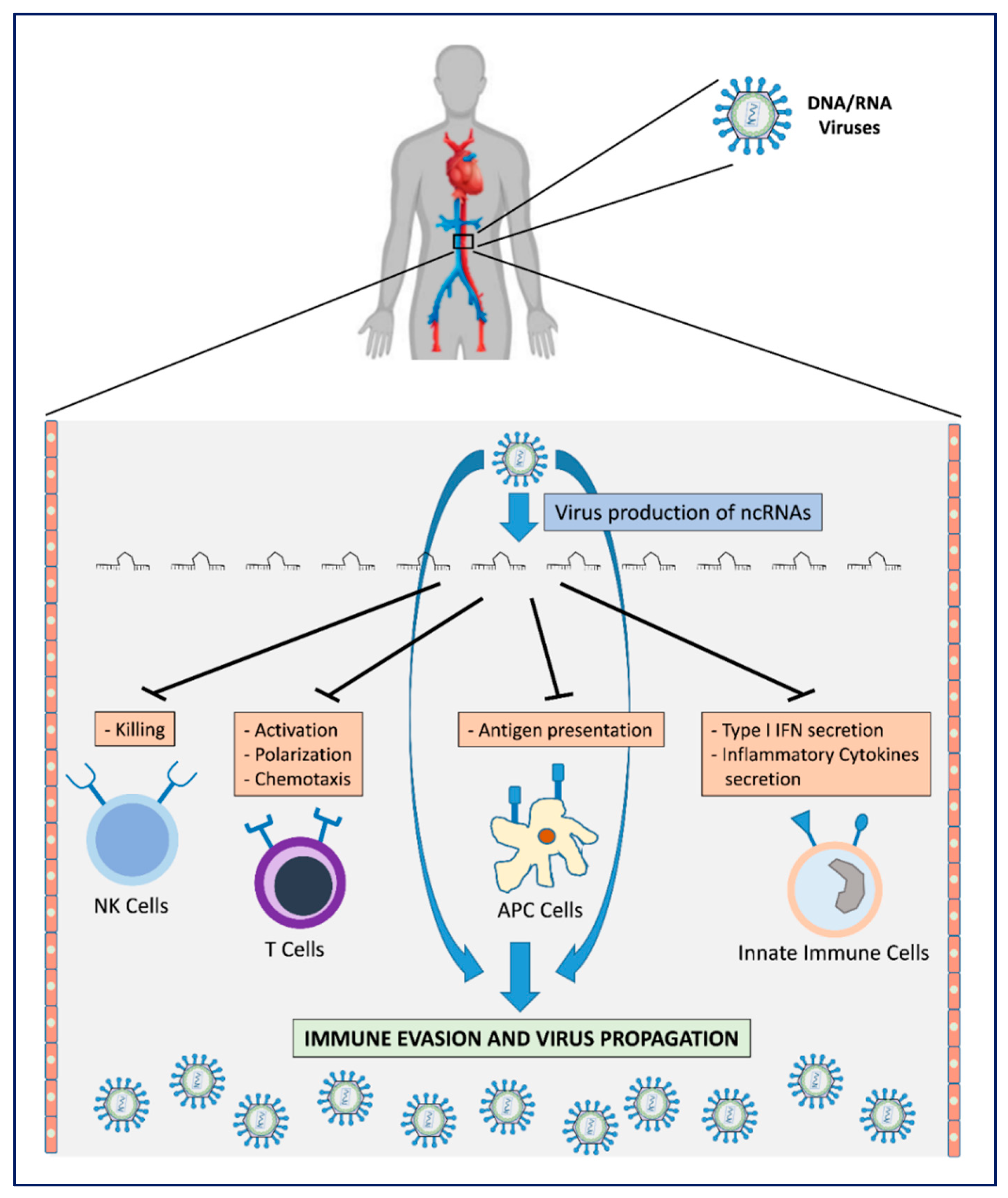
2. Viral Immune Evasion Strategies
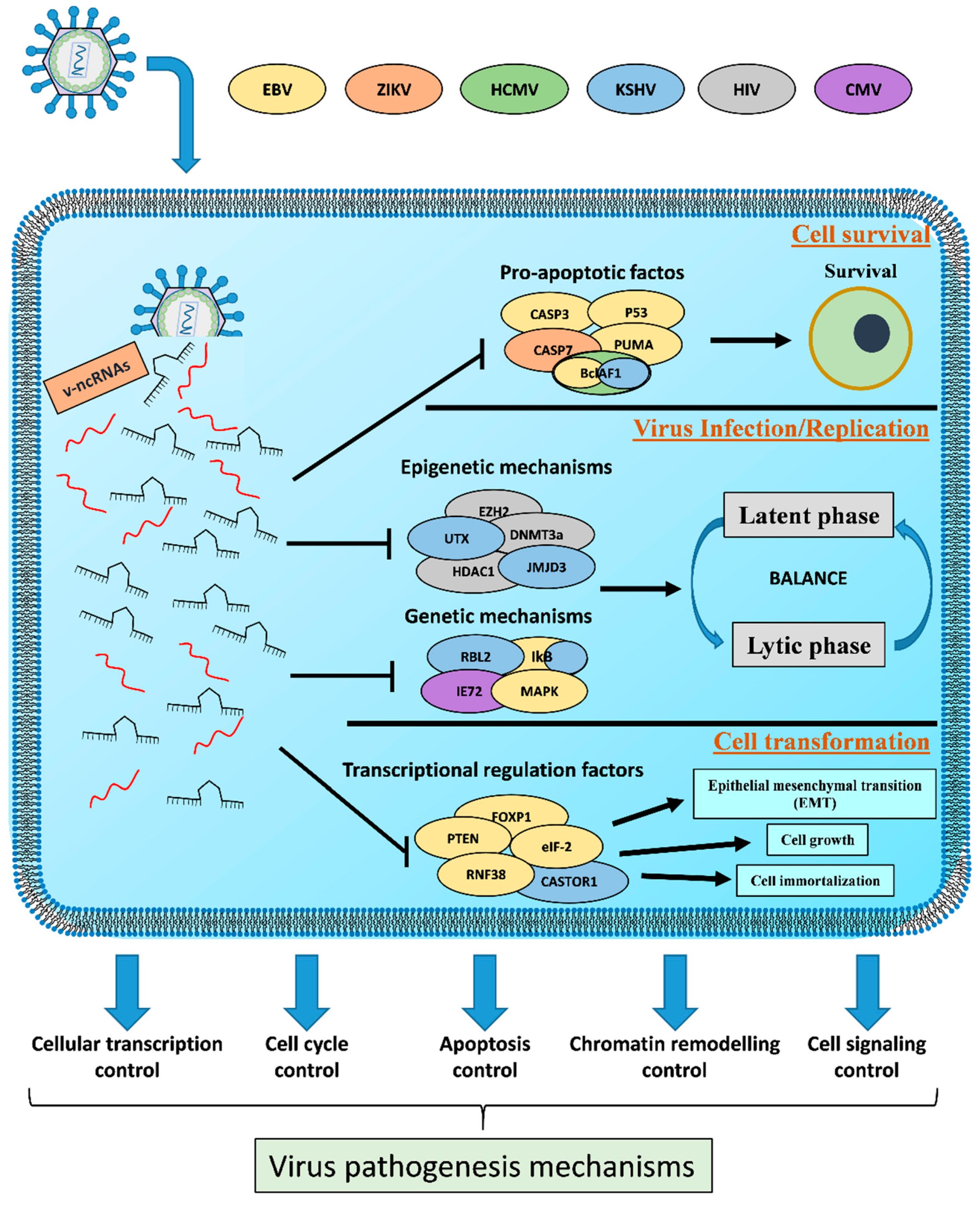
3. Viral Non-Coding RNAs as Transcriptional Weapons
3.1. Regulation of Viral and Host Gene Expression
3.2. Host Cell Survival
3.3. Viral Efficient and Persistent Infection Regulation
3.4. Cell Transformation
4. v-ncRNA Host Mimicry
5. Viral Circular RNA
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Plummer, M.; de Martel, C.; Vignat, J.; Ferlay, J.; Bray, F.; Franceschi, S. Global burden of cancers attributable to infections in 2012: A synthetic analysis. Lancet Global Health 2016, 4, e609–e616. [Google Scholar] [CrossRef]
- Dhama, K.; Khan, S.; Tiwari, R.; Sircar, S.; Bhat, S.; Malik, Y.S.; Singh, K.P.; Chaicumpa, W.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J. Coronavirus Disease 2019-COVID-19. Clin. Microbiol. Rev. 2020, 33. [Google Scholar] [CrossRef] [PubMed]
- DiMaio, D. Viruses, masters at downsizing. Cell Host Microbe 2012, 11, 560–561. [Google Scholar] [CrossRef] [PubMed]
- Harwig, A.; Landick, R.; Berkhout, B. The Battle of RNA Synthesis: Virus versus Host. Viruses 2017, 9, 309. [Google Scholar] [CrossRef] [PubMed]
- Cohen, F.S. How Viruses Invade Cells. Biophys. J. 2016, 110, 1028–1032. [Google Scholar] [CrossRef]
- Mazzon, M.; Marsh, M. Targeting viral entry as a strategy for broad-spectrum antivirals. F1000Research 2019, 8. [Google Scholar] [CrossRef]
- Kobiler, O.; Weitzman, M.D. Herpes simplex virus replication compartments: From naked release to recombining together. PLoS Pathog. 2019, 15, e1007714. [Google Scholar] [CrossRef]
- Tycowski, K.T.; Guo, Y.E.; Lee, N.; Moss, W.N.; Vallery, T.K.; Xie, M.; Steitz, J.A. Viral noncoding RNAs: More surprises. Genes Dev. 2015, 29, 567–584. [Google Scholar] [CrossRef]
- Withers, J.B.; Mondol, V.; Pawlica, P.; Rosa-Mercado, N.A.; Tycowski, K.T.; Ghasempur, S.; Torabi, S.F.; Steitz, J.A. Idiosyncrasies of Viral Noncoding RNAs Provide Insights into Host Cell Biology. Ann. Rev. Virol. 2019, 6, 297–317. [Google Scholar] [CrossRef]
- Mahmoudabadi, G.; Phillips, R. A comprehensive and quantitative exploration of thousands of viral genomes. eLife 2018, 7, e31955. [Google Scholar] [CrossRef]
- Damas, N.D.; Fossat, N.; Scheel, T.K.H. Functional Interplay between RNA Viruses and Non-Coding RNA in Mammals. Non-Coding RNA 2019, 5, 7. [Google Scholar] [CrossRef]
- Jones, K.E.; Patel, N.G.; Levy, M.A.; Storeygard, A.; Balk, D.; Gittleman, J.L.; Daszak, P. Global trends in emerging infectious diseases. Nature 2008, 451, 990–993. [Google Scholar] [CrossRef]
- Charley, P.A.; Wilusz, J. Standing your ground to exoribonucleases: Function of Flavivirus long non-coding RNAs. Virus Res. 2016, 212, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Choi, E.J.; Lee, I.; Lee, Y.S.; Bao, X. Non-Coding RNAs and Their Role in Respiratory Syncytial Virus (RSV) and Human Metapneumovirus (hMPV) Infections. Viruses 2020, 12, 345. [Google Scholar] [CrossRef] [PubMed]
- Amin, N.; McGrath, A.; Chen, Y.-P.P. Evaluation of deep learning in non-coding RNA classification. Nat. Mach. Intell. 2019, 1, 246–256. [Google Scholar] [CrossRef]
- Hombach, S.; Kretz, M. Non-coding RNAs: Classification, Biology and Functioning. Adv. Exp. Med. Biol. 2016, 937, 3–17. [Google Scholar] [CrossRef]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, X.; Wang, Y.Z.; Tian, H.; Xu, S. Research progress of circular RNAs in lung cancer. Cancer Biol. Ther. 2019, 20, 123–129. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Wilczynska, A.; Bushell, M. The complexity of miRNA-mediated repression. Cell Death Differ. 2015, 22, 22–33. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Vishnubalaji, R.; Shaath, H.; Alajez, N.M. Protein Coding and Long Noncoding RNA (lncRNA) Transcriptional Landscape in SARS-CoV-2 Infected Bronchial Epithelial Cells Highlight a Role for Interferon and Inflammatory Response. Genes 2020, 11, 760. [Google Scholar] [CrossRef] [PubMed]
- Lilley, B.N.; Ploegh, H.L. Viral modulation of antigen presentation: Manipulation of cellular targets in the ER and beyond. Immunol. Rev. 2005, 207, 126–144. [Google Scholar] [CrossRef]
- Carriere, J.; Rao, Y.; Liu, Q.; Lin, X.; Zhao, J.; Feng, P. Post-translational Control of Innate Immune Signaling Pathways by Herpesviruses. Front. Microbiol. 2019, 10, 2647. [Google Scholar] [CrossRef] [PubMed]
- Tagawa, T.; Serquina, A.; Kook, I.; Ziegelbauer, J. Viral non-coding RNAs: Stealth strategies in the tug-of-war between humans and herpesviruses. Semin. Cell Dev. Biol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Sadler, A.J.; Williams, B.R. Interferon-inducible antiviral effectors. Nat. Rev. Immun. 2008, 8, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Rossetto, C.C.; Pari, G.S. Kaposi’s sarcoma-associated herpesvirus noncoding polyadenylated nuclear RNA interacts with virus- and host cell-encoded proteins and suppresses expression of genes involved in immune modulation. J. Virol. 2011, 85, 13290–13297. [Google Scholar] [CrossRef]
- Pallares, H.; Costa Navarro, G.S.; Villordo, S.; Merwaiss, F.; de Borba, L.; Ledesma, M.M.G.L.; Ojeda, D.S.; Henrion-Lacritick, A.; Morales, M.A.; Fabri, C.; et al. Zika Virus sfRNA Generation Requires Cooperativity between Duplicated RNA Structures that Are Essential for Productive Infection in Human Cells. J. Virol. 2020. [Google Scholar] [CrossRef]
- Cumberworth, S.L.; Clark, J.J.; Kohl, A.; Donald, C.L. Inhibition of type I interferon induction and signalling by mosquito-borne flaviviruses. Cell. Microbiol. 2017, 19. [Google Scholar] [CrossRef]
- Qiu, L.; Wang, T.; Tang, Q.; Li, G.; Wu, P.; Chen, K. Long Non-coding RNAs: Regulators of Viral Infection and the Interferon Antiviral Response. Front. Microbiol. 2018, 9, 1621. [Google Scholar] [CrossRef]
- Lu, Y.; Qin, Z.; Wang, J.; Zheng, X.; Lu, J.; Zhang, X.; Wei, L.; Peng, Q.; Zheng, Y.; Ou, C.; et al. Epstein-Barr Virus miR-BART6-3p Inhibits the RIG-I Pathway. J. Innate Immun. 2017, 9, 574–586. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.M.; Yu, Y.; Zhao, H.P. EBVBART63p and cellular microRNA197 compromise the immune defense of host cells in EBVpositive Burkitt lymphoma. Mol. Med. Rep. 2017, 15, 1877–1883. [Google Scholar] [CrossRef] [PubMed]
- Hooykaas, M.J.G.; van Gent, M.; Soppe, J.A.; Kruse, E.; Boer, I.G.J.; van Leenen, D.; Koerkamp, M.J.A.G.; Holstege, F.C.P.; Ressing, M.E.; Wiertz, E.; et al. EBV MicroRNA BART16 Suppresses Type I IFN Signaling. J. Immunol. 2017, 198, 4062–4073. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Gao, Y.; Lin, X.; He, Z.; Zhao, Q.; Deng, Q.; Lan, K. A human herpesvirus miRNA attenuates interferon signaling and contributes to maintenance of viral latency by targeting IKKepsilon. Cell Res. 2011, 21, 793–806. [Google Scholar] [CrossRef]
- Abend, J.R.; Ramalingam, D.; Kieffer-Kwon, P.; Uldrick, T.S.; Yarchoan, R.; Ziegelbauer, J.M. Kaposi’s sarcoma-associated herpesvirus microRNAs target IRAK1 and MYD88, two components of the toll-like receptor/interleukin-1R signaling cascade, to reduce inflammatory-cytokine expression. J. Virol. 2012, 86, 11663–11674. [Google Scholar] [CrossRef]
- Skinner, C.M.; Ivanov, N.S.; Barr, S.A.; Chen, Y.; Skalsky, R.L. An Epstein-Barr Virus MicroRNA Blocks Interleukin-1 (IL-1) Signaling by Targeting IL-1 Receptor 1. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Haneklaus, M.; Gerlic, M.; Kurowska-Stolarska, M.; Rainey, A.A.; Pich, D.; McInnes, I.B.; Hammerschmidt, W.; O’Neill, L.A.; Masters, S.L. Cutting edge: miR-223 and EBV miR-BART15 regulate the NLRP3 inflammasome and IL-1beta production. J. Immunol. 2012, 189, 3795–3799. [Google Scholar] [CrossRef]
- Tagawa, T.; Albanese, M.; Bouvet, M.; Moosmann, A.; Mautner, J.; Heissmeyer, V.; Zielinski, C.; Lutter, D.; Hoser, J.; Hastreiter, M.; et al. Epstein-Barr viral miRNAs inhibit antiviral CD4+ T cell responses targeting IL-12 and peptide processing. J. Exp. Med. 2016, 213, 2065–2080. [Google Scholar] [CrossRef]
- Skalsky, R.L.; Corcoran, D.L.; Gottwein, E.; Frank, C.L.; Kang, D.; Hafner, M.; Nusbaum, J.D.; Feederle, R.; Delecluse, H.J.; Luftig, M.A.; et al. The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS Pathog. 2012, 8, e1002484. [Google Scholar] [CrossRef]
- Akhbari, P.; Tobin, D.; Poterlowicz, K.; Roberts, W.; Boyne, J.R. MCV-miR-M1 Targets the Host-Cell Immune Response Resulting in the Attenuation of Neutrophil Chemotaxis. J. Investig. Dermatol. 2018, 138, 2343–2354. [Google Scholar] [CrossRef]
- Qian, K.; Pietila, T.; Ronty, M.; Michon, F.; Frilander, M.J.; Ritari, J.; Tarkkanen, J.; Paulin, L.; Auvinen, P.; Auvinen, E. Identification and validation of human papillomavirus encoded microRNAs. PLoS ONE 2013, 8, e70202. [Google Scholar] [CrossRef]
- Xia, T.; O’Hara, A.; Araujo, I.; Barreto, J.; Carvalho, E.; Sapucaia, J.B.; Ramos, J.C.; Luz, E.; Pedroso, C.; Manrique, M.; et al. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 2008, 68, 1436–1442. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.; Kumar, A.; Ingle, H.; Kumar, H. The Interplay Between Viral-Derived miRNAs and Host Immunity During Infection. Front. Immunol. 2019, 10, 3079. [Google Scholar] [CrossRef] [PubMed]
- Bet, A.; Maze, E.A.; Bansal, A.; Sterrett, S.; Gross, A.; Graff-Dubois, S.; Samri, A.; Guihot, A.; Katlama, C.; Theodorou, I.; et al. The HIV-1 antisense protein (ASP) induces CD8 T cell responses during chronic infection. Retrovirology 2015, 12, 15. [Google Scholar] [CrossRef] [PubMed]
- Saayman, S.; Ackley, A.; Turner, A.W.; Famiglietti, M.; Bosque, A.; Clemson, M.; Planelles, V.; Morris, K.V. An HIV-encoded antisense long noncoding RNA epigenetically regulates viral transcription. Mol. Ther. J. Am. Soc. Gene Ther. 2014, 22, 1164–1175. [Google Scholar] [CrossRef] [PubMed]
- Aguado, L.C.; tenOever, B. RNA virus building blocks-miRNAs not included. PLoS Pathog. 2018, 14, e1006963. [Google Scholar] [CrossRef]
- Chen, Z.; Liang, H.; Chen, X.; Ke, Y.; Zhou, Z.; Yang, M.; Zen, K.; Yang, R.; Liu, C.; Zhang, C.Y. An Ebola virus-encoded microRNA-like fragment serves as a biomarker for early diagnosis of Ebola virus disease. Cell Res. 2016, 26, 380–383. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, J.; Zhang, H.; Wang, M.; Gao, G.F.; Li, X. Ebola virus encodes a miR-155 analog to regulate importin-alpha5 expression. Cell. Mol. Life Sci. CMLS 2016, 73, 3733–3744. [Google Scholar] [CrossRef]
- Stern-Ginossar, N.; Elefant, N.; Zimmermann, A.; Wolf, D.G.; Saleh, N.; Biton, M.; Horwitz, E.; Prokocimer, Z.; Prichard, M.; Hahn, G.; et al. Host immune system gene targeting by a viral miRNA. Science 2007, 317, 376–381. [Google Scholar] [CrossRef]
- Nachmani, D.; Lankry, D.; Wolf, D.G.; Mandelboim, O. The human cytomegalovirus microRNA miR-UL112 acts synergistically with a cellular microRNA to escape immune elimination. Nat. Immunol. 2010, 11, 806–813. [Google Scholar] [CrossRef]
- Boss, I.W.; Renne, R. Viral miRNAs and immune evasion. Biochim. Biophys. Acta 2011, 1809, 708–714. [Google Scholar] [CrossRef] [PubMed]
- Bauman, Y.; Nachmani, D.; Vitenshtein, A.; Tsukerman, P.; Drayman, N.; Stern-Ginossar, N.; Lankry, D.; Gruda, R.; Mandelboim, O. An identical miRNA of the human JC and BK polyoma viruses targets the stress-induced ligand ULBP3 to escape immune elimination. Cell Host Microbe 2011, 9, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Raab-Traub, N. Novel mechanisms of EBV-induced oncogenesis. Curr. Opin. Virol. 2012, 2, 453–458. [Google Scholar] [CrossRef] [PubMed]
- Rossetto, C.C.; Tarrant-Elorza, M.; Verma, S.; Purushothaman, P.; Pari, G.S. Regulation of viral and cellular gene expression by Kaposi’s sarcoma-associated herpesvirus polyadenylated nuclear RNA. J. Virol. 2013, 87, 5540–5553. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Segerman, B.; Zhou, X.; Akusjarvi, G. Adenovirus virus-associated RNAII-derived small RNAs are efficiently incorporated into the rna-induced silencing complex and associate with polyribosomes. J. Virol. 2007, 81, 10540–10549. [Google Scholar] [CrossRef]
- Hutzinger, R.; Feederle, R.; Mrazek, J.; Schiefermeier, N.; Balwierz, P.J.; Zavolan, M.; Polacek, N.; Delecluse, H.J.; Huttenhofer, A. Expression and processing of a small nucleolar RNA from the Epstein-Barr virus genome. PLoS Pathog. 2009, 5, e1000547. [Google Scholar] [CrossRef]
- Wu, Y.; Maruo, S.; Yajima, M.; Kanda, T.; Takada, K. Epstein-Barr virus (EBV)-encoded RNA 2 (EBER2) but not EBER1 plays a critical role in EBV-induced B-cell growth transformation. J. Virol. 2007, 81, 11236–11245. [Google Scholar] [CrossRef]
- Deng, J.; Ptashkin, R.N.; Wang, Q.; Liu, G.; Zhang, G.; Lee, I.; Lee, Y.S.; Bao, X. Human metapneumovirus infection induces significant changes in small noncoding RNA expression in airway epithelial cells. Molecular Ther. Nucl. Acids 2014, 3, e163. [Google Scholar] [CrossRef]
- Grundhoff, A.; Sullivan, C.S. Virus-encoded microRNAs. Virology 2011, 411, 325–343. [Google Scholar] [CrossRef]
- Wang, P. The Opening of Pandora’s Box: An Emerging Role of Long Noncoding RNA in Viral Infections. Front. Immunol. 2018, 9, 3138. [Google Scholar] [CrossRef]
- Zanotto, P.M.; Gibbs, M.J.; Gould, E.A.; Holmes, E.C. A reevaluation of the higher taxonomy of viruses based on RNA polymerases. J. Virol. 1996, 70, 6083–6096. [Google Scholar] [CrossRef] [PubMed]
- Stevens, J.G.; Wagner, E.K.; Devi-Rao, G.B.; Cook, M.L.; Feldman, L.T. RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science 1987, 235, 1056–1059. [Google Scholar] [CrossRef] [PubMed]
- Kang, W.; Mukerjee, R.; Gartner, J.J.; Hatzigeorgiou, A.G.; Sandri-Goldin, R.M.; Fraser, N.W. Characterization of a spliced exon product of herpes simplex type-1 latency-associated transcript in productively infected cells. Virology 2006, 356, 106–114. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Perez, J.T.; Zlatev, I.; Aggarwal, S.; Subramanian, S.; Sachidanandam, R.; Kim, B.; Manoharan, M.; tenOever, B.R. A small-RNA enhancer of viral polymerase activity. J. Virol. 2012, 86, 13475–13485. [Google Scholar] [CrossRef]
- Kurilla, M.G.; Cabradilla, C.D.; Holloway, B.P.; Keene, J.D. Nucleotide sequence and host La protein interactions of rabies virus leader RNA. J. Virol. 1984, 50, 773–778. [Google Scholar] [CrossRef]
- Aparicio, O.; Carnero, E.; Abad, X.; Razquin, N.; Guruceaga, E.; Segura, V.; Fortes, P. Adenovirus VA RNA-derived miRNAs target cellular genes involved in cell growth, gene expression and DNA repair. Nucleic Acids Res. 2010, 38, 750–763. [Google Scholar] [CrossRef]
- Andersson, M.G.; Haasnoot, P.C.; Xu, N.; Berenjian, S.; Berkhout, B.; Akusjarvi, G. Suppression of RNA interference by adenovirus virus-associated RNA. J. Virol. 2005, 79, 9556–9565. [Google Scholar] [CrossRef]
- Albrecht, J.C.; Fleckenstein, B. Nucleotide sequence of HSUR 6 and HSUR 7, two small RNAs of herpesvirus saimiri. Nucleic Acids Res. 1992, 20, 1810. [Google Scholar] [CrossRef]
- Ensser, A.; Fleckenstein, B. T-cell transformation and oncogenesis by gamma2-herpesviruses. Adv. Cancer Res. 2005, 93, 91–128. [Google Scholar] [CrossRef]
- Cazalla, D.; Yario, T.; Steitz, J.A. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science 2010, 328, 1563–1566. [Google Scholar] [CrossRef]
- Lee, N.; Moss, W.N.; Yario, T.A.; Steitz, J.A. EBV noncoding RNA binds nascent RNA to drive host PAX5 to viral DNA. Cell 2015, 160, 607–618. [Google Scholar] [CrossRef] [PubMed]
- Thompson, C.B. Apoptosis in the pathogenesis and treatment of disease. Science 1995, 267, 1456–1462. [Google Scholar] [CrossRef] [PubMed]
- Forch, P.; Valcarcel, J. Molecular mechanisms of gene expression regulation by the apoptosis-promoting protein TIA-1. Apoptosis Int. J. Program. Cell Death 2001, 6, 463–468. [Google Scholar] [CrossRef] [PubMed]
- O’Malley, R.P.; Duncan, R.F.; Hershey, J.W.; Mathews, M.B. Modification of protein synthesis initiation factors and the shut-off of host protein synthesis in adenovirus-infected cells. Virology 1989, 168, 112–118. [Google Scholar] [CrossRef]
- Gartner, J.J.; Sethupathy, P.; Hatzigeorgiou, A.G.; Fraser, N.W. Anti-apoptotic function of a microRNA encoded by the HSV-1 latency-associated transcript. Nature 2008, 451, 600. [Google Scholar] [CrossRef]
- Umbach, J.L.; Kramer, M.F.; Jurak, I.; Karnowski, H.W.; Coen, D.M.; Cullen, B.R. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 2008, 454, 780–783. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, H.L.; Zhong, F.F.; Fan, J.Y. Anti-apoptotic function of herpes simplex virus -2 latency-associated transcript RL1 sequence and screening of its encoded microRNAs. Clin. Exp. Dermatol. 2016, 41, 782–791. [Google Scholar] [CrossRef]
- Feederle, R.; Linnstaedt, S.D.; Bannert, H.; Lips, H.; Bencun, M.; Cullen, B.R.; Delecluse, H.J. A viral microRNA cluster strongly potentiates the transforming properties of a human herpesvirus. PLoS Pathog. 2011, 7, e1001294. [Google Scholar] [CrossRef]
- Feederle, R.; Haar, J.; Bernhardt, K.; Linnstaedt, S.D.; Bannert, H.; Lips, H.; Cullen, B.R.; Delecluse, H.J. The members of an Epstein-Barr virus microRNA cluster cooperate to transform B lymphocytes. J. Virol. 2011, 85, 9801–9810. [Google Scholar] [CrossRef]
- Qiu, J.; Cosmopoulos, K.; Pegtel, M.; Hopmans, E.; Murray, P.; Middeldorp, J.; Shapiro, M.; Thorley-Lawson, D.A. A novel persistence associated EBV miRNA expression profile is disrupted in neoplasia. PLoS Pathog. 2011, 7, e1002193. [Google Scholar] [CrossRef]
- Vereide, D.T.; Seto, E.; Chiu, Y.F.; Hayes, M.; Tagawa, T.; Grundhoff, A.; Hammerschmidt, W.; Sugden, B. Epstein-Barr virus maintains lymphomas via its miRNAs. Oncogene 2014, 33, 1258–1264. [Google Scholar] [CrossRef] [PubMed]
- Nachmani, D.; Stern-Ginossar, N.; Sarid, R.; Mandelboim, O. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe 2009, 5, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.M.; Kong, Y.L.; Wang, L.; Zhu, H.Y.; Wu, J.Z.; Xia, Y.; Li, Y.; Qin, S.C.; Fan, L.; Li, J.Y.; et al. EBV-miR-BHRF1-1 Targets p53 Gene: Potential Role in Epstein-Barr Virus Associated Chronic Lymphocytic Leukemia. Cancer Res. Treat. Off. J. Korean Cancer Assoc. 2020, 52, 492–504. [Google Scholar] [CrossRef]
- Suffert, G.; Malterer, G.; Hausser, J.; Viiliainen, J.; Fender, A.; Contrant, M.; Ivacevic, T.; Benes, V.; Gros, F.; Voinnet, O.; et al. Kaposi’s sarcoma herpesvirus microRNAs target caspase 3 and regulate apoptosis. PLoS Pathog. 2011, 7, e1002405. [Google Scholar] [CrossRef] [PubMed]
- Gottwein, E.; Cullen, B.R. A human herpesvirus microRNA inhibits p21 expression and attenuates p21-mediated cell cycle arrest. J. Virol. 2010, 84, 5229–5237. [Google Scholar] [CrossRef]
- Reeves, M.B.; Davies, A.A.; McSharry, B.P.; Wilkinson, G.W.; Sinclair, J.H. Complex I binding by a virally encoded RNA regulates mitochondria-induced cell death. Science 2007, 316, 1345–1348. [Google Scholar] [CrossRef] [PubMed]
- Slonchak, A.; Hugo, L.E.; Freney, M.E.; Hall-Mendelin, S.; Amarilla, A.A.; Torres, F.J.; Setoh, Y.X.; Peng, N.Y.G.; Sng, J.D.J.; Hall, R.A.; et al. Zika virus noncoding RNA suppresses apoptosis and is required for virus transmission by mosquitoes. Nat. Commun. 2020, 11, 2205. [Google Scholar] [CrossRef]
- Lee, S.H.; Kalejta, R.F.; Kerry, J.; Semmes, O.J.; O’Connor, C.M.; Khan, Z.; Garcia, B.A.; Shenk, T.; Murphy, E. BclAF1 restriction factor is neutralized by proteasomal degradation and microRNA repression during human cytomegalovirus infection. Proc. Natl. Acad. Sci. USA 2012, 109, 9575–9580. [Google Scholar] [CrossRef]
- Riley, K.J.; Rabinowitz, G.S.; Yario, T.A.; Luna, J.M.; Darnell, R.B.; Steitz, J.A. EBV and human microRNAs co-target oncogenic and apoptotic viral and human genes during latency. EMBO J. 2012, 31, 2207–2221. [Google Scholar] [CrossRef]
- Ziegelbauer, J.M.; Sullivan, C.S.; Ganem, D. Tandem array-based expression screens identify host mRNA targets of virus-encoded microRNAs. Nat. Genet. 2009, 41, 130–134. [Google Scholar] [CrossRef]
- Kincaid, R.P.; Sullivan, C.S. Virus-encoded microRNAs: An overview and a look to the future. PLoS Pathog. 2012, 8, e1003018. [Google Scholar] [CrossRef] [PubMed]
- Ray, R.M.; Morris, K.V. Long Non-coding RNAs Mechanisms of Action in HIV-1 Modulation and the Identification of Novel Therapeutic Targets. Non-Coding RNA 2020, 6, 12. [Google Scholar] [CrossRef] [PubMed]
- Cliffe, A.R.; Garber, D.A.; Knipe, D.M. Transcription of the herpes simplex virus latency-associated transcript promotes the formation of facultative heterochromatin on lytic promoters. J. Virol. 2009, 83, 8182–8190. [Google Scholar] [CrossRef] [PubMed]
- Inman, M.; Perng, G.C.; Henderson, G.; Ghiasi, H.; Nesburn, A.B.; Wechsler, S.L.; Jones, C. Region of herpes simplex virus type 1 latency-associated transcript sufficient for wild-type spontaneous reactivation promotes cell survival in tissue culture. J. Virol. 2001, 75, 3636–3646. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, T.M.; Kulesza, C.A. Stability determinants of murine cytomegalovirus long noncoding RNA7.2. J. Virol. 2014, 88, 11630–11633. [Google Scholar] [CrossRef][Green Version]
- Moss, W.N.; Steitz, J.A. Genome-wide analyses of Epstein-Barr virus reveal conserved RNA structures and a novel stable intronic sequence RNA. BMC Genom. 2013, 14, 543. [Google Scholar] [CrossRef]
- Kelly, G.L.; Long, H.M.; Stylianou, J.; Thomas, W.A.; Leese, A.; Bell, A.I.; Bornkamm, G.W.; Mautner, J.; Rickinson, A.B.; Rowe, M. An Epstein-Barr virus anti-apoptotic protein constitutively expressed in transformed cells and implicated in burkitt lymphomagenesis: The Wp/BHRF1 link. PLoS Pathog. 2009, 5, e1000341. [Google Scholar] [CrossRef]
- Sun, R.; Lin, S.F.; Gradoville, L.; Miller, G. Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 1996, 93, 11883–11888. [Google Scholar] [CrossRef]
- Campbell, M.; Izumiya, Y. PAN RNA: Transcriptional exhaust from a viral engine. J. Biomed. Sci. 2020, 27, 41. [Google Scholar] [CrossRef]
- Rossetto, C.C.; Pari, G. KSHV PAN RNA associates with demethylases UTX and JMJD3 to activate lytic replication through a physical interaction with the virus genome. PLoS Pathog. 2012, 8, e1002680. [Google Scholar] [CrossRef]
- Campbell, M.; Kim, K.Y.; Chang, P.C.; Huerta, S.; Shevchenko, B.; Wang, D.H.; Izumiya, C.; Kung, H.J.; Izumiya, Y. A lytic viral long noncoding RNA modulates the function of a latent protein. J. Virol. 2014, 88, 1843–1848. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, A.R.; Shango, J.; Seal, A.; Shukla, D.; Nares, S. Viral miRNAs Alter Host Cell miRNA Profiles and Modulate Innate Immune Responses. Front. Immunol. 2018, 9, 433. [Google Scholar] [CrossRef] [PubMed]
- Grey, F.; Meyers, H.; White, E.A.; Spector, D.H.; Nelson, J. A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog. 2007, 3, e163. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhu, J.; Zhang, W.; Chen, Y.; Zhang, K.; Popescu, L.M.; Ma, X.; Lau, W.B.; Rong, R.; Yu, X.; et al. Signature microRNA expression profile of essential hypertension and its novel link to human cytomegalovirus infection. Circulation 2011, 124, 175–184. [Google Scholar] [CrossRef]
- Shen, K.; Xu, L.; Chen, D.; Tang, W.; Huang, Y. Human cytomegalovirus-encoded miR-UL112 contributes to HCMV-mediated vascular diseases by inducing vascular endothelial cell dysfunction. Virus Genes 2018, 54, 172–181. [Google Scholar] [CrossRef]
- Moody, R.; Zhu, Y.; Huang, Y.; Cui, X.; Jones, T.; Bedolla, R.; Lei, X.; Bai, Z.; Gao, S.J. KSHV microRNAs mediate cellular transformation and tumorigenesis by redundantly targeting cell growth and survival pathways. PLoS Pathog. 2013, 9, e1003857. [Google Scholar] [CrossRef]
- Lin, C.; Zong, J.; Lin, W.; Wang, M.; Xu, Y.; Zhou, R.; Lin, S.; Guo, Q.; Chen, H.; Ye, Y.; et al. EBV-miR-BART8-3p induces epithelial-mesenchymal transition and promotes metastasis of nasopharyngeal carcinoma cells through activating NF-kappaB and Erk1/2 pathways. J. Exp. Clin. Cancer Res. CR 2018, 37, 283. [Google Scholar] [CrossRef]
- Xu, Y.J.; Zhou, R.; Zong, J.F.; Lin, W.S.; Tong, S.; Guo, Q.J.; Lin, C.; Lin, S.J.; Chen, Y.X.; Chen, M.R.; et al. Epstein-Barr virus-coded miR-BART13 promotes nasopharyngeal carcinoma cell growth and metastasis via targeting of the NKIRAS2/NF-kappaB pathway. Cancer Lett. 2019, 447, 33–40. [Google Scholar] [CrossRef]
- Qiu, J.; Thorley-Lawson, D.A. EBV microRNA BART 18-5p targets MAP3K2 to facilitate persistence in vivo by inhibiting viral replication in B cells. Proc. Natl. Acad. Sci. USA 2014, 111, 11157–11162. [Google Scholar] [CrossRef]
- Lei, X.; Bai, Z.; Ye, F.; Xie, J.; Kim, C.G.; Huang, Y.; Gao, S.J. Regulation of NF-kappaB inhibitor IkappaBalpha and viral replication by a KSHV microRNA. Nat. Cell Biol. 2010, 12, 193–199. [Google Scholar] [CrossRef]
- Lu, F.; Stedman, W.; Yousef, M.; Renne, R.; Lieberman, P.M. Epigenetic regulation of Kaposi’s sarcoma-associated herpesvirus latency by virus-encoded microRNAs that target Rta and the cellular Rbl2-DNMT pathway. J. Virol. 2010, 84, 2697–2706. [Google Scholar] [CrossRef] [PubMed]
- Gregorovic, G.; Bosshard, R.; Karstegl, C.E.; White, R.E.; Pattle, S.; Chiang, A.K.; Dittrich-Breiholz, O.; Kracht, M.; Russ, R.; Farrell, P.J. Cellular gene expression that correlates with EBER expression in Epstein-Barr Virus-infected lymphoblastoid cell lines. J. Virol. 2011, 85, 3535–3545. [Google Scholar] [CrossRef] [PubMed]
- Clarke, P.A.; Sharp, N.A.; Clemens, M.J. Translational control by the Epstein-Barr virus small RNA EBER-1. Reversal of the double-stranded RNA-induced inhibition of protein synthesis in reticulocyte lysates. Eur. J. Biochem. 1990, 193, 635–641. [Google Scholar] [CrossRef]
- Song, Y.; Li, Q.; Liao, S.; Zhong, K.; Jin, Y.; Zeng, T. Epstein-Barr virus-encoded miR-BART11 promotes tumor-associated macrophage-induced epithelial-mesenchymal transition via targeting FOXP1 in gastric cancer. Virology 2020, 548, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.M.; Lyu, X.M.; Luo, W.R.; Cui, X.F.; Ye, Y.F.; Yuan, C.C.; Peng, Q.X.; Wu, D.H.; Liu, T.F.; Wang, E.; et al. EBV-miR-BART7-3p promotes the EMT and metastasis of nasopharyngeal carcinoma cells by suppressing the tumor suppressor PTEN. Oncogene 2015, 34, 2156–2166. [Google Scholar] [CrossRef]
- Wang, L.; Damania, B. Kaposi’s sarcoma-associated herpesvirus confers a survival advantage to endothelial cells. Cancer Res. 2008, 68, 4640–4648. [Google Scholar] [CrossRef]
- Li, T.; Ju, E.; Gao, S.J. Kaposi sarcoma-associated herpesvirus miRNAs suppress CASTOR1-mediated mTORC1 inhibition to promote tumorigenesis. J. Clin. Investig. 2019, 129, 3310–3323. [Google Scholar] [CrossRef]
- Cheng, Y.; Geng, L.; Zhao, L.; Zuo, P.; Wang, J. Human papillomavirus E6-regulated microRNA-20b promotes invasion in cervical cancer by targeting tissue inhibitor of metalloproteinase 2. Mol. Med. Rep. 2017, 16, 5464–5470. [Google Scholar] [CrossRef]
- Zhu, Y.K.; Cheng, N.; Hu, Y.; Cen, Y.Z. The role of microRNAs in the pathogenesis of cervical cancer and its relationship to HPV. Sheng Li Ke Xue Jin Zhan Prog. Physiol. 2012, 43, 251–256. [Google Scholar]
- Amodio, N.; Cantafio, M.E.G.; Botta, C.; Agosti, V.; Federico, C.; Caracciolo, D.; Ronchetti, D.; Rossi, M.; Driessen, C.; Neri, A.; et al. Replacement of miR-155 Elicits Tumor Suppressive Activity and Antagonizes Bortezomib Resistance in Multiple Myeloma. Cancers 2019, 11, 236. [Google Scholar] [CrossRef]
- Boss, I.W.; Nadeau, P.E.; Abbott, J.R.; Yang, Y.; Mergia, A.; Renne, R. A Kaposi’s sarcoma-associated herpesvirus-encoded ortholog of microRNA miR-155 induces human splenic B-cell expansion in NOD/LtSz-scid IL2Rgammanull mice. J. Virol. 2011, 85, 9877–9886. [Google Scholar] [CrossRef]
- Dahlke, C.; Maul, K.; Christalla, T.; Walz, N.; Schult, P.; Stocking, C.; Grundhoff, A. A microRNA encoded by Kaposi sarcoma-associated herpesvirus promotes B-cell expansion in vivo. PLoS ONE 2012, 7, e49435. [Google Scholar] [CrossRef] [PubMed]
- Morrison, K.; Manzano, M.; Chung, K.; Schipma, M.J.; Bartom, E.T.; Gottwein, E. The Oncogenic Kaposi’s Sarcoma-Associated Herpesvirus Encodes a Mimic of the Tumor-Suppressive miR-15/16 miRNA Family. Cell Rep. 2019, 29, 2961–2969. [Google Scholar] [CrossRef] [PubMed]
- Kincaid, R.P.; Chen, Y.; Cox, J.E.; Rethwilm, A.; Sullivan, C.S. Noncanonical microRNA (miRNA) biogenesis gives rise to retroviral mimics of lymphoproliferative and immunosuppressive host miRNAs. mBio 2014, 5, e00074. [Google Scholar] [CrossRef] [PubMed]
- Chi, J.Q.; Teng, M.; Yu, Z.H.; Xu, H.; Su, J.W.; Zhao, P.; Xing, G.X.; Liang, H.D.; Deng, R.G.; Qu, L.H.; et al. Marek’s disease virus-encoded analog of microRNA-155 activates the oncogene c-Myc by targeting LTBP1 and suppressing the TGF-beta signaling pathway. Virology 2015, 476, 72–84. [Google Scholar] [CrossRef]
- Kincaid, R.P.; Burke, J.M.; Sullivan, C.S. RNA virus microRNA that mimics a B-cell oncomiR. Proc. Natl. Acad. Sci. USA 2012, 109, 3077–3082. [Google Scholar] [CrossRef]
- You, X.; Zhang, Z.; Fan, J.; Cui, Z.; Zhang, X.E. Functionally orthologous viral and cellular microRNAs studied by a novel dual-fluorescent reporter system. PLoS ONE 2012, 7, e36157. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Nigro, J.M.; Cho, K.R.; Fearon, E.R.; Kern, S.E.; Ruppert, J.M.; Oliner, J.D.; Kinzler, K.W.; Vogelstein, B. Scrambled exons. Cell 1991, 64, 607–613. [Google Scholar] [CrossRef]
- Chen, L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016, 17, 205–211. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Toptan, T.; Abere, B.; Nalesnik, M.A.; Swerdlow, S.H.; Ranganathan, S.; Lee, N.; Shair, K.H.; Moore, P.S.; Chang, Y. Circular DNA tumor viruses make circular RNAs. Proc. Natl. Acad. Sci. USA 2018, 115, E8737–E8745. [Google Scholar] [CrossRef] [PubMed]
- Tagawa, T.; Gao, S.; Koparde, V.N.; Gonzalez, M.; Spouge, J.L.; Serquina, A.P.; Lurain, K.; Ramaswami, R.; Uldrick, T.S.; Yarchoan, R.; et al. Discovery of Kaposi’s sarcoma herpesvirus-encoded circular RNAs and a human antiviral circular RNA. Proc. Natl. Acad. Sci. USA 2018, 115, 12805–12810. [Google Scholar] [CrossRef] [PubMed]
- Ungerleider, N.; Concha, M.; Lin, Z.; Roberts, C.; Wang, X.; Cao, S.; Baddoo, M.; Moss, W.N.; Yu, Y.; Seddon, M.; et al. The Epstein Barr virus circRNAome. PLoS Pathog. 2018, 14, e1007206. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.T.; Chen, J.N.; Gong, L.P.; Bi, Y.H.; Liang, J.; Zhou, L.; He, D.; Shao, C.K. Identification of virus-encoded circular RNA. Virology 2019, 529, 144–151. [Google Scholar] [CrossRef]
- Liu, Q.; Shuai, M.; Xia, Y. Knockdown of EBV-encoded circRNA circRPMS1 suppresses nasopharyngeal carcinoma cell proliferation and metastasis through sponging multiple miRNAs. Cancer Manag. Res. 2019, 11, 8023–8031. [Google Scholar] [CrossRef]
| Mechanisms Influenced by v-ncRNAs | Target Genes | Molecular Mechanism | v-ncRNAs | References |
|---|---|---|---|---|
| PKR, Dicer | PKR and Dicer binding and competitive inhibition | EBERs (EBV) VAI, VA2 (Adenoviruses) TAR-RNA (HIV) | [9] | |
| IRF4 | Downregulation of IRF4-responsive promoter | PAN-RNA (KSHV) | [27] | |
| IFN Type I Secretion | STAT2 | STAT2 binding and depletion via proteasomal degradation | sfRNA1, sfRNA2 (Zika) | [28,29] |
| ISGs | viral RNA-binding ISGs binding and competitive inhibition | sfRNAs (JCV, WNV, Dengue) | [30] | |
| RIG-1 | 3′ UTR RIG-I mRNA binding and inhibition of protein expression | ebv-miR-BART6-3p (EBV) | [31] | |
| CREBBP | 3′ UTR CREBBP mRNA binding and degradation induction | ebv-miR-BART16 (EBV) | [33] | |
| IKKε | 3′ UTR IKKε mRNA binding and inhibition of protein expression | kshv-miR-K12-11 (KSHV) | [34] | |
| IL-6R | mRNA binding and degradation induction | ebv-miR-BART6-3p (EBV) [+ host hsa-miR-197] | [32] | |
| TWEAKR | 3′ UTR TWEAKR mRNA binding and degradation induction | kshv-miR-K12-10 (KSHV) | [35] | |
| Cytokines Production Impairment | IL-1R | mRNA binding and and degradation induction | ebv-miR-BHRF-1-2-5p (EBV) | [36] |
| IL-1β pathway | 3′ UTR NLRP3 mRNA binding | ebv-miR-BART15 (EBV) | [37] | |
| MyD88 | 3′ UTR MyD88 mRNA binding and inhibition of protein expression | kshv-miR-K12-5 (KSHV) | [35] | |
| IRAK1 | 3′ UTR IRAK1 mRNA binding and inhibition of protein expression | kshv-miR-K12-9 (KSHV) | [35] | |
| CTSB (MHC-1) | 3′ UTR CTSB mRNA binding and degradation induction | ebv-miR-BART2m (EBV) | [38] | |
| TAP2 (MHC-1) | 3′ UTR TAP2 mRNA binding and inhibition of protein expression | ebv-miR-BHRF1-3 (EBV) | [38] | |
| Antigen Presentation | LY75 (MHC-1) | 3′ UTR LY75 mRNA binding and inhibition of protein expression | ebv-miR-BART1-5p (EBV) | [39] |
| IFI30 (MHC-II) | 3′ UTR IFI30 mRNA binding and degradation induction | ebv-miR-BART1 (EBV) | [38] | |
| LGMN (MHC-II) | 3′ UTR LGMN mRNA binding and degradation induction | ebv-miR-BART2 (EBV) | [38] | |
| Neutrophils Chemotaxis | SP100 (CXCL8) | 3′ UTR SP100 mRNA binding and translational repression | MCV-miR-M1-5p (MCPyV) | [40] |
| BCL11A, CHD7, ITGAM, RAG-1, TCEA1 | In silico analysis | HPV16-miR-H1-1 (HPV) | [41] | |
| SP3, XRCC4, JAK2, PKNOX1, FOXP1 | In silico analysis | HPV16-miR-H2 (HPV) | [41] | |
| IL12B | 3′ UTR IL12B mRNA binding and degradation induction | ebv-miR-BART1, -miR-BART2, -miR-BART10, -miR-BART22, -miR-BHRF1 (EBV) | [38,39] | |
| T Cells Behaviour | CXCL11 | CXCL11 mRNA binding and inhibition of protein expression | ebv-miR-BHRF1-3 (EBV) | [42] |
| Viral T Antigen | ncRNA sequence binding | sv40-miR-S1, jcv-miR-J1 (JCV, BKV, SV40, MCPyV) | [43] | |
| ASP | Noncoding promoter silencing | HIV-ncRNAs | [45] | |
| Importin-α5 | 3′ UTR Importin-α5 mRNA binding and inhibition of protein expression | EBOV-miR-1-5p (Ebola) | [48] | |
| NK Cells Evasion | MICB | 3′ UTR MICB mRNA binding and inhibition of protein expression | cmv-miR-UL112-1 (CMV), ebv-miR-BART2-5p (EBV), kshv-miR-K12-7 (KSHV) | [49,50,51] |
| ULBP3 | 3′ UTR ULBP3 mRNA binding and protein translation inhibition | Jcv-miR-J1-3p (JCV, BKV) | [52] |
| Mechanisms Influenced by v-ncRNAs | Target Genes | v-ncRNAs | Reference |
|---|---|---|---|
| TIA-1 | mivaRNAI-138 (Adenovirus) | [73] | |
| PKR/eIF-2 | VAI-RNA (Adenovirus) | [74] | |
| TGFβ1, SMAD3 | LAT (HSV-1) | [75] | |
| ActD | miR-H3, miR-H4-3p, miR-H4-5p, miR-H24, miR-H19 (HSV-2) | [77] | |
| Cell Survival (Pro-Apoptotic Factors) | CASP3, PUMA, p53 | EBV-miRNAs | [81,82,83] |
| CASP3 | kshv-miR-K12-1, -miR-K12-3, -miR-K12-4-3p (KSHV) | [84] | |
| p21 | kshv-miR-K12-1 (KSHV) | [85] | |
| Mithocondrial Complex -1 | β-2.7 (MCPyV) | [86] | |
| CASP7 | sfRNAs (Zika) | [87] | |
| BclAF1 | cmv-miR-UL112-1 (CMV), ebv-miR-BART-17-5p (EBV), kshv-miR-K5 (KSHV) | [88] | |
| EZH2, DNMT3a, HDAC1 | HIV1-ncRNAs | [45,92] | |
| HSV1-sisRNAs | [93,94] | ||
| CMV-sisRNAs | [95] | ||
| EBV-sisRNA-1 | [97] | ||
| JMJD3, UTX, LANA | PAN-RNA (KSHV) | [100,101] | |
| MAPK | cmv-miR-UL70 (CMV) | [102] | |
| Virus Infection/Replication | IE72 | cmv-miR-UL112 (CMV) | [103] |
| MAPK, TSPYL2, FXYD2, TAOK2, ST7L, TP73 | cmv-miR-UL112 (CMV) | [104,105] | |
| RNF38, NKIRAS2 | ebv-miR-BART8-3p, -miR-BART13 (EBV) | [106,107,108] | |
| MAP3K2 | ebv-miR-BART18-5p (EBV) | [109] | |
| IkB, NFkB | kshv-miR-K1, -miR-K12-1 (KSHV) | [110] | |
| Rbl2 | kshv-miR-K12-4-5p (KSHV) | [111] | |
| PAN-RNA (KSHV) | [54] | ||
| eIF-2 kinase | EBER1, EBER2 (EBV) | [57,113] | |
| Cell Transformation | FOXP1 | ebv-miR-BART11 (EBV) | [114] |
| PTEN | ebv-miR-BART7-3p (EBV) | [115] | |
| RNF38 | ebv-miR-BART8-3p (EBV) | [107] | |
| CASTOR1 | KSHV-miRNAs | [117] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gallo, A.; Bulati, M.; Miceli, V.; Amodio, N.; Conaldi, P.G. Non-Coding RNAs: Strategy for Viruses’ Offensive. Non-Coding RNA 2020, 6, 38. https://doi.org/10.3390/ncrna6030038
Gallo A, Bulati M, Miceli V, Amodio N, Conaldi PG. Non-Coding RNAs: Strategy for Viruses’ Offensive. Non-Coding RNA. 2020; 6(3):38. https://doi.org/10.3390/ncrna6030038
Chicago/Turabian StyleGallo, Alessia, Matteo Bulati, Vitale Miceli, Nicola Amodio, and Pier Giulio Conaldi. 2020. "Non-Coding RNAs: Strategy for Viruses’ Offensive" Non-Coding RNA 6, no. 3: 38. https://doi.org/10.3390/ncrna6030038
APA StyleGallo, A., Bulati, M., Miceli, V., Amodio, N., & Conaldi, P. G. (2020). Non-Coding RNAs: Strategy for Viruses’ Offensive. Non-Coding RNA, 6(3), 38. https://doi.org/10.3390/ncrna6030038






