ncRNAs in Type-2 Immunity
Abstract
1. Introduction
1.1. Innate and Adaptive Collaboration
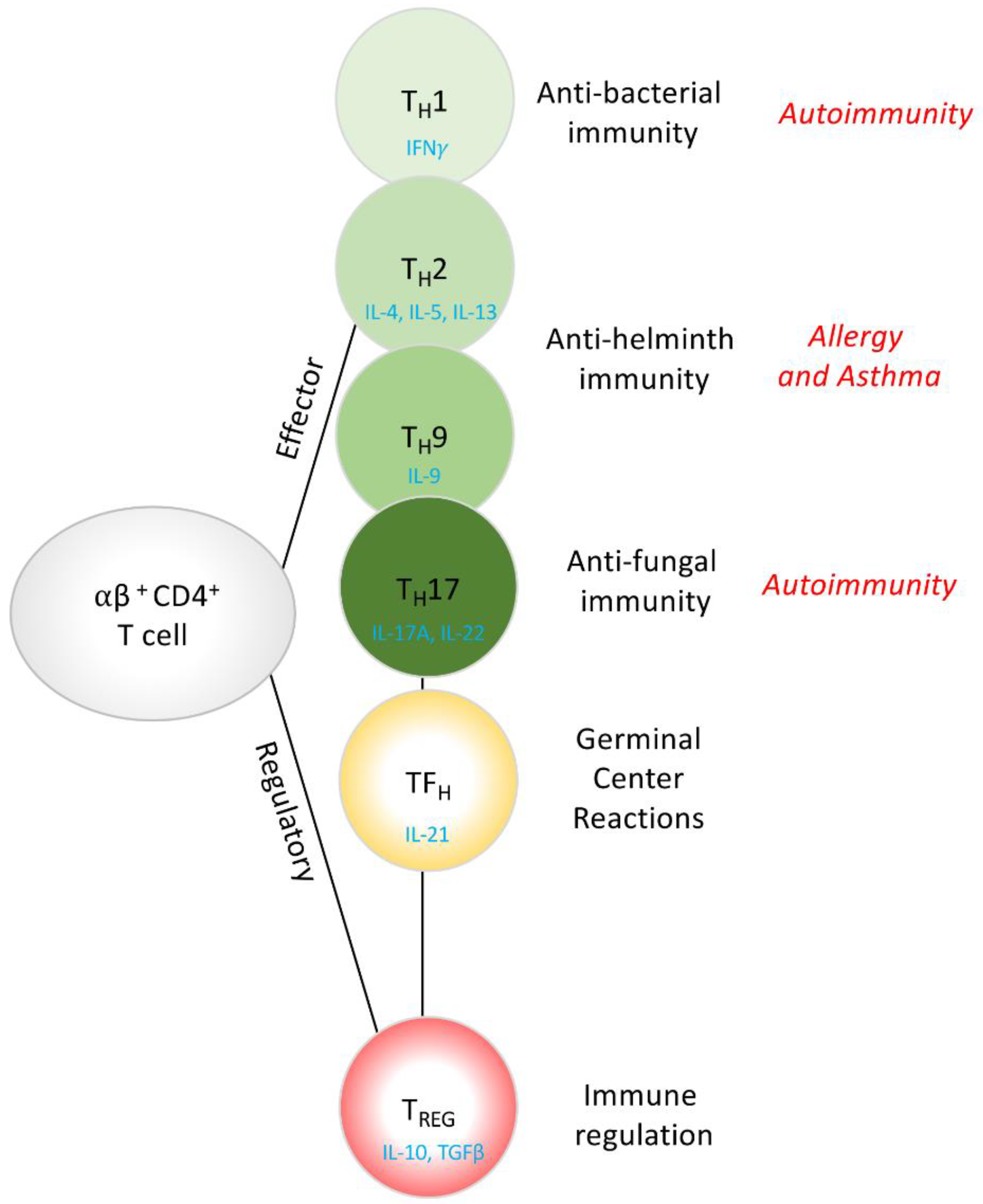
1.2. CD4+ T Cells, Conductors of the Immunology Orchestra
1.3. CD4+ TH2 Cell Differentiation
1.4. TH2 Cell Effector Function and Type-2 Immunity
2. Long Non-Coding RNAs (LncRNA) in Type-2 Immunity
2.1. Cell Type Specificity of lncRNAs
2.2. lncRNA Modes of Action: Nuclear Activity
2.3. lncRNA Cytoplasmic Activity
3. MicroRNAs in Type-2 Immunity
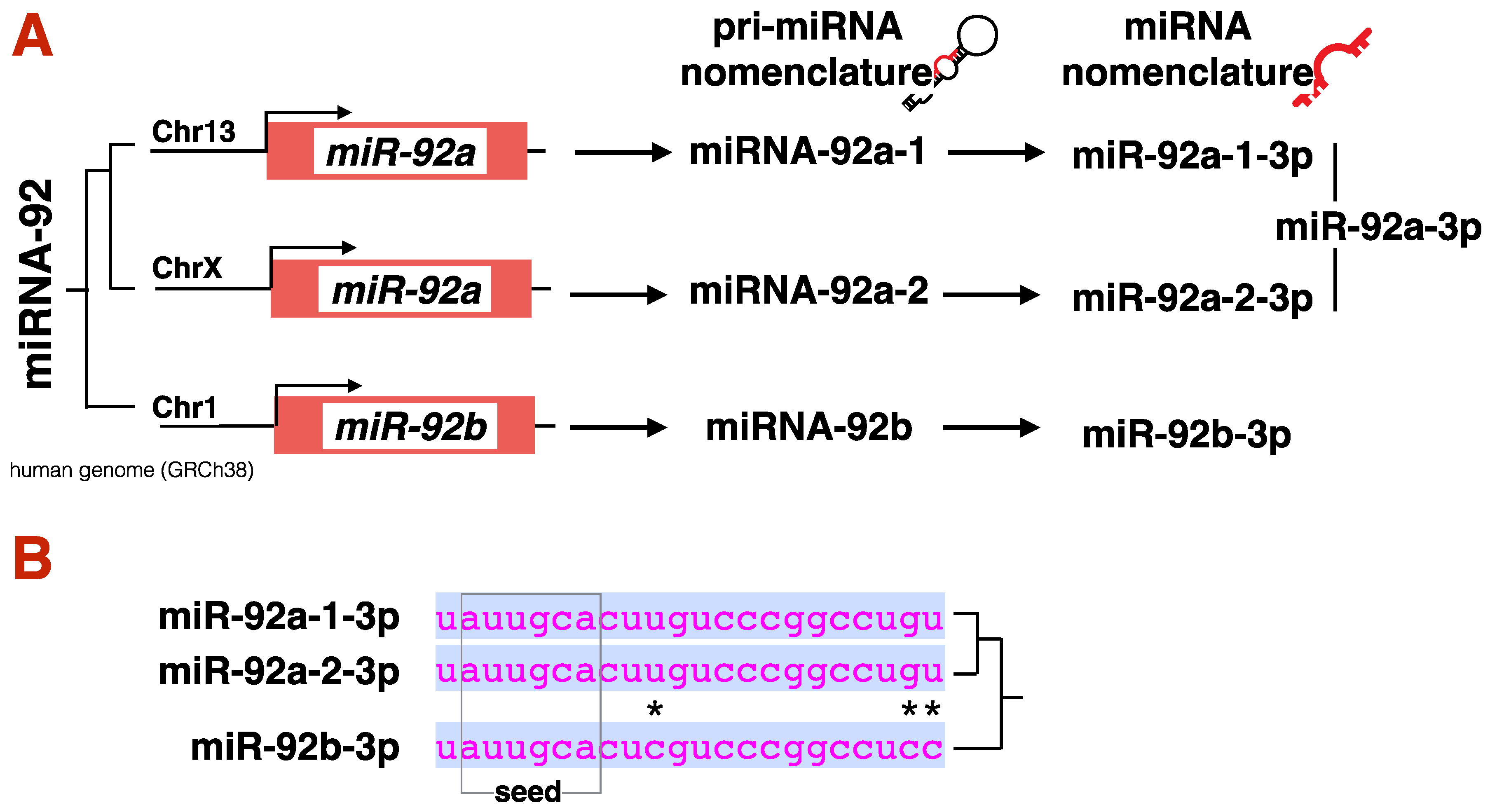
3.1. miRNA Biogenesis
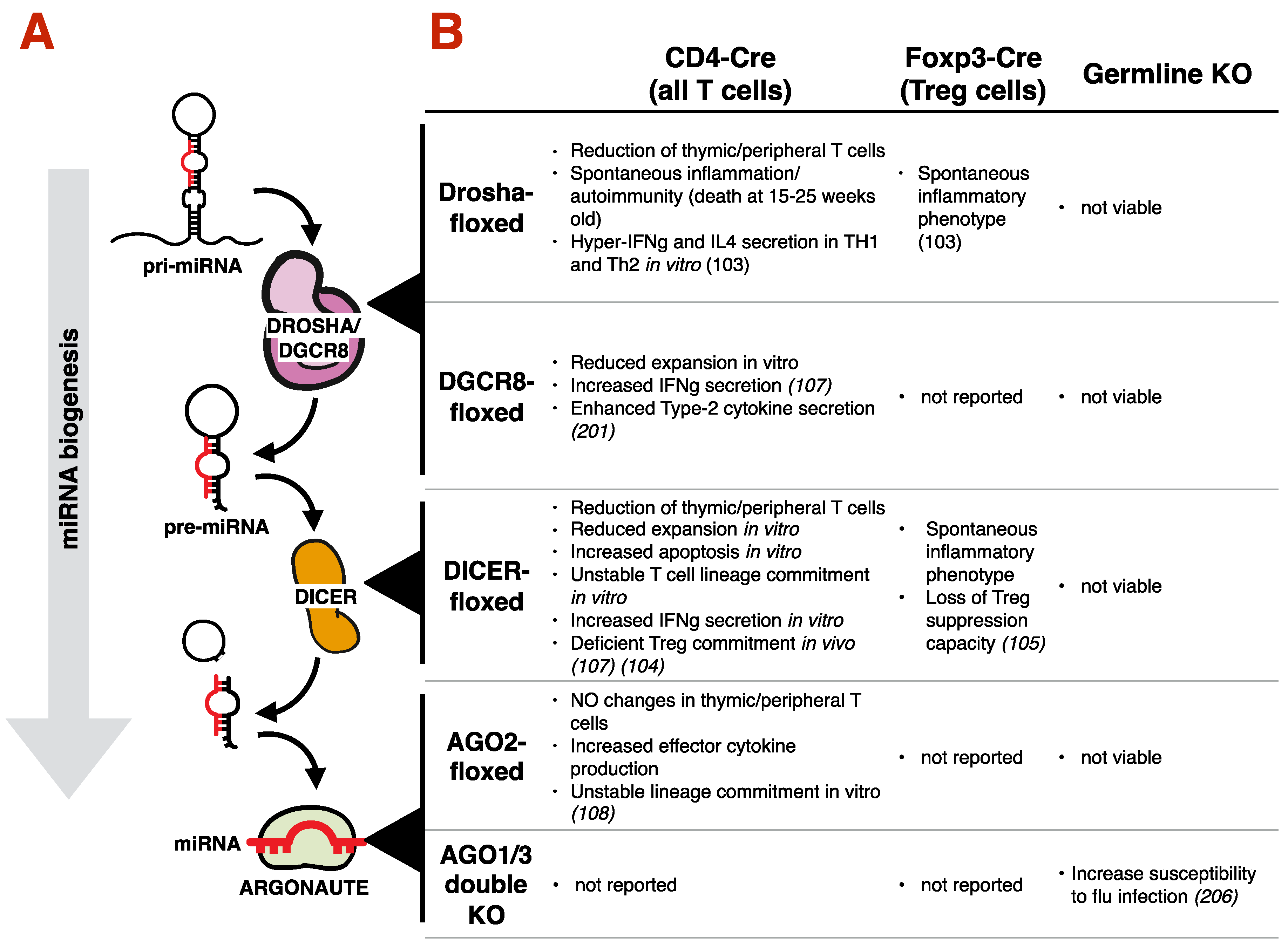
3.2. Requirement of the miRNA Machinery for T Cell Development, Differentiation and Type-2 Immunity
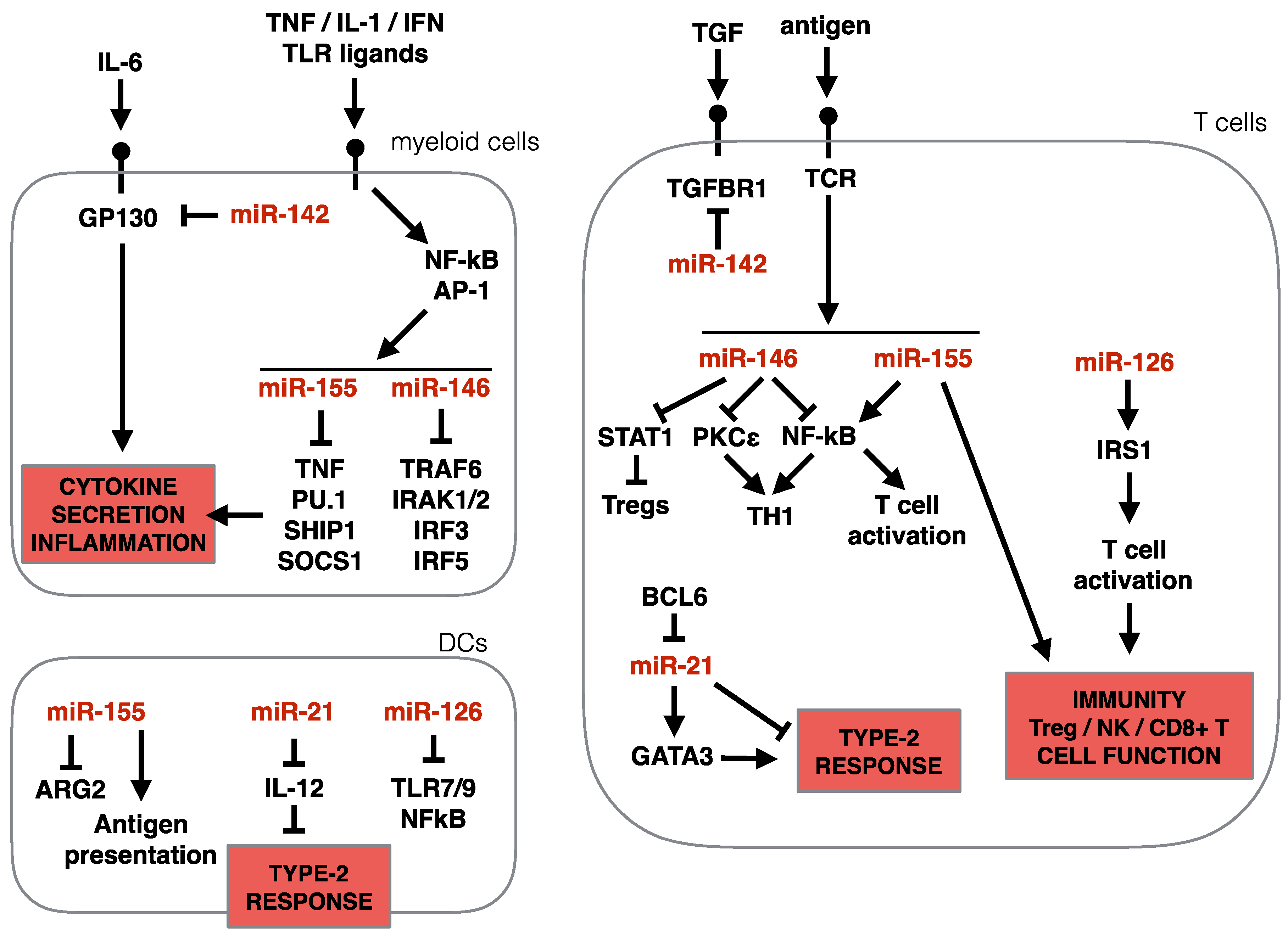
3.3. Impact of Specific miRNAs on Type-2 Immunity
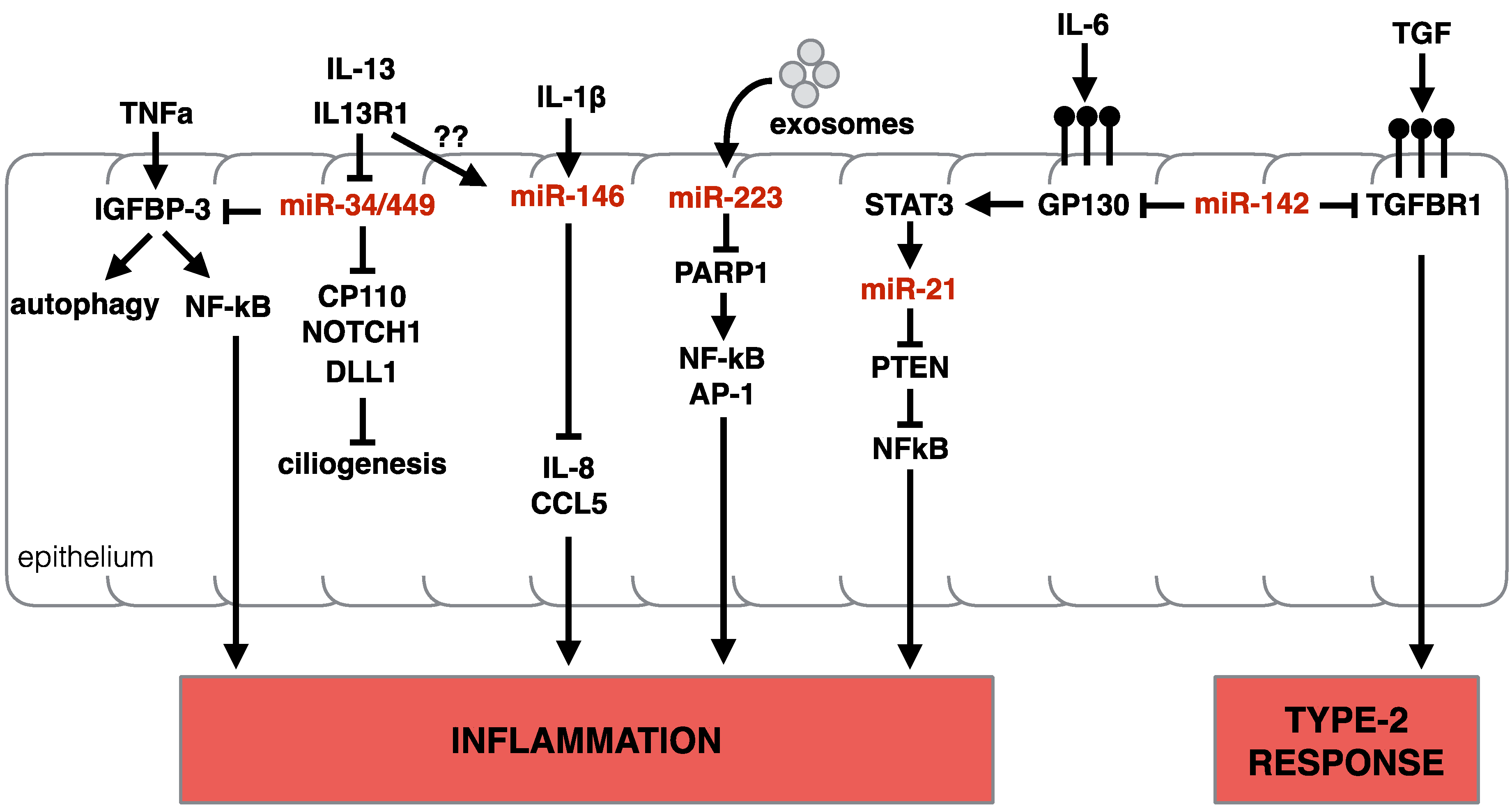
3.3.1. miR-34/449
3.3.2. miR-223
3.3.3. miR-142
3.3.4. miR-146 and miR-155
3.3.5. miR-21
3.3.6. miR-126
3.3.7. miR-375
3.3.8. miR-17~92 Cluster
3.3.9. miR-23~27~24 Cluster
4. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Dardalhon, V.; Awasthi, A.; Kwon, H.; Galileos, G.; Gao, W.; Sobel, R.A.; Mitsdoerffer, M.; Strom, T.B.; Elyamaftablen, W.; Ho, I.-C.; et al. IL-4 inhibits TGF-beta-induced Foxp3+ T cells and, together with TGF-beta, generates IL-9+ IL-10+ Foxp3(-) effector T cells. Nat. Immunol. 2008, 9, 1347–1355. [Google Scholar] [CrossRef] [PubMed]
- Hegazy, A.N.; Peine, M.; Helmstetter, C.; Panse, I.; Frohlich, A.; Bergthaler, A.; Flatz, L.; Pinschewer, D.D.; Radbruch, A.; Lohning, M. Interferons direct Th2 cell reprogramming to generate a stable GATA-3(+)T-bet(+) cell subset with combined Th2 and Th1 cell functions. Immunity 2010, 32, 116–128. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.H.; Voo, K.S.; Liu, B.; Chen, C.Y.; Uygungil, B.; Spoede, W.; Bernstein, J.A.; Huston, D.P.; Liu, Y.J. A novel subset of CD4(+) T(H)2 memory/effector cells that produce inflammatory IL-17 cytokine and promote the exacerbation of chronic allergic asthma. J. Exp. Med. 2010, 207, 2479–2491. [Google Scholar] [CrossRef] [PubMed]
- Veldhoen, M.; Uyttenhove, C.; van Snick, J.; Helmby, H.; Westendorf, A.; Buer, J.; Martin, B.; Wilhelm, C.; Stockinger, B. Transforming growth factor-beta ‘reprograms’ the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat. Immunol. 2008, 9, 1341–1346. [Google Scholar] [CrossRef]
- Pelly, V.S.; Coomes, S.M.; Kannan, Y.; Gialitakis, M.; Entwistle, L.J.; Perez-Lloret, J.; Czieso, S.; Okoye, I.S.; Ruckerl, D.; Allen, J.E.; et al. Interleukin 4 promotes the development of ex-Foxp3 Th2 cells during immunity to intestinal helminths. J. Exp. Med. 2017, 214, 1809–1826. [Google Scholar] [CrossRef]
- Zhu, J.; Yamane, H.; Paul, W.E. Differentiation of effector CD4 T cell populations (*). Annu. Rev. Immunol. 2010, 28, 445–489. [Google Scholar] [CrossRef]
- Zhu, J.; Min, B.; Hu-Li, J.; Watson, C.J.; Grinberg, A.; Wang, Q.; Killeen, N.; Urban, J.F., Jr.; Guo, L.; Paul, W.E. Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nat. Immunol. 2004, 5, 1157–1165. [Google Scholar] [CrossRef]
- Katona, I.M.; Urban, J.F., Jr.; Finkelman, F.D. The role of L3T4+ and Lyt-2+ T cells in the IgE response and immunity to Nippostrongylus brasiliensis. J. Immunol. 1988, 140, 3206–3211. [Google Scholar]
- Conti, H.R.; Shen, F.; Nayyar, N.; Stocum, E.; Sun, J.N.; Lindemann, M.J.; Ho, A.W.; Hai, J.H.; Yu, J.J.; Jung, J.W.; et al. Th17 cells and IL-17 receptor signaling are essential for mucosal host defense against oral candidiasis. J. Exp. Med. 2009, 206, 299–311. [Google Scholar] [CrossRef]
- Zelante, T.; De Luca, A.; D’Angelo, C.; Moretti, S.; Romani, L. IL-17/Th17 in anti-fungal immunity: What’s new? Eur. J. Immunol. 2009, 39, 645–648. [Google Scholar] [CrossRef]
- Licona-Limon, P.; Henao-Mejia, J.; Temann, A.U.; Gagliani, N.; Licona-Limon, I.; Ishigame, H.; Hao, L.; Herbert, D.R.; Flavell, R.A. Th9 cells drive host immunity against gastrointestinal worm infection. Immunity 2013, 39, 744–757. [Google Scholar] [CrossRef] [PubMed]
- Crotty, S. T follicular helper cell biology: A decade of discovery and diseases. Immunity 2019, 50, 1132–1148. [Google Scholar] [CrossRef] [PubMed]
- Josefowicz, S.Z.; Lu, L.F.; Rudensky, A.Y. Regulatory T cells: Mechanisms of differentiation and function. Annu. Rev. Immunol. 2012, 30, 531–564. [Google Scholar] [CrossRef] [PubMed]
- Min, B.; Prout, M.; Hu-Li, J.; Zhu, J.; Jankovic, D.; Morgan, E.S.; Urban, J.F., Jr.; Dvorak, A.M.; Finkelman, F.D.; LeGros, G.; et al. Basophils produce IL-4 and accumulate in tissues after infection with a Th2-inducing parasite. J. Exp. Med. 2004, 200, 507–517. [Google Scholar] [CrossRef]
- Sokol, C.L.; Barton, G.M.; Farr, A.G.; Medzhitov, R. A mechanism for the initiation of allergen-induced T helper type 2 responses. Nat. Immunol. 2008, 9, 310–318. [Google Scholar] [CrossRef]
- van Panhuys, N.; Prout, M.; Forbes, E.; Min, B.; Paul, W.E.; Le Gros, G. Basophils are the major producers of IL-4 during primary helminth infection. J. Immunol. 2011, 186, 2719–2728. [Google Scholar] [CrossRef]
- Perrigoue, J.G.; Saenz, S.A.; Siracusa, M.C.; Allenspach, E.J.; Taylor, B.C.; Giacomin, P.R.; Nair, M.G.; Du, Y.; Zaph, C.; van Rooijen, N.; et al. MHC class II-dependent basophil-CD4+ T cell interactions promote T(H)2 cytokine-dependent immunity. Nat. Immunol. 2009, 10, 697–705. [Google Scholar] [CrossRef]
- Sokol, C.L.; Chu, N.Q.; Yu, S.; Nish, S.A.; Laufer, T.M.; Medzhitov, R. Basophils function as antigen-presenting cells for an allergen-induced T helper type 2 response. Nat. Immunol. 2009, 10, 713–720. [Google Scholar] [CrossRef]
- Yoshimoto, T.; Yasuda, K.; Tanaka, H.; Nakahira, M.; Imai, Y.; Fujimori, Y.; Nakanishi, K. Basophils contribute to T(H)2-IgE responses in vivo via IL-4 production and presentation of peptide-MHC class II complexes to CD4+ T cells. Nat. Immunol. 2009, 10, 706–712. [Google Scholar] [CrossRef]
- Ohnmacht, C.; Pullner, A.; King, S.B.; Drexler, I.; Meier, S.; Brocker, T.; Voehringer, D. Constitutive ablation of dendritic cells breaks self-tolerance of CD4 T cells and results in spontaneous fatal autoimmunity. J. Exp. Med. 2009, 206, 549–559. [Google Scholar] [CrossRef]
- Ohnmacht, C.; Schwartz, C.; Panzer, M.; Schiedewitz, I.; Naumann, R.; Voehringer, D. Basophils orchestrate chronic allergic dermatitis and protective immunity against helminths. Immunity 2010, 33, 364–374. [Google Scholar] [CrossRef] [PubMed]
- Phythian-Adams, A.T.; Cook, P.C.; Lundie, R.J.; Jones, L.H.; Smith, K.A.; Barr, T.A.; Hochweller, K.; Anderton, S.M.; Hammerling, G.J.; Maizels, R.M.; et al. CD11c depletion severely disrupts Th2 induction and development in vivo. J. Exp. Med. 2010, 207, 2089–2096. [Google Scholar] [CrossRef]
- Hammad, H.; Plantinga, M.; Deswarte, K.; Pouliot, P.; Willart, M.A.; Kool, M.; Muskens, F.; Lambrecht, B.N. Inflammatory dendritic cells—Not basophils—Are necessary and sufficient for induction of Th2 immunity to inhaled house dust mite allergen. J. Exp. Med. 2010, 207, 2097–2111. [Google Scholar] [CrossRef] [PubMed]
- Kopf, M.; Coyle, A.J.; Schmitz, N.; Barner, M.; Oxenius, A.; Gallimore, A.; Gutierrez-Ramos, J.C.; Bachmann, M.F. Inducible costimulator protein (ICOS) controls T helper cell subset polarization after virus and parasite infection. J. Exp. Med. 2000, 192, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Tivol, E.A.; Borriello, F.; Schweitzer, A.N.; Lynch, W.P.; Bluestone, J.A.; Sharpe, A.H. Loss of CTLA-4 leads to massive lymphoproliferation and fatal multiorgan tissue destruction, revealing a critical negative regulatory role of CTLA-4. Immunity 1995, 3, 541–547. [Google Scholar] [CrossRef]
- Oosterwegel, M.A.; Mandelbrot, D.A.; Boyd, S.D.; Lorsbach, R.B.; Jarrett, D.Y.; Abbas, A.K.; Sharpe, A.H. The role of CTLA-4 in regulating Th2 differentiation. J. Immunol. 1999, 163, 2634–2639. [Google Scholar]
- Ubaldi, V.; Gatta, L.; Pace, L.; Doria, G.; Pioli, C. CTLA-4 engagement inhibits Th2 but not Th1 cell polarisation. Clin. Dev. Immunol. 2003, 10, 13–17. [Google Scholar] [CrossRef]
- Khattri, R.; Auger, J.A.; Griffin, M.D.; Sharpe, A.H.; Bluestone, J.A. Lymphoproliferative disorder in CTLA-4 knockout mice is characterized by CD28-regulated activation of Th2 responses. J. Immunol. 1999, 162, 5784–5791. [Google Scholar]
- Lee, D.U.; Rao, A. Molecular analysis of a locus control region in the T helper 2 cytokine gene cluster: A target for STAT6 but not GATA3. Proc. Natl. Acad. Sci. USA 2004, 101, 16010–16015. [Google Scholar] [CrossRef]
- Cote-Sierra, J.; Foucras, G.; Guo, L.; Chiodetti, L.; Young, H.A.; Hu-Li, J.; Zhu, J.; Paul, W.E. Interleukin 2 plays a central role in Th2 differentiation. Proc. Natl. Acad. Sci. USA 2004, 101, 3880–3885. [Google Scholar] [CrossRef]
- Zheng, W.; Flavell, R.A. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 1997, 89, 587–596. [Google Scholar] [CrossRef]
- Pai, S.Y.; Truitt, M.L.; Ho, I.C. GATA-3 deficiency abrogates the development and maintenance of T helper type 2 cells. Proc. Natl. Acad. Sci. USA 2004, 101, 1993–1998. [Google Scholar] [CrossRef] [PubMed]
- Ansel, K.M.; Djuretic, I.; Tanasa, B.; Rao, A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu. Rev. Immunol. 2006, 24, 607–656. [Google Scholar] [CrossRef] [PubMed]
- Yagi, R.; Zhu, J.; Paul, W.E. An updated view on transcription factor GATA3-mediated regulation of Th1 and Th2 cell differentiation. Int. Immunol. 2011, 23, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Soumelis, V.; Reche, P.A.; Kanzler, H.; Yuan, W.; Edward, G.; Homey, B.; Gilliet, M.; Ho, S.; Antonenko, S.; Lauerma, A.; et al. Human epithelial cells trigger dendritic cell mediated allergic inflammation by producing TSLP. Nat. Immunol. 2002, 3, 673–680. [Google Scholar] [CrossRef]
- Yu, H.S.; Angkasekwinai, P.; Chang, S.H.; Chung, Y.; Dong, C. Protease allergens induce the expression of IL-25 via Erk and p38 MAPK pathway. J. Korean Med. Sci. 2010, 25, 829–834. [Google Scholar] [CrossRef]
- Angkasekwinai, P.; Park, H.; Wang, Y.H.; Chang, S.H.; Corry, D.B.; Liu, Y.J.; Zhu, Z.; Dong, C. Interleukin 25 promotes the initiation of proallergic type 2 responses. J. Exp. Med. 2007, 204, 1509–1517. [Google Scholar] [CrossRef]
- Moon, P.D.; Kim, H.M. Thymic stromal lymphopoietin is expressed and produced by caspase-1/NF-kappaB pathway in mast cells. Cytokine 2011, 54, 239–243. [Google Scholar] [CrossRef]
- Kashyap, M.; Rochman, Y.; Spolski, R.; Samsel, L.; Leonard, W.J. Thymic stromal lymphopoietin is produced by dendritic cells. J. Immunol. 2011, 187, 1207–1211. [Google Scholar] [CrossRef]
- Zhou, B.; Comeau, M.R.; De Smedt, T.; Liggitt, H.D.; Dahl, M.E.; Lewis, D.B.; Gyarmati, D.; Aye, T.; Campbell, D.J.; Ziegler, S.F. Thymic stromal lymphopoietin as a key initiator of allergic airway inflammation in mice. Nat. Immunol. 2005, 6, 1047–1053. [Google Scholar] [CrossRef]
- Wang, Y.H.; Liu, Y.J. Thymic stromal lymphopoietin, OX40-ligand, and interleukin-25 in allergic responses. Clin. Exp. Allergy 2009, 39, 798–806. [Google Scholar] [CrossRef]
- Neill, D.R.; Wong, S.H.; Bellosi, A.; Flynn, R.J.; Daly, M.; Langford, T.K.; Bucks, C.; Kane, C.M.; Fallon, P.G.; Pannell, R.; et al. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature 2010, 464, 1367–1370. [Google Scholar] [CrossRef] [PubMed]
- Saenz, S.A.; Siracusa, M.C.; Perrigoue, J.G.; Spencer, S.P.; Urban, J.F., Jr.; Tocker, J.E.; Budelsky, A.L.; Kleinschek, M.A.; Kastelein, R.A.; Kambayashi, T.; et al. IL25 elicits a multipotent progenitor cell population that promotes T(H)2 cytokine responses. Nature 2010, 464, 1362–1366. [Google Scholar] [CrossRef] [PubMed]
- Moro, K.; Yamada, T.; Tanabe, M.; Takeuchi, T.; Ikawa, T.; Kawamoto, H.; Furusawa, J.; Ohtani, M.; Fujii, H.; Koyasu, S. Innate production of T(H)2 cytokines by adipose tissue-associated c-Kit(+)Sca-1(+) lymphoid cells. Nature 2010, 463, 540–544. [Google Scholar] [CrossRef] [PubMed]
- Fort, M.M.; Cheung, J.; Yen, D.; Li, J.; Zurawski, S.M.; Lo, S.; Menon, S.; Clifford, T.; Hunte, B.; Lesley, R.; et al. IL-25 induces IL-4, IL-5, and IL-13 and Th2-associated pathologies in vivo. Immunity 2001, 15, 985–995. [Google Scholar] [CrossRef]
- Kondo, Y.; Yoshimoto, T.; Yasuda, K.; Futatsugi-Yumikura, S.; Morimoto, M.; Hayashi, N.; Hoshino, T.; Fujimoto, J.; Nakanishi, K. Administration of IL-33 induces airway hyperresponsiveness and goblet cell hyperplasia in the lungs in the absence of adaptive immune system. Int. Immunol. 2008, 20, 791–800. [Google Scholar] [CrossRef]
- Angkasekwinai, P.; Chang, S.H.; Thapa, M.; Watarai, H.; Dong, C. Regulation of IL-9 expression by IL-25 signaling. Nat. Immunol. 2010, 11, 250–256. [Google Scholar] [CrossRef]
- Zhao, A.; Urban, J.F., Jr.; Sun, R.; Stiltz, J.; Morimoto, M.; Notari, L.; Madden, K.B.; Yang, Z.; Grinchuk, V.; Ramalingam, T.R.; et al. Critical role of IL-25 in nematode infection-induced alterations in intestinal function. J. Immunol. 2010, 185, 6921–6929. [Google Scholar] [CrossRef]
- Neill, D.R.; McKenzie, A.N. Nuocytes and beyond: New insights into helminth expulsion. Trends Parasitol. 2011, 27, 214–221. [Google Scholar] [CrossRef]
- Koyasu, S.; Moro, K. Type 2 innate immune responses and the natural helper cell. Immunology 2011, 132, 475–481. [Google Scholar] [CrossRef]
- Al-Shami, A.; Spolski, R.; Kelly, J.; Fry, T.; Schwartzberg, P.L.; Pandey, A.; Mackall, C.L.; Leonard, W.J. A role for thymic stromal lymphopoietin in CD4(+) T cell development. J. Exp. Med. 2004, 200, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Al-Shami, A.; Spolski, R.; Kelly, J.; Keane-Myers, A.; Leonard, W.J. A role for TSLP in the development of inflammation in an asthma model. J. Exp. Med. 2005, 202, 829–839. [Google Scholar] [CrossRef] [PubMed]
- Komai-Koma, M.; Xu, D.; Li, Y.; McKenzie, A.N.; McInnes, I.B.; Liew, F.Y. IL-33 is a chemoattractant for human Th2 cells. Eur. J. Immunol. 2007, 37, 2779–2786. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.H.; Angkasekwinai, P.; Lu, N.; Voo, K.S.; Arima, K.; Hanabuchi, S.; Hippe, A.; Corrigan, C.J.; Dong, C.; Homey, B.; et al. IL-25 augments type 2 immune responses by enhancing the expansion and functions of TSLP-DC-activated Th2 memory cells. J. Exp. Med. 2007, 204, 1837–1847. [Google Scholar] [CrossRef]
- Mohrs, M.; Blankespoor, C.M.; Wang, Z.E.; Loots, G.G.; Afzal, V.; Hadeiba, H.; Shinkai, K.; Rubin, E.M.; Locksley, R.M. Deletion of a coordinate regulator of type 2 cytokine expression in mice. Nat. Immunol. 2001, 2, 842–847. [Google Scholar] [CrossRef]
- Solymar, D.C.; Agarwal, S.; Bassing, C.H.; Alt, F.W.; Rao, A. A 3’ enhancer in the IL-4 gene regulates cytokine production by Th2 cells and mast cells. Immunity 2002, 17, 41–50. [Google Scholar] [CrossRef]
- Tykocinski, L.O.; Hajkova, P.; Chang, H.D.; Stamm, T.; Sozeri, O.; Lohning, M.; Hu-Li, J.; Niesner, U.; Kreher, S.; Friedrich, B.; et al. A critical control element for interleukin-4 memory expression in T helper lymphocytes. J. Biol. Chem. 2005, 280, 28177–28185. [Google Scholar] [CrossRef]
- de Vries, J.E.; Punnonen, J.; Cocks, B.G.; de Waal Malefyt, R.; Aversa, G. Regulation of the human IgE response by IL4 and IL13. Res. Immunol. 1993, 144, 597–601. [Google Scholar] [CrossRef]
- Lundgren, M.; Persson, U.; Larsson, P.; Magnusson, C.; Smith, C.I.; Hammarstrom, L.; Severinson, E. Interleukin 4 induces synthesis of IgE and IgG4 in human B cells. Eur. J. Immunol. 1989, 19, 1311–1315. [Google Scholar] [CrossRef]
- Herbert, D.R.; Holscher, C.; Mohrs, M.; Arendse, B.; Schwegmann, A.; Radwanska, M.; Leeto, M.; Kirsch, R.; Hall, P.; Mossmann, H.; et al. Alternative macrophage activation is essential for survival during schistosomiasis and downmodulates T helper 1 responses and immunopathology. Immunity 2004, 20, 623–635. [Google Scholar] [CrossRef]
- Motoji, T.; Okada, M.; Takanashi, M.; Masuda, M.; Tanaka, K.; Oshimi, K.; Mizoguchi, H. Induction of eosinophilic colonies by interleukin-5 on acute myeloblastic leukaemic cells. Br. J. Haematol. 1990, 74, 169–172. [Google Scholar] [CrossRef] [PubMed]
- Takatsu, K.; Tominaga, A.; Hamaoka, T. Antigen-induced T cell-replacing factor (TRF). I. Functional characterization of a TRF-producing helper T cell subset and genetic studies on TRF production. J. Immunol. 1980, 124, 2414–2422. [Google Scholar] [PubMed]
- Wynn, T.A. IL-13 effector functions. Annu. Rev. Immunol. 2003, 21, 425–456. [Google Scholar] [CrossRef] [PubMed]
- Woodruff, P.G.; Modrek, B.; Choy, D.F.; Jia, G.; Abbas, A.R.; Ellwanger, A.; Koth, L.L.; Arron, J.R.; Fahy, J.V. T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am. J. Respir. Crit Care Med. 2009, 180, 388–395. [Google Scholar] [CrossRef]
- Levine, S.J.; Wenzel, S.E. Narrative review: The role of Th2 immune pathway modulation in the treatment of severe asthma and its phenotypes. Ann. Intern. Med. 2010, 152, 232–237. [Google Scholar] [CrossRef]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef]
- Kopp, F.; Mendell, J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef]
- Engreitz, J.M.; Haines, J.E.; Perez, E.M.; Munson, G.; Chen, J.; Kane, M.; McDonel, P.E.; Guttman, M.; Lander, E.S. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 2016, 539, 452–455. [Google Scholar] [CrossRef]
- Mattioli, K.; Volders, P.J.; Gerhardinger, C.; Lee, J.C.; Maass, P.G.; Mele, M.; Rinn, J.L. High-throughput functional analysis of lncRNA core promoters elucidates rules governing tissue specificity. Genome Res. 2019, 29, 344–355. [Google Scholar] [CrossRef]
- Mele, M.; Mattioli, K.; Mallard, W.; Shechner, D.M.; Gerhardinger, C.; Rinn, J.L. Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs. Genome Res. 2017, 27, 27–37. [Google Scholar] [CrossRef]
- Necsulea, A.; Soumillon, M.; Warnefors, M.; Liechti, A.; Daish, T.; Zeller, U.; Baker, J.C.; Grutzner, F.; Kaessmann, H. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 2014, 505, 635–640. [Google Scholar] [CrossRef] [PubMed]
- Ulitsky, I.; Bartel, D.P. lincRNAs: Genomics, evolution, and mechanisms. Cell 2013, 154, 26–46. [Google Scholar] [CrossRef] [PubMed]
- Pang, K.C.; Frith, M.C.; Mattick, J.S. Rapid evolution of noncoding RNAs: Lack of conservation does not mean lack of function. Trends Genet. 2006, 22, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Gong, Y.; Yan, G.; Wang, D.; Qiao, Y.; Wang, Q.; Liu, B.; Hou, J.; Li, R.; Tang, C. Down-regulation of lncRNA MEG3 promotes hypoxia-induced human pulmonary artery smooth muscle cell proliferation and migration via repressing PTEN by sponging miR-21. Biochem. Biophys. Res. Commun. 2018, 495, 2125–2132. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.J.; Horlbeck, M.A.; Cho, S.W.; Birk, H.S.; Malatesta, M.; He, D.; Attenello, F.J.; Villalta, J.E.; Cho, M.Y.; Chen, Y.; et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 2017, 355. [Google Scholar] [CrossRef]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef]
- Atianand, M.K.; Hu, W.; Satpathy, A.T.; Shen, Y.; Ricci, E.P.; Alvarez-Dominguez, J.R.; Bhatta, A.; Schattgen, S.A.; McGowan, J.D.; Blin, J.; et al. A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell 2016, 165, 1672–1685. [Google Scholar] [CrossRef]
- Kotzin, J.J.; Spencer, S.P.; McCright, S.J.; Kumar, D.B.U.; Collet, M.A.; Mowel, W.K.; Elliott, E.N.; Uyar, A.; Makiya, M.A.; Dunagin, M.C.; et al. The long non-coding RNA Morrbid regulates Bim and short-lived myeloid cell lifespan. Nature 2016, 537, 239–243. [Google Scholar] [CrossRef]
- Han, X.; Huang, S.; Xue, P.; Fu, J.; Liu, L.; Zhang, C.; Yang, L.; Xia, L.; Sun, L.; Huang, S.K.; et al. LncRNA PTPRE-AS1 modulates M2 macrophage activation and inflammatory diseases by epigenetic promotion of PTPRE. Sci. Adv. 2019, 5. [Google Scholar] [CrossRef]
- Zhou, Y.X.; Zhao, W.; Mao, L.W.; Wang, Y.L.; Xia, L.Q.; Cao, M.; Shen, J.; Chen, J. Long non-coding RNA NIFK-AS1 inhibits M2 polarization of macrophages in endometrial cancer through targeting miR-146a. Int. J. Biochem. Cell Biol. 2018, 104, 25–33. [Google Scholar] [CrossRef]
- Hu, G.; Tang, Q.; Sharma, S.; Yu, F.; Escobar, T.M.; Muljo, S.A.; Zhu, J.; Zhao, K. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat. Immunol. 2013, 14, 1190–1198. [Google Scholar] [CrossRef] [PubMed]
- Spurlock, C.F., III; Tossberg, J.T.; Guo, Y.; Collier, S.P.; Crooke, P.S., III; Aune, T.M. Expression and functions of long noncoding RNAs during human T helper cell differentiation. Nat. Commun. 2015, 6, 6932. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Nestor, C.E.; Zhao, S.; Lentini, A.; Bohle, B.; Benson, M.; Wang, H. Profiling of human CD4+ T-cell subsets identifies the TH2-specific noncoding RNA GATA3-AS1. J. Allergy Clin. Immunol. 2013, 132, 1005–1008. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, H.R.; Shaginurova, G.; Kim, L.C.; Chapman, N.; Spurlock, C.F., III; Aune, T.M. Divergent lncRNA GATA3-AS1 Regulates GATA3 Transcription in T-Helper 2 Cells. Front. Immunol. 2018, 9, 2512. [Google Scholar] [CrossRef] [PubMed]
- Willingham, A.T.; Orth, A.P.; Batalov, S.; Peters, E.C.; Wen, B.G.; Aza-Blanc, P.; Hogenesch, J.B.; Schultz, P.G. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 2005, 309, 1570–1573. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Xue, Y.; Han, Y.; Lin, L.; Wu, C.; Xu, S.; Jiang, Z.; Xu, J.; Liu, Q.; Cao, X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 2014, 344, 310–313. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Lee, H.; Han, S.; Kwon, C.S.; Lee, D. Biogenesis and regulation of the let-7 miRNAs and their functional implications. Protein Cell 2016, 7, 100–113. [Google Scholar] [CrossRef]
- Bartel, D.P. Metazoan microRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef]
- Wilson, R.C.; Doudna, J.A. Molecular mechanisms of RNA interference. Annu. Rev. Biophys. 2013, 42, 217–239. [Google Scholar] [CrossRef]
- Eulalio, A.; Huntzinger, E.; Izaurralde, E. Getting to the root of miRNA-mediated gene silencing. Cell 2008, 132, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Eulalio, A.; Huntzinger, E.; Izaurralde, E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat. Struct. Mol. Biol. 2008, 15, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Hannon, G.J.; Hammond, S.M.; Carmell, M.A.; Rivas, F.V.; Marsden, C.G.; Thomson, J.M.; Song, J.-J.; Joshua-Tor, L. Argonaute2 is the catalytic engine of mammalian RNAi. Science 2004, 305, 1437–1441. [Google Scholar] [CrossRef] [PubMed]
- Lazzaretti, D.; Tournier, I.; Izaurralde, E. The C-terminal domains of human TNRC6A, TNRC6B, and TNRC6C silence bound transcripts independently of Argonaute proteins. RNA 2009, 15, 1059–1066. [Google Scholar] [CrossRef]
- Lian, S.L.; Li, S.; Abadal, G.X.; Pauley, B.A.; Fritzler, M.J.; Chan, E.K.L. The C-terminal half of human Ago2 binds to multiple GW-rich regions of GW182 and requires GW182 to mediate silencing. RNA 2009, 15, 804–813. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Cheloufi, S.; Dos Santos, C.O.; Chong, M.M.W.; Hannon, G.J. A dicer-independent miRNA biogenesis pathway that requires ago catalysis. Nature 2010, 465, 584–589. [Google Scholar] [CrossRef]
- Park, M.S.; Phan, H.D.; Busch, F.; Hinckley, S.H.; Brackbill, J.A.; Wysocki, V.H.; Nakanishi, K. Human argonaute3 has slicer activity. Nucleic Acids Res. 2017, 45, 11867–11877. [Google Scholar] [CrossRef]
- Kertesz, M.; Iovino, N.; Unnerstall, U.; Gaul, U.; Segal, E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007, 39, 1278–1284. [Google Scholar] [CrossRef]
- Golden, R.J.; Chen, B.; Li, T.; Braun, J.; Manjunath, H.; Chen, X.; Wu, J.; Schmid, V.; Chang, T.-C.; Kopp, F.; et al. An Argonaute phosphorylation cycle promotes microRNA-mediated silencing. Nature 2017, 542, 197–202. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The biochemical basis of microRNA targeting efficacy. Science 2019, 366. [Google Scholar] [CrossRef] [PubMed]
- Iwakawa, H.-O.; Tomari, Y. The functions of microRNAs: mRNA decay and translational repression. Trends Cell Biol. 2015, 25, 651–665. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.K.-H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Chong, M.M.W.; Littman, D.R.; Rudensky, A.Y.; Rasmussen, J.P.; Rundensky, A.Y. The RNAseIII enzyme drosha is critical in T cells for preventing lethal inflammatory disease. J. Exp. Med. 2008, 205, 2005–2017. [Google Scholar] [CrossRef]
- Muljo, S.A.; Ansel, K.M.; Kanellopoulou, C.; Livingston, D.M.; Rao, A.; Rajewsky, K. Aberrant T cell differentiation in the absence of Dicer. J. Exp. Med. 2005, 202, 261–269. [Google Scholar] [CrossRef]
- Liston, A.; Lu, L.-F.; O’Carroll, D.; Tarakhovsky, A.; Rudensky, A.Y. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J. Exp. Med. 2008, 205, 1993–2004. [Google Scholar] [CrossRef]
- Okoye, I.S.; Coomes, S.M.; Pelly, V.S.; Czieso, S.; Papayannopoulos, V.; Tolmachova, T.; Seabra, M.C.; Wilson, M.S. MicroRNA-containing T-regulatory-cell-derived exosomes suppress pathogenic T helper 1 cells. Immunity 2014, 41, 89–103. [Google Scholar] [CrossRef]
- Steiner, D.F.; Thomas, M.F.; Hu, J.K.; Yang, Z.; Babiarz, J.E.; Allen, C.D.C.; Matloubian, M.; Blelloch, R.; Ansel, K.M. MicroRNA-29 regulates T-box transcription factors and interferon-γ production in helper T cells. Immunity 2011, 35, 169–181. [Google Scholar] [CrossRef]
- Bronevetsky, Y.; Villarino, A.V.; Eisley, C.J.; Barbeau, R.; Barczak, A.J.; Heinz, G.A.; Kremmer, E.; Heissmeyer, V.; McManus, M.T.; Erle, D.J.; et al. T cell activation induces proteasomal degradation of Argonaute and rapid remodeling of the microRNA repertoire. J. Exp. Med. 2013, 210, 417–432. [Google Scholar] [CrossRef] [PubMed]
- Pua, H.H.; Ansel, K.M. MicroRNA regulation of allergic inflammation and asthma. Curr. Opin. Immunol. 2015, 36, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Papi, A.; Brightling, C.; Pedersen, S.E.; Reddel, H.K. Asthma. Lancet 2018, 391, 783–800. [Google Scholar] [CrossRef]
- Solberg, O.D.; Ostrin, E.J.; Love, M.I.; Peng, J.C.; Bhakta, N.R.; Hou, L.; Nguyen, C.; Solon, M.; Nguyen, C.; Barczak, A.J.; et al. Airway epithelial miRNA expression is altered in asthma. Am. J. Respir. Crit. Care Med. 2012, 186, 965–974. [Google Scholar] [CrossRef]
- Wu, X.-B.; Wang, M.-Y.; Zhu, H.-Y.; Tang, S.-Q.; You, Y.-D.; Xie, Y.-Q. Overexpression of microRNA-21 and microRNA-126 in the patients of bronchial asthma. Int. J. Clin. Exp. Med. 2014, 7, 1307–1312. [Google Scholar]
- Sonkoly, E.; Janson, P.; Majuri, M.-L.; Savinko, T.; Fyhrquist, N.; Eidsmo, L.; Xu, N.; Meisgen, F.; Wei, T.; Bradley, M.; et al. MiR-155 is overexpressed in patients with atopic dermatitis and modulates T-cell proliferative responses by targeting cytotoxic T lymphocyte-associated antigen 4. J. Allergy Clin. Immunol. 2010, 126, 581–589. [Google Scholar] [CrossRef]
- Vennegaard, M.T.; Bonefeld, C.M.; Hagedorn, P.H.; Bangsgaard, N.; Løvendorf, M.B.; Odum, N.; Woetmann, A.; Geisler, C.; Skov, L. Allergic contact dermatitis induces upregulation of identical microRNAs in humans and mice. Contact Dermat. 2012, 67, 298–305. [Google Scholar] [CrossRef]
- Kelada, S.; Sethupathy, P.; Okoye, I.S.; Kistasis, E.; Czieso, S.; White, S.D.; Chou, D.; Martens, C.; Ricklefs, S.M.; Virtaneva, K.; et al. miR-182 and miR-10a are key regulators of Treg specialisation and stability during Schistosome and Leishmania-associated inflammation. PLoS Pathog. 2013, 9, e1003451. [Google Scholar] [CrossRef]
- Okoye, I.S.; Czieso, S.; Ktistaki, E.; Roderick, K.; Coomes, S.M.; Pelly, V.S.; Kannan, Y.; Lloret-Perez, J.; Zhao, J.L.; Baltimore, D.; et al. Transcriptomics identified a critical role for Th2 cell-intrinsic miR-155 in mediating allergy and antihelminth immunity. Proc. Natl. Acad. Sci. USA 2014, 111, E3081–E3090. [Google Scholar] [CrossRef]
- Song, R.; Walentek, P.; Sponer, N.; Klimke, A.; Lee, J.S.; Dixon, G.; Harland, R.; Wan, Y.; Lishko, P.; Lize, M.; et al. miR-34/449 miRNAs are required for motile ciliogenesis by repressing cp110. Nature 2014, 510, 115–120. [Google Scholar] [CrossRef]
- Guseh, J.S.; Bores, S.A.; Stanger, B.Z.; Zhou, Q.; Anderson, W.J.; Melton, D.A.; Rajagopal, J. Notch signaling promotes airway mucous metaplasia and inhibits alveolar development. Development 2009, 136, 1751–1759. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.H.; Kim, B.S.; Uhm, T.G.; Lee, S.-H.; Lee, G.R.; Park, C.-S.; Chung, I.Y. Gamma-secretase inhibitor reduces allergic pulmonary inflammation by modulating Th1 and Th2 responses. Am. J. Respir. Crit. Care Med. 2009, 179, 875–882. [Google Scholar] [CrossRef] [PubMed]
- Marcet, B.; Chevalier, B.; Luxardi, G.; Coraux, C.; Zaragosi, L.-E.; Cibois, M.; Robbe-Sermesant, K.; Jolly, T.; Cardinaud, B.; Moreilhon, C.; et al. Control of vertebrate multiciliogenesis by miR-449 through direct repression of the Delta/Notch pathway. Nat. Cell Biol. 2011, 13, 693–699. [Google Scholar] [CrossRef]
- Kim, S.R.; Lee, K.S.; Lee, K.B.; Lee, Y.C. Recombinant IGFBP-3 inhibits allergic lung inflammation, VEGF production, and vascular leak in a mouse model of asthma. Allergy 2012, 67, 869–877. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Zhang, S.; Sun, Y.; Li, S.; Ning, Y.; Dong, Y.; Shang, Y.; Bai, C. MicroRNA-34/449 targets IGFBP-3 and attenuates airway remodeling by suppressing Nur77-mediated autophagy. Cell Death Dis. 2017, 8, e2998. [Google Scholar] [CrossRef]
- Lu, T.X.; Sherrill, J.D.; Wen, T.; Plassard, A.J.; Besse, J.A.; Abonia, J.P.; Franciosi, J.P.; Putnam, P.E.; Eby, M.; Martin, L.J.; et al. MicroRNA signature in patients with eosinophilic esophagitis, reversibility with glucocorticoids, and assessment as disease biomarkers. J. Allergy Clin. Immunol. 2012, 129, 1064–1075. [Google Scholar] [CrossRef]
- Johnnidis, J.B.; Harris, M.H.; Wheeler, R.T.; Stehling-Sun, S.; Lam, M.H.; Kirak, O.; Brummelkamp, T.R.; Fleming, M.D.; Camargo, F.D. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 2008, 451, 1125–1129. [Google Scholar] [CrossRef]
- Lu, T.X.; Lim, E.-J.; Besse, J.A.; Itskovich, S.; Plassard, A.J.; Fulkerson, P.C.; Aronow, B.J.; Rothenberg, M.E. MiR-223 deficiency increases eosinophil progenitor proliferation. J. Immunol. 2013, 190, 1576–1582. [Google Scholar] [CrossRef]
- Zhuang, G.; Meng, C.; Guo, X.; Cheruku, P.S.; Shi, L.; Xu, H.; Li, H.; Wang, G.; Evans, A.R.; Safe, S.; et al. A novel regulator of macrophage activation. Circulation 2012, 125, 2892–2903. [Google Scholar] [CrossRef]
- Draijer, C.; Robbe, P.; Boorsma, C.E.; Hylkema, M.N.; Melgert, B.N. Dual role of YM1 + M2 macrophages in allergic lung inflammation. Sci. Rep. 2018, 8, 5105. [Google Scholar] [CrossRef]
- Yuan, X.; Berg, N.; Lee, J.W.; Le, T.T.; Neudecker, V.; Jing, N.; Eltzschig, H. MicroRNA miR-223 as regulator of innate immunity. J. Leukoc. Biol. 2018, 104, 515–524. [Google Scholar] [CrossRef]
- Zhou, W.; Pal, A.S.; Hsu, A.Y.-H.; Gurol, T.; Zhu, X.; Wirbisky-Hershberger, S.E.; Freeman, J.L.; Kasinski, A.L.; Deng, Q. MicroRNA-223 suppresses the canonical NF-κB pathway in basal keratinocytes to dampen neutrophilic inflammation. Cell Rep. 2018, 22, 1810–1823. [Google Scholar] [CrossRef]
- Neudecker, V.; Brodsky, K.S.; Clambey, E.T.; Schmidt, E.P.; Packard, T.A.; Davenport, B.; Standiford, T.J.; Weng, T.; Fletcher, A.A.; Barthel, L.; et al. Neutrophil transfer of miR-223 to lung epithelial cells dampens acute lung injury in mice. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef]
- Lu, T.X.; Munitz, A.; Rothenberg, M.E. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J. Immunol. 2009, 182, 4994–5002. [Google Scholar] [CrossRef]
- Kasashima, K.; Nakamura, Y.; Kozu, T. Altered expression profiles of microRNAs during TPA-induced differentiation of HL-60 cells. Biochem. Biophys. Res. Commun. 2004, 322, 403–410. [Google Scholar] [CrossRef]
- Sharma, S.; Liu, J.; Wei, J.; Yuan, H.; Zhang, T.; Bishopric, N.H. Repression of miR-142 by p300 and MAPK is required for survival signalling via gp130 during adaptive hypertrophy. EMBO Mol. Med. 2012, 4, 617–632. [Google Scholar] [CrossRef]
- Yokoyama, A.; Kohno, N.; Fujino, S.; Hamada, H.; Inoue, Y.; Fujioka, S.; Ishida, S.; Hiwada, K. Circulating interleukin-6 levels in patients with bronchial asthma. Am. J. Respir. Crit. Care Med. 1995, 151, 1354–1358. [Google Scholar] [CrossRef]
- Rincon, M.; Irvin, C.G. Role of IL-6 in asthma and other inflammatory pulmonary diseases. Int. J. Biol. Sci. 2012, 8, 1281–1290. [Google Scholar] [CrossRef]
- Kramer, N.J.; Wang, W.-L.; Reyes, E.Y.; Kumar, B.; Chen, C.-C.; Ramakrishna, C.; Cantin, E.M.; Vonderfecht, S.L.; Taganov, K.D.; Chau, N.; et al. Altered lymphopoiesis and immunodeficiency in miR-142 null mice. Blood 2015, 125, 3720–3730. [Google Scholar] [CrossRef]
- Bartel, S.; Carraro, G.; Alessandrini, F.; Krauss-Etschmann, S.; Ricciardolo, F.L.M.; Bellusci, S. miR-142-3p is associated with aberrant WNT signaling during airway remodeling in asthma. Am. J. Physiol. Lung Cell. Mol. Physiol. 2018, 315, L328–L333. [Google Scholar] [CrossRef]
- Xu, S.; Wei, J.; Wang, F.; Kong, L.-Y.; Ling, X.-Y.; Nduom, E.; Gabrusiewicz, K.; Doucette, T.; Yang, Y.; Yaghi, N.K.; et al. Effect of miR-142-3p on the M2 macrophage and therapeutic efficacy against murine glioblastoma. J. Natl. Cancer Inst. 2014, 106, 5369. [Google Scholar] [CrossRef]
- Taganov, K.D.; Boldin, M.P.; Chang, K.-J.; Baltimore, D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef]
- Yin, Q.; McBride, J.; Fewell, C.; Lacey, M.; Wang, X.; Lin, Z.; Cameron, J.; Flemington, E.K. MicroRNA-155 is an Epstein-Barr virus-induced gene that modulates Epstein-Barr virus-regulated gene expression pathways. J. Virol. 2008, 82, 5295–5306. [Google Scholar] [CrossRef]
- Testa, U.; Pelosi, E.; Castelli, G.; Labbaye, C. miR-146 and miR-155: Two key modulators of immune response and tumor development. Noncoding RNA 2017, 3, 22. [Google Scholar] [CrossRef]
- Boldin, M.P.; Taganov, K.D.; Rao, D.S.; Yang, L.; Zhao, J.L.; Kalwani, M.; Garcia-Flores, Y.; Luong, M.; Devrekanli, A.; Xu, J.; et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J. Exp. Med. 2011, 208, 1189–1201. [Google Scholar] [CrossRef]
- Rodriguez, A.; Vigorito, E.; Clare, S.; Warren, M.V.; Couttet, P.; Soond, D.R.; van Dongen, S.; Grocock, R.J.; Das, P.P.; Miska, E.A.; et al. Requirement of bic/microRNA-155 for normal immune function. Science 2007, 316, 608–611. [Google Scholar] [CrossRef]
- Yang, M.; Eyers, F.; Xiang, Y.; Guo, M.; Young, I.G.; Rosenberg, H.F.; Foster, P.S. Expression profiling of differentiating eosinophils in bone marrow cultures predicts functional links between microRNAs and their target mRNAs. PLoS ONE 2014, 9, e97537. [Google Scholar] [CrossRef]
- Yang, L.; Boldin, M.P.; Yu, Y.; Liu, C.S.; Ea, C.-K.; Ramakrishnan, P.; Taganov, K.D.; Zhao, J.L.; Baltimore, D. miR-146a controls the resolution of T cell responses in mice. J. Exp. Med. 2012, 209, 1655–1670. [Google Scholar] [CrossRef]
- Möhnle, P.; Schütz, S.V.; van der Heide, V.; Hübner, M.; Luchting, B.; Sedlbauer, J.; Limbeck, E.; Hinske, L.C.; Briegel, J.; Kreth, S. MicroRNA-146a controls Th1-cell differentiation of human CD4+ T lymphocytes by targeting PRKCε. Eur. J. Immunol. 2015, 45, 260–272. [Google Scholar] [CrossRef]
- Lu, L.-F.; Boldin, M.P.; Chaudhry, A.; Lin, L.-L.; Taganov, K.D.; Hanada, T.; Yoshimura, A.; Baltimore, D.; Rudensky, A.Y. Function of miR-146a in controlling treg cell-mediated regulation of Th1 responses. Cell 2010, 142, 914–929. [Google Scholar] [CrossRef]
- Perry, M.M.; Moschos, S.A.; Williams, A.E.; Shepherd, N.J.; Larner-Svensson, H.M.; Lindsay, M.A. Rapid changes in microRNA-146a expression negatively regulate the IL-1beta-induced inflammatory response in human lung alveolar epithelial cells. J. Immunol. 2008, 180, 5689–5698. [Google Scholar] [CrossRef] [PubMed]
- Mashima, R. Physiological roles of miR-155. Immunology 2015, 145, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Costinean, S.; Zanesi, N.; Pekarsky, Y.; Tili, E.; Volinia, S.; Heerema, N.; Croce, C.M. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc. Natl. Acad. Sci. USA 2006, 103, 7024–7029. [Google Scholar] [CrossRef] [PubMed]
- Tili, E.; Michaille, J.-J.; Cimino, A.; Costinean, S.; Dumitru, C.D.; Adair, B.; Fabbri, M.; Alder, H.; Liu, C.G.; Calin, G.A.; et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J. Immunol. 2007, 179, 5082–5089. [Google Scholar] [CrossRef] [PubMed]
- Kueh, H.Y.; Champhekar, A.; Champhekhar, A.; Nutt, S.L.; Elowitz, M.B.; Rothenberg, E.V. Positive feedback between PU.1 and the cell cycle controls myeloid differentiation. Science 2013, 341, 670–673. [Google Scholar] [CrossRef] [PubMed]
- Laslo, P.; Spooner, C.J.; Warmflash, A.; Lancki, D.W.; Lee, H.-J.; Sciammas, R.; Gantner, B.N.; Dinner, A.R.; Singh, H. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell 2006, 126, 755–766. [Google Scholar] [CrossRef]
- Carotta, S.; Dakic, A.; D’Amico, A.; Pang, S.H.M.; Greig, K.T.; Nutt, S.L.; Wu, L. The transcription factor PU.1 controls dendritic cell development and Flt3 cytokine receptor expression in a dose-dependent manner. Immunity 2010, 32, 628–641. [Google Scholar] [CrossRef]
- Rosenbauer, F.; Wagner, K.; Kutok, J.L.; Iwasaki, H.; Le Beau, M.M.; Okuno, Y.; Akashi, K.; Fiering, S.; Tenen, D.G. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat. Genet. 2004, 36, 624–630. [Google Scholar] [CrossRef]
- Dakic, A.; Metcalf, D.; Di Rago, L.; Mifsud, S.; Wu, L.; Nutt, S.L. PU.1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis. J. Exp. Med. 2005, 201, 1487–1502. [Google Scholar] [CrossRef]
- Iwasaki, H.; Somoza, C.; Shigematsu, H.; Duprez, E.A.; Iwasaki-Arai, J.; Mizuno, S.-i.; Arinobu, Y.; Geary, K.; Zhang, P.; Dayaram, T.; et al. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood 2005, 106, 1590–1600. [Google Scholar] [CrossRef]
- Feng, R.; Desbordes, S.C.; Xie, H.; Tillo, E.S.; Pixley, F.; Stanley, E.R.; Graf, T. PU.1 and C/EBPα/β convert fibroblasts into macrophage-like cells. Proc. Natl. Acad. Sci. USA 2008, 105, 6057–6062. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, H.; Mizuno, S.-I.; Arinobu, Y.; Ozawa, H.; Mori, Y.; Shigematsu, H.; Takatsu, K.; Tenen, D.G.; Akashi, K. The order of expression of transcription factors directs hierarchical specification of hematopoietic lineages. Genes Dev. 2006, 20, 3010–3021. [Google Scholar] [CrossRef]
- Vigorito, E.; Perks, K.L.; Abreu-Goodger, C.; Bunting, S.; Xiang, Z.; Kohlhaas, S.; Das, P.P.; Miska, E.A.; Rodriguez, A.; Bradley, A.; et al. MicroRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity 2007, 27, 847–859. [Google Scholar] [CrossRef]
- Carotta, S.; Willis, S.N.; Hasbold, J.; Inouye, M.; Pang, S.H.M.; Emslie, D.; Light, A.; Chopin, M.; Shi, W.; Wang, H.; et al. The transcription factors IRF8 and PU.1 negatively regulate plasma cell differentiation. J. Exp. Med. 2014, 211, 2169–2181. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, H.; Ungerbäck, J.; Wang, X.; Matsumoto, M.; Nakayama, K.I.; Cohen, S.M.; Tanaka, T.; Rothenberg, E.V. Transcription factor PU.1 represses and activates gene expression in early T cells by redirecting partner transcription factor binding. Immunity 2018, 48, 1119–1134. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.-C.; Sehra, S.; Goswami, R.; Yao, W.; Yu, Q.; Stritesky, G.L.; Jabeen, R.; McKinley, C.; Ahyi, A.-N.; Han, L.; et al. The transcription factor PU.1 is required for the development of IL-9-producing T cells and allergic inflammation. Nat. Immunol. 2010, 11, 527–534. [Google Scholar] [CrossRef]
- O’Connell, R.M.; Kahn, D.; Gibson, W.S.J.; Round, J.L.; Scholz, R.L.; Chaudhuri, A.A.; Kahn, M.E.; Rao, D.S.; Baltimore, D. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity 2010, 33, 607–619. [Google Scholar] [CrossRef]
- Murugaiyan, G.; Beynon, V.; Mittal, A.; Joller, N.; Weiner, H.L. Silencing microRNA-155 ameliorates experimental autoimmune encephalomyelitis. J. Immunol. 2011, 187, 2213–2221. [Google Scholar] [CrossRef]
- Johansson, K.; Malmhall, C.; Ramos-Ramirez, P.; Radinger, M. MicroRNA-155 is a critical regulator of type 2 innate lymphoid cells and IL-33 signaling in experimental models of allergic airway inflammation. J. Allergy Clin. Immunol. 2017, 139, 1007–1016. [Google Scholar] [CrossRef]
- Banerjee, A.; Schambach, F.; DeJong, C.S.; Hammond, S.M.; Reiner, S.L. Micro-RNA-155 inhibits IFN-gamma signaling in CD4+ T cells. Eur. J. Immunol. 2010, 40, 225–231. [Google Scholar] [CrossRef]
- Thai, T.-H.; Calado, D.P.; Casola, S.; Ansel, K.M.; Xiao, C.; Xue, Y.; Murphy, A.; Frendewey, D.; Valenzuela, D.; Kutok, J.L.; et al. Regulation of the germinal center response by microRNA-155. Science 2007, 316, 604–608. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.-F.; Gasteiger, G.; Yu, I.-S.; Chaudhry, A.; Hsin, J.-P.; Lu, Y.; Bos, P.D.; Lin, L.-L.; Zawislak, C.L.; Cho, S.; et al. A single miRNA-mRNA interaction affects the immune response in a context- and cell-type-specific manner. Immunity 2015, 43, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Dunand-Sauthier, I.; Irla, M.; Carnesecchi, S.; Seguín-Estévez, Q.; Vejnar, C.E.; Zdobnov, E.M.; Santiago-Raber, M.-L.; Reith, W. Repression of arginase-2 expression in dendritic cells by microRNA-155 is critical for promoting T cell proliferation. J. Immunology 2014, 193, 1690–1700. [Google Scholar] [CrossRef] [PubMed]
- Zech, A.; Ayata, C.K.; Pankratz, F.; Meyer, A.; Baudiß, K.; Cicko, S.; Yegutkin, G.G.; Grundmann, S.; Idzko, M. MicroRNA-155 modulates P2R signaling and Th2 priming of dendritic cells during allergic airway inflammation in mice. Allergy 2015, 70, 1121–1129. [Google Scholar] [CrossRef]
- Singh, P.B.; Pua, H.H.; Happ, H.C.; Schneider, C.; von Moltke, J.; Locksley, R.M.; Baumjohann, D.; Ansel, K.M. MicroRNA regulation of type 2 innate lymphoid cell homeostasis and function in allergic inflammation. J. Exp. Med. 2017, 214, 3627–3643. [Google Scholar] [CrossRef]
- Mattes, J.; Collison, A.; Plank, M.; Phipps, S.; Foster, P.S. Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proc. Natl. Acad. Sci. USA 2009, 106, 18704–18709. [Google Scholar] [CrossRef]
- Collison, A.; Herbert, C.; Siegle, J.S.; Mattes, J.; Foster, P.S.; Kumar, R.K. Altered expression of microRNA in the airway wall in chronic asthma: miR-126 as a potential therapeutic target. BMC Pulm. Med. 2011, 11, 29. [Google Scholar] [CrossRef]
- Lu, T.X.; Hartner, J.; Lim, E.-J.; Fabry, V.; Mingler, M.K.; Cole, E.T.; Orkin, S.H.; Aronow, B.J.; Rothenberg, M.E. MicroRNA-21 limits in vivo immune response-mediated activation of the IL-12/IFN-gamma pathway, Th1 polarization, and the severity of delayed-type hypersensitivity. J. Immunol. 2011, 187, 3362–3373. [Google Scholar] [CrossRef]
- Oswald, I.P.; Caspar, P.; Jankovic, D.; Wynn, T.A.; Pearce, E.J.; Sher, A. IL-12 inhibits Th2 cytokine responses induced by eggs of Schistosoma mansoni. J. Immunol. 1994, 153, 1707–1713. [Google Scholar]
- Sawant, D.V.; Wu, H.; Kaplan, M.H.; Dent, A.L. The Bcl6 target gene microRNA-21 promotes Th2 differentiation by a T cell intrinsic pathway. Mol. Immunol. 2013, 54, 435–442. [Google Scholar] [CrossRef]
- Yamashita, M.; Shinnakasu, R.; Asou, H.; Kimura, M.; Hasegawa, A.; Hashimoto, K.; Hatano, N.; Ogata, M.; Nakayama, T. Ras-ERK MAPK cascade regulates GATA3 stability and Th2 differentiation through ubiquitin-proteasome pathway. J. Biol. Chem. 2005, 280, 29409–29419. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Rao, S.; Tsai, L.M.; Lee, S.K.; He, Y.; Sutcliffe, E.L.; Srivastava, M.; Linterman, M.; Zheng, L.; Simpson, N.; et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity 2009, 31, 457–468. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Lee, H.Y.; Choi, J.Y.; Hur, J.; Kim, I.K.; Kim, Y.K.; Kang, J.Y.; Lee, S.Y. Inhibition of MicroRNA-21 by an antagomir ameliorates allergic inflammation in a mouse model of asthma. Exp. Lung Res. 2017, 43, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Aurora, A.B.; Johnson, B.A.; Qi, X.; McAnally, J.; Hill, J.A.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell 2008, 15, 261–271. [Google Scholar] [CrossRef]
- Lechman, E.R.; Gentner, B.; Ng, S.W.K.; Schoof, E.M.; van Galen, P.; Kennedy, J.A.; Nucera, S.; Ciceri, F.; Kaufmann, K.B.; Takayama, N.; et al. MiR-126 regulates distinct self-renewal outcomes in normal and malignant hematopoietic stem cells. Cancer Cell 2016, 29, 602–606. [Google Scholar] [CrossRef]
- Okuyama, K.; Ikawa, T.; Gentner, B.; Hozumi, K.; Harnprasopwat, R.; Lu, J.; Yamashita, R.; Ha, D.; Toyoshima, T.; Chanda, B.; et al. MicroRNA-126-mediated control of cell fate in B-cell myeloid progenitors as a potential alternative to transcriptional factors. Proc. Natl. Acad. Sci. USA 2013, 110, 13410–13415. [Google Scholar] [CrossRef]
- Zhang, J.; Du, Y.-Y.; Lin, Y.-F.; Chen, Y.-T.; Yang, L.; Wang, H.-J.; Ma, D. The cell growth suppressor, mir-126, targets IRS-1. Biochem. Biophys. Res. Commun. 2008, 377, 136–140. [Google Scholar] [CrossRef]
- Chu, F.; Hu, Y.; Zhou, Y.; Guo, M.; Lu, J.; Zheng, W.; Xu, H.; Zhao, J.; Xu, L. MicroRNA-126 deficiency enhanced the activation and function of CD4+ T cells by elevating IRS-1 pathway. Clin. Exp. Immunol. 2018, 191, 166–179. [Google Scholar] [CrossRef]
- Qin, A.; Wen, Z.; Zhou, Y.; Li, Y.; Li, Y.; Luo, J.; Ren, T.; Xu, L. MicroRNA-126 regulates the induction and function of CD4(+) Foxp3(+) regulatory T cells through PI3K/AKT pathway. J. Cell. Mol. Med. 2013, 17, 252–264. [Google Scholar] [CrossRef]
- Knudsen, L.A.; Petersen, N.; Schwartz, T.W.; Egerod, K.L. The microRNA repertoire in enteroendocrine cells: Identification of miR-375 as a potential regulator of the enteroendocrine lineage. Endocrinology 2015, 156, 3971–3983. [Google Scholar] [CrossRef]
- Biton, M.; Levin, A.; Slyper, M.; Alkalay, I.; Horwitz, E.; Mor, H.; Kredo-Russo, S.; Avnit-Sagi, T.; Cojocaru, G.; Zreik, F.; et al. Epithelial microRNAs regulate gut mucosal immunity via epithelium-T cell crosstalk. Nat. Immunol. 2011, 12, 239–246. [Google Scholar] [CrossRef]
- Shroyer, N.F.; Wallis, D.; Venken, K.J.T.; Bellen, H.J.; Zoghbi, H.Y. Gfi1 functions downstream of Math1 to control intestinal secretory cell subtype allocation and differentiation. Genes Dev. 2005, 19, 2412–2417. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.X.; Lim, E.J.; Wen, T.; Plassard, A.J.; Hogan, S.P.; Martin, L.J.; Aronow, B.J.; Rothenberg, M.E. MiR-375 is downregulated in epithelial cells after IL-13 stimulation and regulates an IL-13-induced epithelial transcriptome. Mucosal Immunol. 2012, 5, 388–396. [Google Scholar] [CrossRef]
- Simpson, L.J.; Patel, S.; Bhakta, N.R.; Choy, D.F.; Brightbill, H.D.; Ren, X.; Wang, Y.; Pua, H.H.; Baumjohann, D.; Montoya, M.M.; et al. A microRNA upregulated in asthma airway T cells promotes TH2 cytokine production. Nat. Immunol. 2014, 15, 1162–1170. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Srinivasan, L.; Calado, D.P.; Patterson, H.C.; Zhang, B.; Wang, J.; Henderson, J.M.; Kutok, J.L.; Rajewsky, K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat. Immunol. 2008, 9, 405–414. [Google Scholar] [CrossRef]
- de Kouchkovsky, D.; Esensten, J.H.; Rosenthal, W.L.; Morar, M.M.; Bluestone, J.A.; Jeker, L.T. MicroRNA-17–92 regulates IL-10 production by regulatory T cells and control of experimental autoimmune encephalomyelitis. J. Immunol. 2013, 191, 1594–1605. [Google Scholar] [CrossRef]
- Liu, S.-Q.; Jiang, S.; Li, C.; Zhang, B.; Li, Q.-J. MiR-17-92 cluster targets phosphatase and tensin homology and ikaros family zinc finger 4 to promote TH17-mediated inflammation. J. Biol. Chem. 2014, 289. [Google Scholar] [CrossRef]
- Chen, L.; Guo, D. The functions of tumor suppressor PTEN in innate and adaptive immunity. Cell. Mol. Immunol. 2017, 14, 581–589. [Google Scholar] [CrossRef]
- Tanzer, A.; Stadler, P.F. Molecular evolution of a microRNA cluster. J. Mol. Biol. 2004, 339, 327–335. [Google Scholar] [CrossRef]
- Kuchen, S.; Resch, W.; Yamane, A.; Kuo, N.; Li, Z.; Chakraborty, T.; Wei, L.; Laurence, A.; Yasuda, T.; Peng, S.; et al. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity 2010, 32, 828–839. [Google Scholar] [CrossRef]
- Guo, Y.E.; Riley, K.J.; Iwasaki, A.; Steitz, J.A. Alternative capture of noncoding RNAs or protein-coding genes by herpesviruses to alter host T cell function. Mol. Cell 2014, 54, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Chandran, P.A.; Keller, A.; Weinmann, L.; Seida, A.A.; Braun, M.; Andreev, K.; Fischer, B.; Horn, E.; Schwinn, S.; Junker, M.; et al. The TGF-β-inducible miR-23a cluster attenuates IFN-γ levels and antigen-specific cytotoxicity in human CD8⁺ T cells. J. Leukoc. Biol. 2014, 96, 633–645. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Wu, C.-J.; Yasuda, T.; Cruz, L.O.; Khan, A.A.; Lin, L.-L.; Nguyen, D.T.; Miller, M.; Lee, H.-M.; Kuo, M.-L.; et al. MiR-23∼27∼24 clusters control effector T cell differentiation and function. J. Exp. Med. 2016, 213, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Pua, H.H.; Steiner, D.F.; Patel, S.; Gonzalez, J.R.; Ortiz-Carpena, J.F.; Kageyama, R.; Chiou, N.-T.; Gallman, A.; de Kouchkovsky, D.; Jeker, L.T.; et al. MicroRNAs 24 and 27 suppress allergic inflammation and target a network of regulators of T helper 2 cell-associated cytokine production. Immunity 2016, 44, 821–832. [Google Scholar] [CrossRef]
- Wenzel, S.; Ford, L.; Pearlman, D.; Spector, S.; Sher, L.; Skobieranda, F.; Wang, L.; Kirkesseli, S.; Rocklin, R.; Bock, B.; et al. Dupilumab in persistent asthma with elevated eosinophil levels. N. Engl. J. Med. 2013, 368, 2455–2466. [Google Scholar] [CrossRef]
- Marone, G.; Spadaro, G.; Braile, M.; Poto, R.; Criscuolo, G.; Pahima, H.; Loffredo, S.; Levi-Schaffer, F.; Varricchi, G. Tezepelumab: A novel biological therapy for the treatment of severe uncontrolled asthma. Expert Opin. Investig. Drugs 2019, 28, 931–940. [Google Scholar] [CrossRef]
- Harries, L.W. RNA biology provides new therapeutic targets for human disease. Front. Genet. 2019, 10, 205. [Google Scholar] [CrossRef]

| Human Type-2 Diseases | Mouse Type-2 Diseases | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Skin | Respiratory Tract | Skin | Respiratory Tract | ||||||
| miRNA | Function | Atopic Dermatitis | Psoriasis | EoE * (Esophagus Brushing) | Asthma (Airway Brushing) | Atopic Dermatitis | IL-13Tg # (Whole Lung) | Allergic Asthma % (Whole Lung) | |
| miR-142 | immune-modulator | Sonkoly, E. Vennegaard, M.T. | Lu, T. X. | Vennegaard, M.T. | Lu, T.X. | UP-REGULATED | |||
| miR-223 | anti-inflammatory | Sonkoly, E. Vennegaard, M.T. | Lu, T. X. | Vennegaard, M.T. | |||||
| miR-21 | oncogene / pro-inflammatory | Vennegaard, M.T. | Joyce, C.E. | Lu, T.X. | Wu, X.-B | Vennegaard, M.T. | Lu, T.X. | Mattes, J. | |
| miR-126 | pro-inflammatory / T cell suppr. | Wu, X.-B | Collison, A | ||||||
| miR-135 | unclear | Sonkoly, E. | Joyce, C.E. | ||||||
| miR-146 | anti-inflammatory | Vennegaard, M.T. | Lu, T.X. | Vennegaard, M.T. | Lu, T.X. | ||||
| miR-92 | anti-apoptotic | Lu, T.X. | Solberg, O.D. | ||||||
| miR-1268 | unclear | Joyce, C.E. | Solberg, O.D. | ||||||
| miR-222 | oncogene / DC maturation | Joyce, C.E. | Lu, T.X. | ||||||
| miR-31 | unclear | Sonkoly, E. | Joyce, C.E. | ||||||
| miR-149 | anti-inflammatory | Solberg, O.D. | |||||||
| miR-33 | unclear | Joyce, C.E. | |||||||
| miR-155 | oncogene / modulator | Sonkoly, E. | Okoye, I | ||||||
| mR-149 | anti-inflammatory | Sonkoly, E. Vennegaard, M.T. | Lu, T.X. | Vennegaard, M.T. | |||||
| miR-193 | anti-tumorigenic anti-inflammatory | Sonkoly, E. Vennegaard, M.T. | Vennegaard, M.T. | DOWN-REGULATED | |||||
| miR-30 | unclear | Sonkoly, E. | Lu, T.X. | Solberg, O.D. | |||||
| miR-33 | unclear | Vennegaard, M.T. | Vennegaard, M.T. | ||||||
| miR-181 | anti-inflammatory | Vennegaard, M.T. | Vennegaard, M.T. | ||||||
| miR-34/449 | cilia differentiation | Solberg, O.D. | Yin, H. | ||||||
| miR-99 | anti-inflammatory | Sonkoly, E. | Solberg, O.D. | ||||||
| miR-195 | anti-inflammatory | Sonkoly, E. | Lu, T.X. | ||||||
| miR-26 | anti-inflammatory | Sonkoly, E. | Solberg, O.D. | ||||||
| miR-101 | pro-inflammatory | Sonkoly, E. | Lu, T.X. | ||||||
| miR-365 | anti-inflammatory | Sonkoly, E. | Lu, T.X. | ||||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guidi, R.; Wedeles, C.J.; Wilson, M.S. ncRNAs in Type-2 Immunity. Non-Coding RNA 2020, 6, 10. https://doi.org/10.3390/ncrna6010010
Guidi R, Wedeles CJ, Wilson MS. ncRNAs in Type-2 Immunity. Non-Coding RNA. 2020; 6(1):10. https://doi.org/10.3390/ncrna6010010
Chicago/Turabian StyleGuidi, Riccardo, Christopher J. Wedeles, and Mark S. Wilson. 2020. "ncRNAs in Type-2 Immunity" Non-Coding RNA 6, no. 1: 10. https://doi.org/10.3390/ncrna6010010
APA StyleGuidi, R., Wedeles, C. J., & Wilson, M. S. (2020). ncRNAs in Type-2 Immunity. Non-Coding RNA, 6(1), 10. https://doi.org/10.3390/ncrna6010010





