Small RNA Species and microRNA Profiles are Altered in Severe Asthma Nanovesicles from Broncho Alveolar Lavage and Associate with Impaired Lung Function and Inflammation
Abstract
1. Introduction
2. Methods
2.1. Subjects and Sampling
2.2. Ethics
2.3. Nanovesicle Small RNA-Sequencing
2.3.1. Extraction of Nanovesicle RNA from BAL
2.3.2. Small RNA-Sequencing
2.4. Pathway Analysis for Differentially Expressed microRNAs in Severe Asthmatic Nanovesicles
2.5. Western Blotting
2.6. Validation of Small-RNA Sequencing Using qPCR
3. Statistics
3.1. Statistical Analysis
3.2. Power Calculation
4. Results
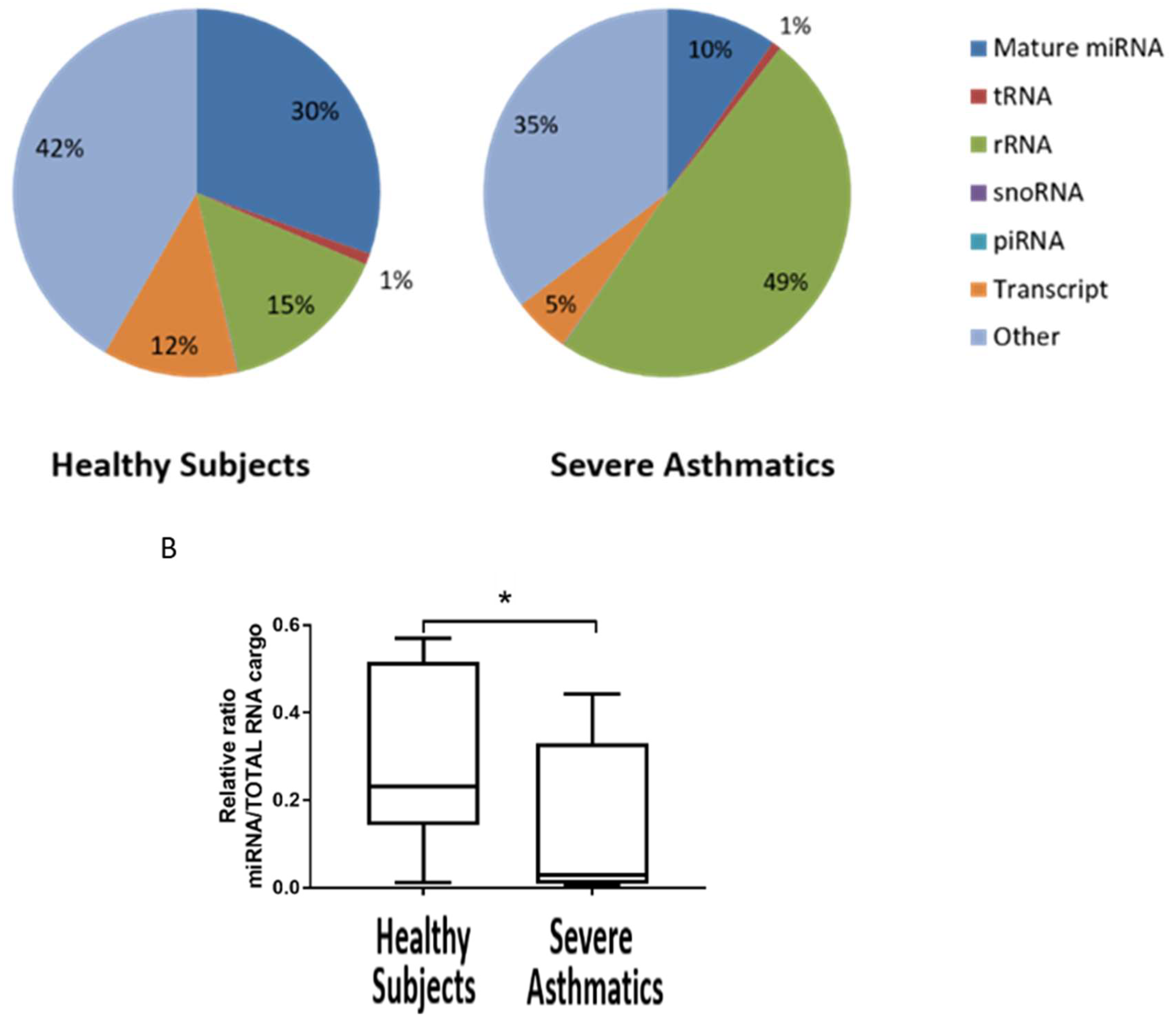
4.1. Small RNA Cargo Is Altered in Severe Asthma Nanovesicles
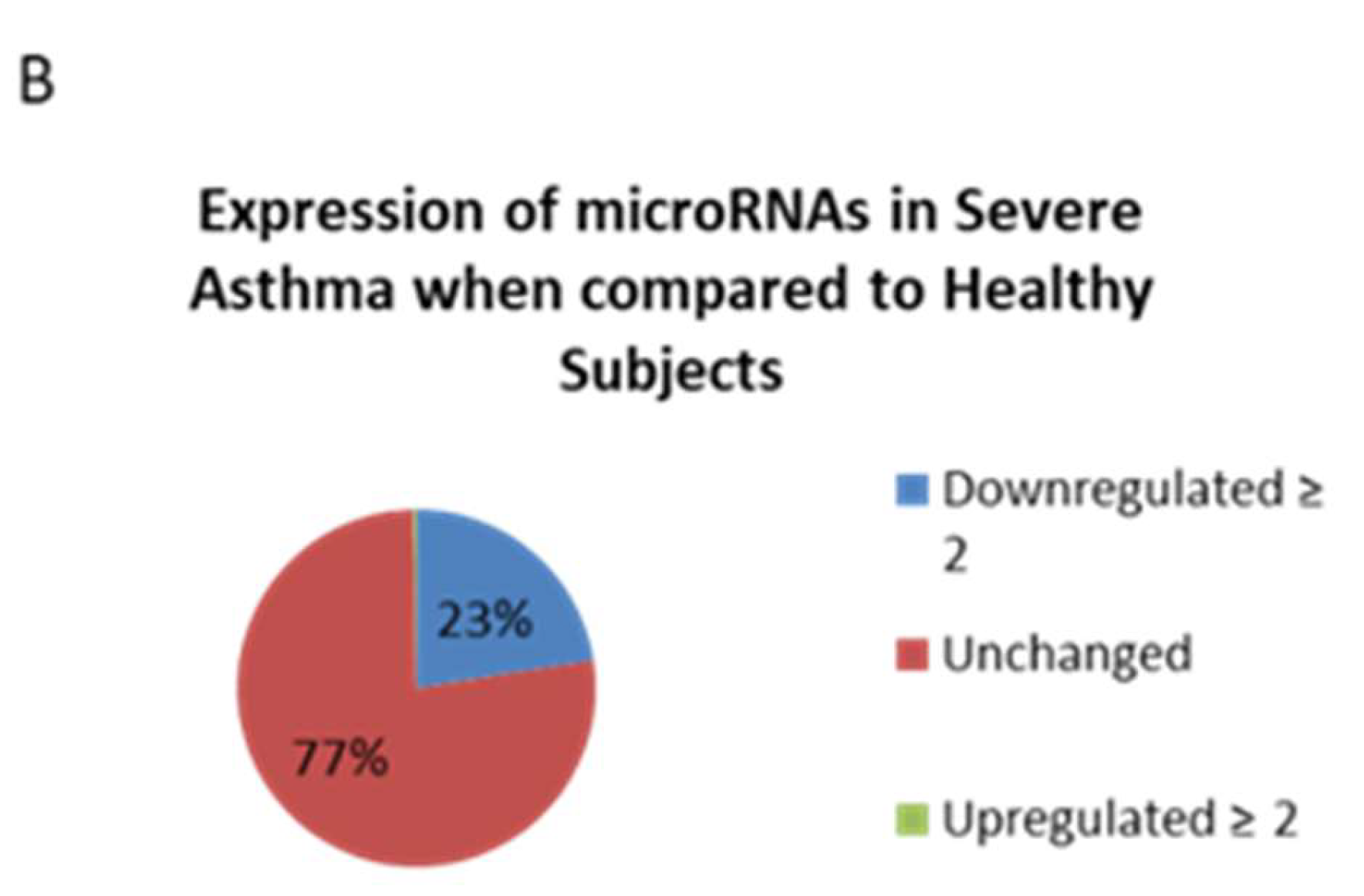
4.2. Reduced Load of microRNAs in Nanovesicles from Severe Asthmatics Patients
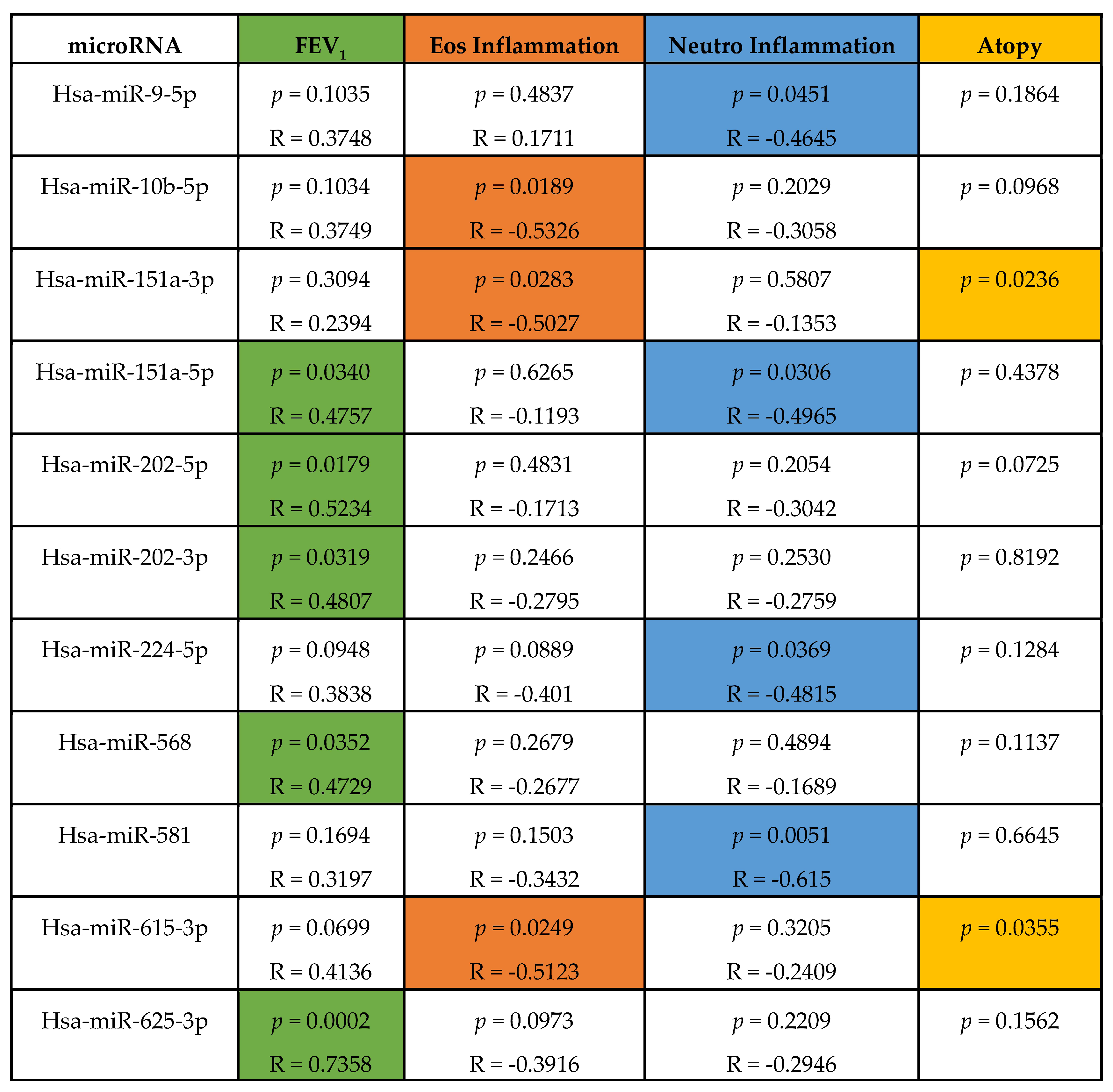
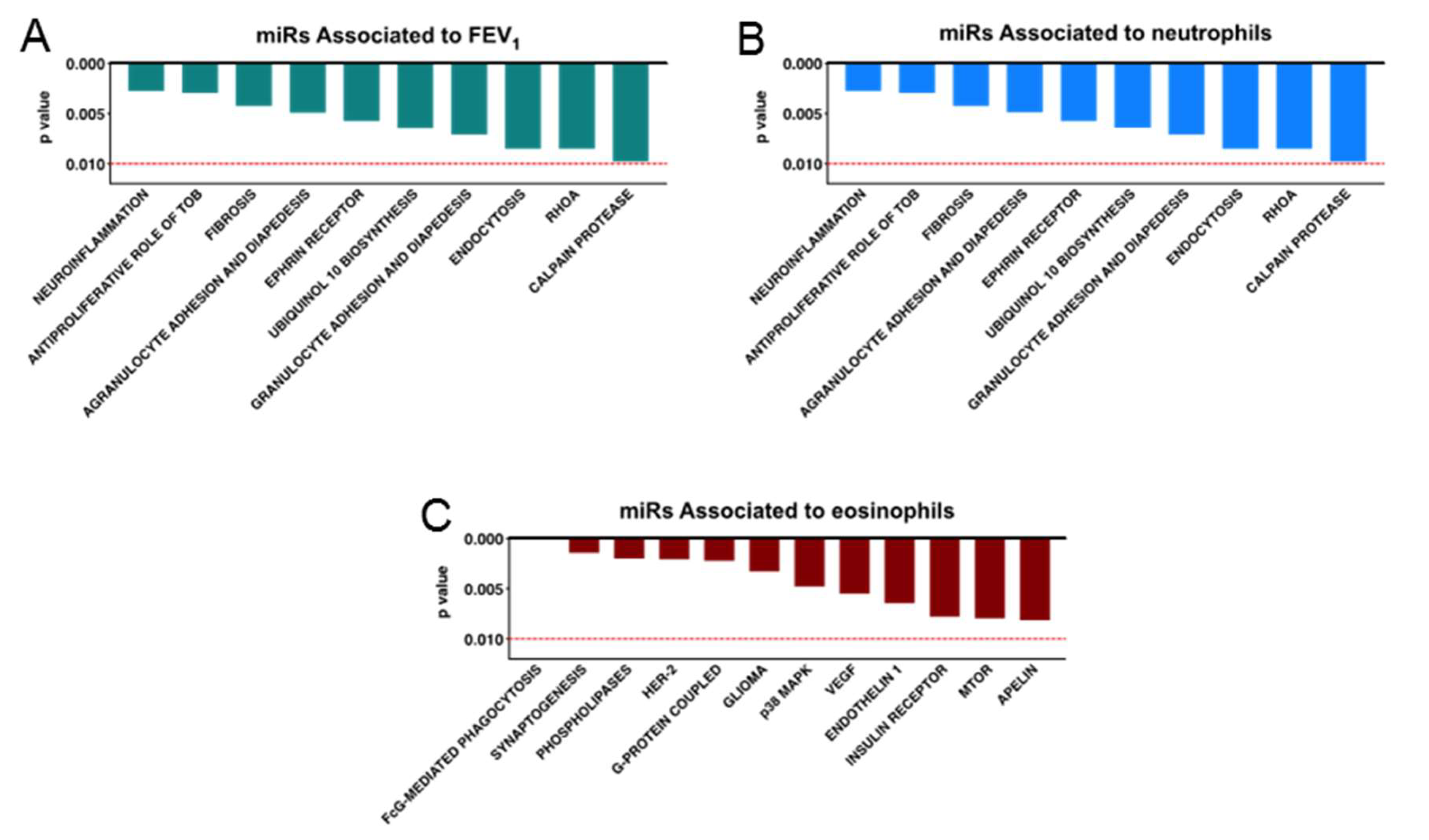
4.3. Altered microRNA Content of Severe Asthmatic Nanovesicles Affects Cellular Pathways Involved in Airway Inflammation and Remodeling, and Correlates with FEV1 and Differential Eosinophil and Neutrophil Infiltration
4.4. MicroRNAs Associated with FEV1, Neutrophil, and Eosinophil Infiltration Regulate Pathways Relevant to Asthma Pathology
5. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Trams, E.G.; Lauter, C.J.; Salem, N., Jr.; Heine, U. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim. Biophys. Acta. 1981, 645, 63–70. [Google Scholar]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Rupani, H.; Martinez-Nunez, R.T.; Dennison, P.; Lau, L.C.; Jayasekera, N.; Havelock, T.; Francisco-Garcia, A.S.; Grainge, C.; Howarth, P.H.; Sanchez-Elsner, T. Toll-like Receptor 7 Is Reduced in Severe Asthma and Linked to an Altered MicroRNA Profile. Am. J. Respir. Crit. Care Med. 2016, 194, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Nunez, R.T.; Bondanese, V.P.; Louafi, F.; Francisco-Garcia, A.S.; Rupani, H.; Bedke, N.; Holgate, S.; Howarth, P.H.; Davies, D.E.; Sanchez-Elsner, T. A microRNA network dysregulated in asthma controls IL-6 production in bronchial epithelial cells. PLoS ONE 2014, 9, e111659. [Google Scholar] [CrossRef] [PubMed]
- Sinha, A.; Yadav, A.K.; Chakraborty, S.; Kabra, S.K.; Lodha, R.; Kumar, M.; Kulshreshtha, A.; Sethi, T.; Pandey, R.; Malik, G.; et al. Exosome-enclosed microRNAs in exhaled breath hold potential for biomarker discovery in patients with pulmonary diseases. J. Allergy Clin. Immunol. 2013, 132, 219–222. [Google Scholar] [CrossRef] [PubMed]
- Levanen, B.; Bhakta, N.R.; Torregrosa Paredes, P.; Barbeau, R.; Hiltbrunner, S.; Pollack, J.L.; Sköld, C.M.; Svartengren, M.; Grunewald, J.; Gabrielsson, S.; et al. Altered microRNA profiles in bronchoalveolar lavage fluid exosomes in asthmatic patients. J. Allergy Clin. Immunol. 2013, 131, 894–903. [Google Scholar] [CrossRef] [PubMed]
- Maes, T.; Cobos, F.A.; Schleich, F.; Sorbello, V.; Henket, M.; De Preter, K.; Bracke, K.R.; Conickx, G.; Mesnil, C.; Vandesompele, J.; et al. Asthma inflammatory phenotypes show differential microRNA expression in sputum. J. Allergy Clin. Immunol. 2016, 137, 1433–1446. [Google Scholar] [CrossRef] [PubMed]
- Hinks, T.S.C.; Brown, T.; Lau, L.C.K.; Rupani, H.; Barber, C.; Elliott, S.; Ward, J.A.; Ono, J.; Ohta, S.; Izuhara, K.; et al. Multidimensional endotyping in patients with severe asthma reveals inflammatory heterogeneity in matrix metalloproteinases and chitinase 3-like protein 1. J. Allergy Clin. Immunol. 2016, 138, 61–75. [Google Scholar] [CrossRef] [PubMed]
- Lab, H. FASTX-Toolkit. Available online: http://hannonlab.cshl.edu/fastx_toolkit/ (accessed on 2 April 2014).
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Method. 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Kirov, S.; Snoddy, J. WebGestalt: An integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005, 33, W741–W748. [Google Scholar] [CrossRef] [PubMed]
- Faul, F.; Erdfelder, E.; Lang, A.G.; Buchner, A. G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods 2007, 39, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Huaux, F.; Gharaee-Kermani, M.; Liu, T.; Morel, V.; McGarry, B.; Ullenbruch, M.; Kunkel, S.L.; Wang, J.; Xing, Z.; Phan, S.H. Role of Eotaxin-1 (CCL11) and CC chemokine receptor 3 (CCR3) in bleomycin-induced lung injury and fibrosis. Am. J. Pathol. 2005, 167, 1485–1496. [Google Scholar] [CrossRef]
- Nouailles, G.; Dorhoi, A.; Koch, M.; Zerrahn, J.; Weiner, J., 3rd; Faé, K.C.; Arrey, F.; Kuhlmann, S.; Bandermann, S.; Loewe, D.; et al. CXCL5-secreting pulmonary epithelial cells drive destructive neutrophilic inflammation in tuberculosis. J. Clin. Investig. 2014, 124, 1268–1282. [Google Scholar] [CrossRef] [PubMed]
- Amatngalim, G.D.; van Wijck, Y.; de Mooij-Eijk, Y.; Verhoosel, R.M.; Harder, J.; Lekkerkerker, A.N.; Janssen, R.A.; Hiemstra, P.S. Basal cells contribute to innate immunity of the airway epithelium through production of the antimicrobial protein RNase 7. J. Immunol. 2015, 194, 3340–3350. [Google Scholar] [CrossRef] [PubMed]
- Singhania, A.; Rupani, H.; Jayasekera, N.; Lumb, S.; Hales, P.; Gozzard, N.; Davies, D.E.; Woelk, C.H.; Howarth, P.H. Altered Epithelial Gene Expression in Peripheral Airways of Severe Asthma. PLoS ONE 2017, 12, e0168680. [Google Scholar] [CrossRef] [PubMed]

| ID | Sex | Age | Atopy | FEV1% | Eos % | Neut % | ACQ | GINA Step |
|---|---|---|---|---|---|---|---|---|
| H1 | F | 51 | N | 124 | 0.25 | 2.5 | 0 | 0 |
| H2 | M | 50 | N | 94 | 0 | 0.5 | 0 | 0 |
| H3 | F | 44 | N | 88 | 0 | 1.8 | 0 | 0 |
| H4 | F | 38 | N | 97 | 0.5 | 3.5 | 0 | 0 |
| H5 | F | 58 | N | 137 | 0.25 | 4.5 | 0 | 0 |
| H6 | M | 29 | N | 85.3 | 0.25 | 4.3 | 0 | 0 |
| H7 | M | 26 | N | 104 | 0 | 1.25 | 0 | 0 |
| H8 | F | 31 | Y | 110 | 1 | 0.75 | 0 | 0 |
| SA1 | M | 62 | N | 53 | 9.3 | 10 | 4.2 | 4 |
| SA2 | F | 31 | N | 57 | 0 | 2.8 | 4 | 5 |
| SA3 | M | 59 | N | 47.5 | 0.5 | 3.3 | 4.29 | 5 |
| SA4 | F | 21 | Y | 46 | 0 | 8.3 | 3.6 | 5 |
| SA5 | F | 63 | N | 77.8 | 2 | 10 | 3.29 | 4 |
| SA6 | F | 43 | Y | 46.5 | 0.3 | 3.3 | 3.57 | 4 |
| SA7 | F | 26 | N | 71.5 | 0 | 4.3 | 1.71 | 4 |
| SA8 | M | 61 | Y | 88 | 4.3 | 28.3 | 3.3 | 4 |
| SA9 | F | 58 | N | 56 | 1.25 | 4.5 | 2.72 | 4 |
| SA10 | F | 45 | Y | 94 | 0.25 | 7.8 | 2.14 | 4 |
| SA11 | F | 37 | Y | 77 | 1.5 | 1 | 4.2 | 4 |
| SA12 | F | 51 | Y | 77 | 0 | 6.5 | 2.86 | 4 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Francisco-Garcia, A.S.; Garrido-Martín, E.M.; Rupani, H.; Lau, L.C.K.; Martinez-Nunez, R.T.; Howarth, P.H.; Sanchez-Elsner, T. Small RNA Species and microRNA Profiles are Altered in Severe Asthma Nanovesicles from Broncho Alveolar Lavage and Associate with Impaired Lung Function and Inflammation. Non-Coding RNA 2019, 5, 51. https://doi.org/10.3390/ncrna5040051
Francisco-Garcia AS, Garrido-Martín EM, Rupani H, Lau LCK, Martinez-Nunez RT, Howarth PH, Sanchez-Elsner T. Small RNA Species and microRNA Profiles are Altered in Severe Asthma Nanovesicles from Broncho Alveolar Lavage and Associate with Impaired Lung Function and Inflammation. Non-Coding RNA. 2019; 5(4):51. https://doi.org/10.3390/ncrna5040051
Chicago/Turabian StyleFrancisco-Garcia, Ana S., Eva M. Garrido-Martín, Hitasha Rupani, Laurie C. K. Lau, Rocio T. Martinez-Nunez, Peter H. Howarth, and Tilman Sanchez-Elsner. 2019. "Small RNA Species and microRNA Profiles are Altered in Severe Asthma Nanovesicles from Broncho Alveolar Lavage and Associate with Impaired Lung Function and Inflammation" Non-Coding RNA 5, no. 4: 51. https://doi.org/10.3390/ncrna5040051
APA StyleFrancisco-Garcia, A. S., Garrido-Martín, E. M., Rupani, H., Lau, L. C. K., Martinez-Nunez, R. T., Howarth, P. H., & Sanchez-Elsner, T. (2019). Small RNA Species and microRNA Profiles are Altered in Severe Asthma Nanovesicles from Broncho Alveolar Lavage and Associate with Impaired Lung Function and Inflammation. Non-Coding RNA, 5(4), 51. https://doi.org/10.3390/ncrna5040051






