Vesiculated Long Non-Coding RNAs: Offshore Packages Deciphering Trans-Regulation between Cells, Cancer Progression and Resistance to Therapies
Abstract
:1. Introduction
2. The ncRNA Precursor’s Incorporation into EVs
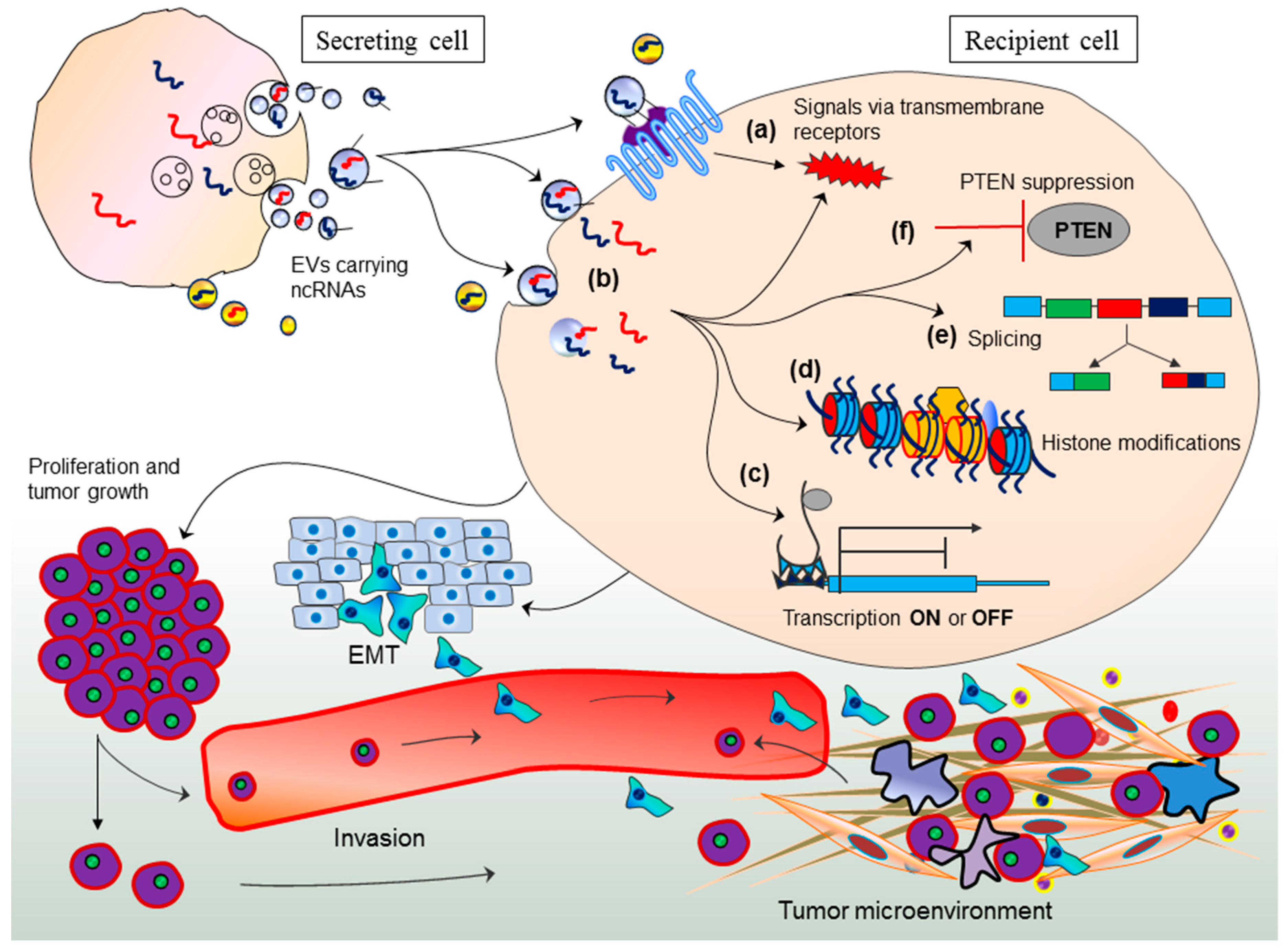
3. EV-Mediated miRNA Transport and Epigenetic Regulation in Recipient Cells
4. EV-Mediated Long Non-Coding RNA Transport: A Novel Source of Epigenetic Regulation
5. EV-Associated ncRNAs: Conveyers of Genomic Instability and Tumor Progression
6. EV-ncRNAs Are Extended Messages in Regulating Responses to Chemotherapy
7. Roles in Tumor Inhibition
8. EV-Based RNA Interference in Targeted Cancer Cells: Vehicle of Gene Therapy
9. EV-Encapsulated ncRNAs as Diagnostic and Prognostic Biomarkers: At a Glance
10. Concluding Remarks and Clinical Implications
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Nawaz, M.; Camussi, G.; Valadi, H.; Nazarenko, I.; Ekstrom, K.; Wang, X.; Principe, S.; Shah, N.; Ashraf, NM.; Fatima, F.; et al. The emerging role of extracellular vesicles as biomarkers for urogenital cancers. Nat. Rev. Urol. 2014, 11, 688–701. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.M.; Abdelmohsen, K.; Mustapic, M.; Kapogiannis, D.; Gorospe, M. RNA in extracellular vesicles. Wiley Interdiscip. Rev.: RNA 2017. [Google Scholar] [CrossRef] [PubMed]
- Keerthikumar, S.; Chisanga, D.; Ariyaratne, D.; Al Saffar, H.; Anand, S.; Zhao, K.; Samuel, M.; Pathan, M.; Jois, M.; Chilamkurti, N.; et al. ExoCarta: A web-based compendium of exosomal cargo. J. Mol. Biol. 2016, 428, 688–692. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Mathivanan, S.; Ji, H.; Simpson, R.J. Exosomes: Extracellular organelles important in intercellular communication. J. Proteom. 2010, 73, 1907–1920. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, M.; Fatima, F.; Vallabhaneni, K.C.; Penfornis, P.; Valadi, H.; Ekstrom, K.; Kholia, S.; Whitt, J.D.; Fernandes, J.D.; Pochampally, R.; et al. Extracellular vesicles: evolving factors in stem cell biology. Stem Cells Int. 2016, 2016, 1073140. [Google Scholar] [CrossRef] [PubMed]
- Fatima, F.; Nawaz, M. Stem cell-derived exosomes: Roles in stromal remodeling, tumor progression, and cancer immunotherapy. Chin. J. Cancer 2015, 34, 541–653. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 925–933. [Google Scholar] [CrossRef] [PubMed]
- Taft, R.J.; Pang, K.C.; Mercer, T.R.; Dinger, M.; Mattick, J.S. Non-coding RNAs: Regulators of disease. J. Pathol. 2010, 220, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar]
- Consortium, E.P.; Birney, E.; Stamatoyannopoulos, J.A.; Dutta, A.; Guigo, R.; Gingeras, T.R.; Margulies, E.H.; Weng, Z.; Snyder, M.; Dermitzakis, E.T.; et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007, 447, 799–816. [Google Scholar]
- Lee, J.T. Epigenetic regulation by long noncoding RNAs. Science 2012, 338, 1435–1439. [Google Scholar] [CrossRef] [PubMed]
- Consortium, E.P. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 2004, 306, 636–640. [Google Scholar] [CrossRef] [PubMed]
- Doolittle, W.F. Is junk DNA bunk? A critique of ENCODE. Proc. Natl. Acad. Sci. USA 2013, 110, 5294–5300. [Google Scholar] [CrossRef] [PubMed]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Grosshans, H.; Filipowicz, W. Proteomics joins the search for microRNA targets. Cell 2008, 134, 560–562. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Pasquinelli, A.E.; Ruvkun, G. Control of developmental timing by micrornas and their targets. Annu. Rev. Cell Dev. Biol. 2002, 18, 495–513. [Google Scholar] [CrossRef] [PubMed]
- Lim, L.P.; Lau, N.C.; Weinstein, E.G.; Abdelhakim, A.; Yekta, S.; Rhoades, M.W.; Burge, C.B.; Bartel, D.P. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003, 17, 991–1008. [Google Scholar] [CrossRef] [PubMed]
- Carrington, J.C.; Ambros, V. Role of microRNAs in plant and animal development. Science 2003, 301, 336–338. [Google Scholar] [CrossRef] [PubMed]
- Ambros, V. MicroRNA pathways in flies and worms: Growth, death, fat, stress, and timing. Cell 2003, 113, 673–676. [Google Scholar] [CrossRef]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Sempere, L.F.; Freemantle, S.; Pitha-Rowe, I.; Moss, E.; Dmitrovsky, E.; Ambros, V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol 2004, 5, R13. [Google Scholar] [CrossRef] [PubMed]
- Ivey, K.N.; Srivastava, D. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell 2010, 7, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- Banfai, B.; Jia, H.; Khatun, J.; Wood, E.; Risk, B.; Gundling, W.E., Jr.; Kundaje, A.; Gunawardena, H.P.; Yu, Y.; Xie, L.; et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Biol. 2012, 22, 1646–1657. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y.; et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Mattick, J.S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013, 20, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.M.; Guttman, M.; Huarte, M.; Garber, M.; Raj, A.; Rivea Morales, D.; Thomas, K.; Presser, A.; Bernstein, B.E.; van Oudenaarden, A.; et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 11667–11672. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Wapinski, O.; Chang, H.Y. Long noncoding RNAs and human disease. Trends Cell Biol. 2011, 21, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Spizzo, R.; Almeida, M.I.; Colombatti, A.; Calin, G.A. Long non-coding RNAs and cancer: A new frontier of translational research? Oncogene 2012, 31, 4577–4587. [Google Scholar] [CrossRef] [PubMed]
- Gibb, E.A.; Brown, C.J.; Lam, W.L. The functional role of long non-coding RNA in human carcinomas. Mol. Cancer 2011, 10, 38. [Google Scholar] [CrossRef] [PubMed]
- Amicone, L.; Citarella, F.; Cicchini, C. Epigenetic regulation in hepatocellular carcinoma requires long noncoding RNAs. BioMed Res. Int. 2015, 2015, 473942. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.C.; Spitale, R.C.; Chang, H.Y. Long intergenic noncoding RNAs: New links in cancer progression. Cancer Res. 2011, 71, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Gutschner, T.; Hammerle, M.; Diederichs, S. MALAT1—A paradigm for long noncoding RNA function in cancer. J. Mol. Med. 2013, 91, 791–801. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Bi, J.; Xue, X.; Zheng, L.; Zhi, K.; Hua, J.; Fang, G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012, 279, 3159–3165. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.L.; Wang, Y.; Jin, B.; Chen, H.; Xie, B.S.; Mao, Z.B. Senescence-associated Long Non-coding RNA (SALNR) Delays Oncogene-induced Senescence through NF90 Regulation. J. Biol. Chem. 2015, 290, 30175–30192. [Google Scholar] [CrossRef] [PubMed]
- Abdelmohsen, K.; Panda, A.; Kang, M.J.; Xu, J.; Selimyan, R.; Yoon, J.H.; Martindale, J.L.; De, S.; Wood, W.H.; Becker, K.G.; et al. Senescence-associated lncRNAs: Senescence-associated long noncoding RNAs. Aging Cell 2013, 12, 890–900. [Google Scholar] [CrossRef] [PubMed]
- Askarian-Amiri, M.E.; Leung, E.; Finlay, G.; Baguley, B.C. The regulatory role of long noncoding RNAs in Cancer Drug Resistance. Methods Mol. Biol. 2016, 1395, 207–227. [Google Scholar] [PubMed]
- Deng, H.; Zhang, J.; Shi, J.; Guo, Z.; He, C.; Ding, L.; Tang, J.H.; Hou, Y. Role of long non-coding RNA in tumor drug resistance. Tumour Biol. 2016, 37, 11623–11631. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, V.B.; Ovsepian, S.V.; Carrascosa, L.G.; Buske, F.A.; Radulovic, V.; Niyazi, M.; Moertl, S.; Trau, M.; Atkinson, M.J.; Anastasov, N. PARTICLE, a triplex-forming long ncRNA, regulates locus-specific methylation in response to low-dose irradiation. Cell Rep. 2015, 11, 474–485. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [PubMed]
- Savelyeva, A.V.; Baryakin, D.N.; Chikova, E.D.; Kuligina, E.V.; Richter, V.A.; Semenov, D.V. Vesicular and extra-eesicular RNAs of human blood plasma. Adv. Exp. Med. Biol. 2016, 924, 117–119. [Google Scholar] [PubMed]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Vickers, K.C.; Remaley, A.T. Lipid-based carriers of microRNAs and intercellular communication. Curr. Opin. Lipidol. 2012, 23, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Cortez, M.A.; Bueso-Ramos, C.; Ferdin, J.; Lopez-Berestein, G.; Sood, A.K.; Calin, G.A. MicroRNAs in body fluids—The mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 2011, 8, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Chevillet, J.R.; Kang, Q.; Ruf, I.K.; Briggs, H.A.; Vojtech, L.N.; Hughes, S.M.; Cheng, H.H.; Arroyo, J.D.; Meredith, E.K.; Gallichotte, E.N.; et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc. Natl. Acad. Sci. USA 2014, 111, 14888–14893. [Google Scholar] [CrossRef] [PubMed]
- Turchinovich, A.; Weiz, L.; Langheinz, A.; Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011, 39, 7223–7233. [Google Scholar] [CrossRef] [PubMed]
- Gourzones, C.; Ferrand, F.R.; Amiel, C.; Verillaud, B.; Barat, A.; Guerin, M.; Gattolliat, C.H.; Gelin, A.; Klibi, J.; Chaaben, A.B.; et al. Consistent high concentration of the viral microRNA BART17 in plasma samples from nasopharyngeal carcinoma patients—Evidence of non-exosomal transport. Virol. J. 2013, 10, 119. [Google Scholar] [CrossRef] [PubMed]
- Mateescu, B.; Kowal, E.; Balkom, B.; Bartel, S.; Bhattacharyya, S.; Buzas, E.; Buck, A.; Candia, P.; Chow, F.; Das, S.; et al. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA. J. Extracell. Vesicles 2017, 6. [Google Scholar] [CrossRef]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Radmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Jeon, K.; Lee, J.T.; Kim, S.; Kim, V.N. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002, 21, 4663–4670. [Google Scholar] [CrossRef] [PubMed]
- Lund, E.; Guttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear export of microRNA precursors. Science 2004, 303, 95–98. [Google Scholar] [CrossRef] [PubMed]
- Yi, R.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.S.; Lai, R.C.; Lee, M.M.; Choo, A.B.; Lee, C.N.; Lim, S.K. Mesenchymal stem cell secretes microparticles enriched in pre-microRNAs. Nucleic Acids Res. 2010, 38, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Gibbings, D.J.; Ciaudo, C.; Erhardt, M.; Voinnet, O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 2009, 11, 1143–1149. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Pressman, S.; Andress, A.P.; Kim, K.; White, J.L.; Cassidy, J.J.; Li, X.; Lubell, K.; Lim, D.H.; Cho, I.S.; et al. Silencing by small RNAs is linked to endosomal trafficking. Nat. Cell Biol. 2009, 11, 1150–1156. [Google Scholar] [CrossRef] [PubMed]
- Squadrito, M.L.; Baer, C.; Burdet, F.; Maderna, C.; Gilfillan, G.D.; Lyle, R.; Ibberson, M.; De Palma, M.; et al. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 2014, 8, 1432–1446. [Google Scholar] [CrossRef] [PubMed]
- Melo, S.A.; Sugimoto, H.; O’Connell, J.T.; Kato, N.; Villanueva, A.; Vidal, A.; Qiu, L.; Vitkin, E.; Perelman, L.T.; Melo, C.A.; et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 2014, 26, 707–721. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Ratajczak, J.; Miekus, K.; Kucia, M.; Zhang, J.; Reca, R.; Dvorak, P.; Ratajczak, M.Z. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: Evidence for horizontal transfer of mRNA and protein delivery. Leukemia 2006, 20, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Aliotta, J.M.; Pereira, M.; Johnson, K.W.; de Paz, N.; Dooner, M.S.; Puente, N.; Ayala, C.; Brilliant, K.; Berz, D.; Lee, D.; et al. Microvesicle entry into marrow cells mediates tissue-specific changes in mRNA by direct delivery of mRNA and induction of transcription. Exp. Hematol. 2010, 38, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Vojtech, L.; Woo, S.; Hughes, S.; Levy, C.; Ballweber, L.; Sauteraud, R.P.; Strobl, J.; Westerberg, K.; Gottardo, R.; Tewari, M.; et al. Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res. 2014, 42, 7290–7304. [Google Scholar] [CrossRef] [PubMed]
- Quesenberry, P.J.; Aliotta, J.; Deregibus, M.C.; Camussi, G. Role of extracellular RNA-carrying vesicles in cell differentiation and reprogramming. Stem Cell Res. Ther. 2015, 6, 153. [Google Scholar] [CrossRef] [PubMed]
- Aliotta, J.M.; Pereira, M.; Li, M.; Amaral, A.; Sorokina, A.; Dooner, M.S.; Sears, E.H.; Brilliant, K.; Ramratnam, B.; Hixson, D.C.; et al. Stable cell fate changes in marrow cells induced by lung-derived microvesicles. J. Extracell. Vesicles 2012, 1, 18163. [Google Scholar] [CrossRef] [PubMed]
- Yuan, A.; Farber, E.L.; Rapoport, A.L.; Tejada, D.; Deniskin, R.; Akhmedov, N.B.; Farber, D.B. Transfer of microRNAs by embryonic stem cell microvesicles. PLoS ONE 2009, 4, e4722. [Google Scholar] [CrossRef] [PubMed]
- Eirin, A.; Riester, S.M.; Zhu, X.Y.; Tang, H.; Evans, J.M.; O’Brien, D.; van Wijnen, A.J.; Lerman, L.O. MicroRNA and mRNA cargo of extracellular vesicles from porcine adipose tissue-derived mesenchymal stem cells. Gene 2014, 551, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Deng, Z.; Dahmane, N.; Tsai, K.; Wang, P.; Williams, D.R.; Kossenkov, A.V.; Showe, L.C.; Zhang, R.; Huang, Q.; et al. Telomeric repeat-containing RNA (TERRA) constitutes a nucleoprotein component of extracellular inflammatory exosomes. Proc. Natl. Acad. Sci. USA 2015, 112, E6293–E6300. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Lieberman, P.M. The crosstalk of telomere dysfunction and inflammation through cell-free TERRA containing exosomes. RNA Biol. 2016, 13, 690–695. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Kim, D.; Han, J.; Kim, Y.; Lee, M.; Jin, E.J. PBMC and exosome-derived Hotair is a critical regulator and potent marker for rheumatoid arthritis. Clin. Exp. Med. 2015, 15, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Berrondo, C.; Flax, J.; Kucherov, V.; Siebert, A.; Osinski, T.; Rosenberg, A.; Fucile, C.; Richheimer, S.; Beckham, C.J. Expression of the long non-coding RNA HOTAIR correlates with disease progression in bladder cancer and is contained in bladder cancer patient urinary exosomes. PLoS ONE 2016, 11, e0147236. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhou, Y.; Lu, J.; Sun, Y.; Xiao, H.; Liu, M.; Tian, L. Combined detection of serum exosomal miR-21 and HOTAIR as diagnostic and prognostic biomarkers for laryngeal squamous cell carcinoma. Med. Oncol. 2014, 31, 148. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Cai, Y.; Wu, G.; Chen, X.; Liu, Y.; Wang, X.; Yu, J.; Li, C.; Chen, X.; Jose, P.A.; et al. Plasma long non-coding RNA, CoroMarker, a novel biomarker for diagnosis of coronary artery disease. Clin. Sci. 2015, 129, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Gezer, U.; Ozgur, E.; Cetinkaya, M.; Isin, M.; Dalay, N. Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol. Int. 2014, 38, 1076–1079. [Google Scholar] [CrossRef] [PubMed]
- Ahadi, A.; Khoury, S.; Losseva, M.; Tran, N. A comparative analysis of lncRNAs in prostate cancer exosomes and their parental cell lines. Genom. Data 2016, 9, 7–9. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.J.; Bikle, D.D. LncRNA: A new player in 1alpha, 25(OH)2 vitamin D3/VDR protection against skin cancer formation. Exp. Dermatol. 2014, 23, 147–150. [Google Scholar] [CrossRef] [PubMed]
- Gallo, A.; Vella, S.; Miele, M.; Timoneri, F.; Di Bella, M.; Bosi, S.; Sciveres, M.; Conaldi, P.G. Global profiling of viral and cellular non-coding RNAs in Epstein-Barr virus-induced lymphoblastoid cell lines and released exosome cargos. Cancer Lett. 2017, 388, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Du, C.; Xie, H.; Zang, R.; Shen, Z.; Li, H.; Chen, P.; Xu, X.; Xia, Y.; Tang, W. Apoptotic neuron-secreted HN12 inhibits cell apoptosis in Hirschsprung’s disease. Int. J. Nanomed. 2016, 11, 5871–5881. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, Y.; Tsujimoto, M.; Yanoshita, R. Next-generation sequencing of protein-coding and long non-protein-coding RNAs in two types of exosomes derived from human whole saliva. Biol. Pharm. Bull. 2016, 39, 1496–1507. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Yao, J.; Wu, D.C.; Nottingham, R.M.; Mohr, S.; Hunicke-Smith, S.; Lambowitz, A.M. High-throughput sequencing of human plasma RNA by using thermostable group II intron reverse transcriptases. RNA 2016, 22, 111–128. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Liu, C.G.; Sevignani, C.; Ferracin, M.; Felli, N.; Dumitru, C.D.; Shimizu, M.; Cimmino, A.; Zupo, S.; Dono, M.; et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc. Natl. Acad. Sci. USA 2004, 101, 11755–11760. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Ferracin, M.; Cimmino, A.; Di Leva, G.; Shimizu, M.; Wojcik, S.E.; Iorio, M.V.; Visone, R.; Sever, N.I.; Fabbri, M.; et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005, 353, 1793–1801. [Google Scholar] [CrossRef]
- Janowska-Wieczorek, A.; Wysoczynski, M.; Kijowski, J.; Marquez-Curtis, L.; Machalinski, B.; Ratajczak, J.; Ratajczak, M.Z. Microvesicles derived from activated platelets induce metastasis and angiogenesis in lung cancer. Int. J. Cancer 2005, 113, 752–760. [Google Scholar] [CrossRef] [PubMed]
- Costa-Silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Zhang, H.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H.; et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015, 17, 816–826. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Costa-Silva, B.; Shen, T.L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Lima, L.G.; Chammas, R.; Monteiro, R.Q.; Moreira, M.E.; Barcinski, M.A. Tumor-derived microvesicles modulate the establishment of metastatic melanoma in a phosphatidylserine-dependent manner. Cancer Lett. 2009, 283, 168–175. [Google Scholar] [CrossRef]
- Greening, D.W.; Gopal, S.K.; Mathias, R.A.; Liu, L.; Sheng, J.; Zhu, H.J.; Simpson, R.J. Emerging roles of exosomes during epithelial-mesenchymal transition and cancer progression. Semin. Cell Dev. Biol. 2015, 40, 60–71. [Google Scholar] [CrossRef] [PubMed]
- Taverna, S.; Amodeo, V.; Saieva, L.; Russo, A.; Giallombardo, M.; De Leo, G.; Alessandro, R. Exosomal shuttling of miR-126 in endothelial cells modulates adhesive and migratory abilities of chronic myelogenous leukemia cells. Mol. Cancer 2014, 13, 169. [Google Scholar] [CrossRef] [PubMed]
- Peinado, H.; Aleckovic, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; Garcia-Santos, G.; Ghajar, C.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012, 18, 883–891. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, M.; Fatima, F.; Nazarenko, I.; Ekstrom, K.; Murtaza, I.; Anees, M.; Sultan, A.; Neder, L.; Camussi, G.; Valadi, H.; et al. Extracellular vesicles in ovarian cancer: Applications to tumor biology, immunotherapy and biomarker discovery. Expert Rev. Proteom. 2016, 13, 395–409. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, S.; Yao, J.; Lowery, F.J.; Zhang, Q.; Huang, W.C.; Li, P.; Li, M.; Wang, X.; Zhang, C.; et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature 2015, 527, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yan, I.K.; Haga, H.; Patel, T. Modulation of hypoxia-signaling pathways by extracellular linc-RoR. J. Cell Sci. 2014, 127, 1585–1594. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Pochampally, R.; Watabe, K.; Lu, Z.; Mo, Y.Y. Exosome-mediated transfer of miR-10b promotes cell invasion in breast cancer. Mol. Cancer 2014, 13, 256. [Google Scholar] [CrossRef] [PubMed]
- Fong, M.Y.; Zhou, W.; Liu, L.; Alontaga, A.Y.; Chandra, M.; Ashby, J.; Chow, A.; O’Connor, S.T.; Li, S.; Chin, A.R.; et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 2015, 17, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Felicetti, F.; De Feo, A.; Coscia, C.; Puglisi, R.; Pedini, F.; Pasquini, L.; Bellenghi, M.; Errico, M.C.; Pagani, E.; Care, A. Exosome-mediated transfer of miR-222 is sufficient to increase tumor malignancy in melanoma. J. Transl. Med. 2016, 14, 56. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Liu, R.; Shi, Y.J.; Yin, L.H.; Pu, Y.P. Exosome-shuttling microRNA-21 promotes cell migration and invasion-targeting PDCD4 in esophageal cancer. Int. J. Oncol. 2016, 48, 2567–2579. [Google Scholar] [CrossRef] [PubMed]
- Bigagli, E.; Luceri, C.; Guasti, D.; Cinci, L. Exosomes secreted from human colon cancer cells influence the adhesion of neighboring metastatic cells: Role of microRNA-210. Cancer Biol. Ther. 2016, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kogure, T.; Lin, W.L.; Yan, I.K.; Braconi, C.; Patel, T. Intercellular nanovesicle-mediated microRNA transfer: A mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology 2011, 54, 1237–1248. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Chen, J.; Su, F.; Yu, B.; Su, F.; Lin, L.; Liu, Y. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol. Cancer 2011, 10, 117. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Ochiya, T. Unraveling the mystery of cancer by secretory microRNA: Horizontal microRNA transfer between living cells. Front Genet. 2011, 2, 97. [Google Scholar] [CrossRef] [PubMed]
- Sakha, S.; Muramatsu, T.; Ueda, K.; Inazawa, J. Exosomal microRNA miR-1246 induces cell motility and invasion through the regulation of DENND2D in oral squamous cell carcinoma. Sci. Rep. 2016, 6, 38750. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, C.A.; Andahur, E.I.; Valenzuela, R.; Castellon, E.A.; Fulla, J.A.; Ramos, C.G.; Trivino, J.C. Exosomes from bulk and stem cells from human prostate cancer have a differential microRNA content that contributes cooperatively over local and pre-metastatic niche. Oncotarget 2015, 7, 3993–4008. [Google Scholar]
- Umezu, T.; Tadokoro, H.; Azuma, K.; Yoshizawa, S.; Ohyashiki, K.; Ohyashiki, J.H. Exosomal miR-135b shed from hypoxic multiple myeloma cells enhances angiogenesis by targeting factor-inhibiting HIF-1. Blood 2014, 124, 3748–3757. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Luo, F.; Wang, B.; Li, H.; Xu, Y.; Liu, X.; Shi, L.; Lu, X.; Xu, W.; Lu, L.; et al. STAT3-regulated exosomal miR-21 promotes angiogenesis and is involved in neoplastic processes of transformed human bronchial epithelial cells. Cancer Lett. 2016, 370, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Horiguchi, H.; Kobune, M.; Kikuchi, S.; Yoshida, M.; Murata, M.; Murase, K.; Iyama, S.; Takada, K.; Sato, T.; Ono, K.; et al. Extracellular vesicle miR-7977 is involved in hematopoietic dysfunction of mesenchymal stromal cells via poly(rC) binding protein 1 reduction in myeloid neoplasms. Haematologica 2016. [Google Scholar] [CrossRef] [PubMed]
- Roccaro, A.M.; Sacco, A.; Maiso, P.; Azab, A.K.; Tai, Y.T.; Reagan, M.; Azab, F.; Flores, L.M.; Campigotto, F.; Weller, E.; et al. BM mesenchymal stromal cell-derived exosomes facilitate multiple myeloma progression. J. Clin. Investig. 2013, 123, 1542–1555. [Google Scholar] [CrossRef] [PubMed]
- De Veirman, K.; Wang, J.; Xu, S.; Leleu, X.; Himpe, E.; Maes, K.; De Bruyne, E.; Van Valckenborgh, E.; Vanderkerken, K.; Menu, E.; et al. Induction of miR-146a by multiple myeloma cells in mesenchymal stromal cells stimulates their pro-tumoral activity. Cancer Lett. 2016, 377, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, X.; Zhu, R.; Han, Q.; Zhao, R.C. Lung cancer exosomes initiate global long non-coding RNA changes in mesenchymal stem cells. Int. J. Oncol. 2016, 48, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Kogure, T.; Yan, I.K.; Lin, W.L.; Patel, T. Extracellular vesicle-mediated transfer of a novel long noncoding RNA TUC339: A mechanism of intercellular signaling in human hepatocellular cancer. Genes Cancer 2013, 4, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Conigliaro, A.; Costa, V.; Lo Dico, A.; Saieva, L.; Buccheri, S.; Dieli, F.; Manno, M.; Raccosta, S.; Mancone, C.; Tripodi, M.; et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol. Cancer 2015, 14, 155. [Google Scholar] [CrossRef] [PubMed]
- Iempridee, T. Long non-coding RNA H19 enhances cell proliferation and anchorage-independent growth of cervical cancer cell lines. Exp. Biol. Med. 2016, 242, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Ahadi, A.; Brennan, S.; Kennedy, P.J.; Hutvagner, G.; Tran, N. Long non-coding RNAs harboring miRNA seed regions are enriched in prostate cancer exosomes. Sci. Rep. 2016, 6, 24922. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Lin, W.; Qi, P.; Xu, M.D.; Wu, X.; Ni, S.; Huang, D.; Weng, W.W.; Tan, C.; Sheng, W.; et al. Circulating long RNAs in serum extracellular vesicles: Their characterization and potential application as biomarkers for diagnosis of colorectal cancer. Cancer Epidemiol. Biomark. Prev. 2016, 25, 1158–1166. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Xu, R.; Ji, H.; Greening, D.W.; Rai, A.; Izumikawa, K.; Ishikawa, H.; Takahashi, N.; Simpson, R.J. Transcriptome and long noncoding RNA sequencing of three extracellular vesicle subtypes released from the human colon cancer LIM1863 cell line. Sci. Rep. 2016, 6, 38397. [Google Scholar] [CrossRef] [PubMed]
- Salton, M.; Misteli, T. Small molecule modulators of pre-mRNA splicing in cancer therapy. Trends Mol. Med. 2016, 22, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Douglas, A.G.; Wood, M.J. RNA splicing: Disease and therapy. Brief. Funct. Genom. 2011, 10, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Dvinge, H.; Kim, E.; Abdel-Wahab, O.; Bradley, R.K. RNA splicing factors as oncoproteins and tumour suppressors. Nat. Rev. Cancer 2016, 16, 413–430. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Cooper, T.A. Pre-mRNA splicing in disease and therapeutics. Trends Mol. Med. 2012, 18, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yan, I.K.; Wood, J.; Haga, H.; Patel, T. Involvement of extracellular vesicle long noncoding RNA (linc-VLDLR) in tumor cell responses to chemotherapy. Mol. Cancer Res. 2014, 12, 1377–1387. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yan, I.K.; Kogure, T.; Haga, H.; Patel, T. Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio. 2014, 4, 458–467. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.G.; Yang, M.F.; Ren, Y.Q.; Wu, C.H.; Wang, L.Q. Exosomes mediated transfer of lncRNA UCA1 results in increased tamoxifen resistance in breast cancer cells. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 4362–4368. [Google Scholar] [PubMed]
- Santos, J.C.; Ribeiro, M.L.; Sarian, L.O.; Ortega, M.M.; Derchain, S.F. Exosomes-mediate microRNAs transfer in breast cancer chemoresistance regulation. Am. J. Cancer Res. 2016, 6, 2129–2139. [Google Scholar] [PubMed]
- Chen, W.X.; Cai, Y.Q.; Lv, M.M.; Chen, L.; Zhong, S.L.; Ma, T.F.; Zhao, J.H.; Tang, J.H. Exosomes from docetaxel-resistant breast cancer cells alter chemosensitivity by delivering microRNAs. Tumour Biol. 2014, 35, 9649–9659. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.X.; Liu, X.M.; Lv, M.M.; Chen, L.; Zhao, J.H.; Zhong, S.L.; Ji, M.H.; Hu, Q.; Luo, Z.; Wu, J.Z.; et al. Exosomes from drug-resistant breast cancer cells transmit chemoresistance by a horizontal transfer of microRNAs. PLoS ONE 2014, 9, e95240. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Lai, X.; Yu, S.; Chen, S.; Ma, Y.; Zhang, Y.; Li, H.; Zhu, X.; Yao, L.; Zhang, J. Exosomal miR-221/222 enhances tamoxifen resistance in recipient ER-positive breast cancer cells. Breast Cancer Res. Treat. 2014, 147, 423–431. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, K.; Lowry, M.C.; Corcoran, C.; Martinez, V.G.; Daly, M.; Rani, S.; Gallagher, W.M.; Radomski, M.W.; MacLeod, R.A.; O'Driscoll, L. miR-134 in extracellular vesicles reduces triple-negative breast cancer aggression and increases drug sensitivity. Oncotarget 2015, 6, 32774–32789. [Google Scholar] [PubMed]
- Senfter, D.; Holzner, S.; Kalipciyan, M.; Staribacher, A.; Walzl, A.; Huttary, N.; Krieger, S.; Brenner, S.; Jager, W.; Krupitza, G.; et al. Loss of miR-200 family in 5-fluorouracil resistant colon cancer drives lymphendothelial invasiveness in vitro. Hum. Mol. Genet. 2015, 24, 3689–3698. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Hosokawa, M.; Ueda, K.; Iwakawa, S. Effects of decitabine on invasion and exosomal expression of miR-200c and miR-141 in oxaliplatin-resistant colorectal cancer cells. Biol. Pharm. Bull. 2015, 38, 1272–1279. [Google Scholar] [CrossRef] [PubMed]
- Corcoran, C.; Rani, S.; O’Driscoll, L. miR-34a is an intracellular and exosomal predictive biomarker for response to docetaxel with clinical relevance to prostate cancer progression. Prostate 2014, 74, 1320–1334. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yuan, T.; Liang, M.; Du, M.; Xia, S.; Dittmar, R.; Wang, D.; See, W.; Costello, B.A.; Quevedo, F.; et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur. Urol. 2015, 67, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Pan, L.; Xiang, B.; Zhu, H.; Wu, Y.; Chen, M.; Guan, P.; Zou, X.; Valencia, C.A.; Dong, B.; et al. Potential role of exosome-associated microRNA panels and in vivo environment to predict drug resistance for patients with multiple myeloma. Oncotarget 2016, 7, 30876–30891. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Cui, Y.; Li, Z.; Jiao, Z.; Zhang, Y.; He, Y.; Chen, G.; Zhou, Q.; Wang, W.; Zhou, X.; et al. Radiation-induced miR-208a increases the proliferation and radioresistance by targeting p21 in human lung cancer cells. J. Exp. Clin. Cancer Res. 2016, 35, 7. [Google Scholar] [CrossRef] [PubMed]
- Dinh, T.K.; Fendler, W.; Chalubinska-Fendler, J.; Acharya, S.S.; O'Leary, C.; Deraska, P.V.; D’Andrea, A.D.; Chowdhury, D.; Kozono, D. Circulating miR-29a and miR-150 correlate with delivered dose during thoracic radiation therapy for non-small cell lung cancer. Radiat. Oncol. 2016, 11, 61. [Google Scholar] [CrossRef] [PubMed]
- Au Yeung, C.L.; Co, N.N.; Tsuruga, T.; Yeung, T.L.; Kwan, S.Y.; Leung, C.S.; Li, Y.; Lu, E.S.; Kwan, K.; Wong, K.K.; et al. Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nat. Commun. 2016, 7, 11150. [Google Scholar] [CrossRef] [PubMed]
- Lou, G.; Song, X.; Yang, F.; Wu, S.; Wang, J.; Chen, Z.; Liu, Y. Exosomes derived from miR-122-modified adipose tissue-derived MSCs increase chemosensitivity of hepatocellular carcinoma. J. Hematol. Oncol. 2015, 8, 122. [Google Scholar] [CrossRef] [PubMed]
- Bliss, S.A.; Sinha, G.; Sandiford, O.A.; Williams, L.M.; Engelberth, D.J.; Guiro, K.; Isenalumhe, L.L.; Greco, S.J.; Ayer, S.; Bryan, M.; et al. Mesenchymal stem cell-derived exosomes stimulate cycling quiescence and early breast cancer dormancy in bone marrow. Cancer Res. 2016, 76, 5832–5844. [Google Scholar] [CrossRef] [PubMed]
- Ono, M.; Kosaka, N.; Tominaga, N.; Yoshioka, Y.; Takeshita, F.; Takahashi, R.U.; Yoshida, M.; Tsuda, H.; Tamura, K.; Ochiya, T. Exosomes from bone marrow mesenchymal stem cells contain a microRNA that promotes dormancy in metastatic breast cancer cells. Sci. Signal. 2014, 7, ra63. [Google Scholar] [CrossRef] [PubMed]
- Weiner-Gorzel, K.; Dempsey, E.; Milewska, M.; McGoldrick, A.; Toh, V.; Walsh, A.; Lindsay, S.; Gubbins, L.; Cannon, A.; Sharpe, D.; et al. Overexpression of the microRNA miR-433 promotes resistance to paclitaxel through the induction of cellular senescence in ovarian cancer cells. Cancer Med. 2015, 4, 745–758. [Google Scholar] [CrossRef] [PubMed]
- Pink, R.C.; Samuel, P.; Massa, D.; Caley, D.P.; Brooks, S.A.; Carter, D.R. The passenger strand, miR-21-3p, plays a role in mediating cisplatin resistance in ovarian cancer cells. Gynecol. Oncol. 2015, 137, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Adi Harel, S.; Bossel Ben-Moshe, N.; Aylon, Y.; Bublik, D.R.; Moskovits, N.; Toperoff, G.; Azaiza, D.; Biagoni, F.; Fuchs, G.; Wilder, S.; et al. Reactivation of epigenetically silenced miR-512 and miR-373 sensitizes lung cancer cells to cisplatin and restricts tumor growth. Cell Death Differ. 2015, 22, 1328–1340. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Iguchi, H.; Yoshioka, Y.; Hagiwara, K.; Takeshita, F.; Ochiya, T. Competitive interactions of cancer cells and normal cells via secretory microRNAs. J. Biol. Chem. 2012, 287, 1397–1405. [Google Scholar] [CrossRef] [PubMed]
- Zaharie, F.; Muresan, M.S.; Petrushev, B.; Berce, C.; Gafencu, G.A.; Selicean, S.; Jurj, A.; Cojocneanu-Petric, R.; Lisencu, C.I.; Pop, L.A.; et al. Exosome-carried microRNA-375 inhibits cell progression and dissemination via Bcl-2 blocking in colon cancer. J. Gastroint. Liver Dis. 2015, 24, 435–443. [Google Scholar]
- Takahara, K.; Ii, M.; Inamoto, T.; Nakagawa, T.; Ibuki, N.; Yoshikawa, Y.; Tsujino, T.; Uchimoto, T.; Saito, K.; Takai, T.; et al. microRNA-145 mediates the inhibitory effect of adipose tissue-derived stromal cells on prostate cancer. Stem Cells Dev. 2016, 25, 1290–1298. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.D.; Wu, X.H.; Fan, Y.R.; Tan, B.; Quan, Z.; Luo, C.L. Exosome-derived microRNA-29c induces apoptosis of BIU-87 cells by down regulating BCL-2 and MCL-1. Asian Pac. J. Cancer Prev. 2014, 15, 3471–3476. [Google Scholar] [CrossRef] [PubMed]
- Rivoltini, L.; Chiodoni, C.; Squarcina, P.; Tortoreto, M.; Villa, A.; Vergani, B.; Burdek, M.; Botti, L.; Arioli, I.; Cova, A.; et al. TNF-related apoptosis-inducing ligand (TRAIL)-armed exosomes deliver proapoptotic signals to tumor site. Clin. Cancer Res. 2016, 22, 3499–3512. [Google Scholar] [CrossRef] [PubMed]
- Leverson, J.D.; Phillips, D.C.; Mitten, M.J.; Boghaert, E.R.; Diaz, D.; Tahir, S.K.; Belmont, L.D.; Nimmer, P.; Xiao, Y.; Ma, X.M.; et al. Exploiting selective BCL-2 family inhibitors to dissect cell survival dependencies and define improved strategies for cancer therapy. Sci. Transl. Med. 2015, 7, 279ra40. [Google Scholar] [CrossRef] [PubMed]
- Wong, H.K.; Fatimy, R.E.; Onodera, C.; Wei, Z.; Yi, M.; Mohan, A.; Gowrisankaran, S.; Karmali, P.; Marcusson, E.; Wakimoto, H.; et al. The Cancer Genome Atlas analysis predicts microRNA for targeting cancer growth and vascularization in glioblastoma. Mol. Ther. 2015, 23, 1234–1247. [Google Scholar] [CrossRef] [PubMed]
- Reza, A.M.; Choi, Y.J.; Yasuda, H.; Kim, J.H. Human adipose mesenchymal stem cell-derived exosomal-miRNAs are critical factors for inducing anti-proliferation signalling to A2780 and SKOV-3 ovarian cancer cells. Sci. Rep. 2016, 6, 38498. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yu, J.; Zhang, H.; Wang, B.; Guo, H.; Bai, J.; Wang, J.; Dong, Y.; Zhao, Y.; Wang, Y. Exosomes-derived miR-302b suppresses lung cancer cell proliferation and migration via TGFbetaRII inhibition. Cell Physiol. Biochem. 2016, 38, 1715–1726. [Google Scholar] [CrossRef] [PubMed]
- Katakowski, M.; Buller, B.; Zheng, X.; Lu, Y.; Rogers, T.; Osobamiro, O.; Shu, W.; Jiang, F.; Chopp, M. Exosomes from marrow stromal cells expressing miR-146b inhibit glioma growth. Cancer Lett. 2013, 335, 201–204. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.K.; Park, S.R.; Jung, B.K.; Jeon, Y.K.; Lee, Y.S.; Kim, M.K.; Kim, Y.G.; Jang, J.Y.; Kim, C.W. Exosomes derived from mesenchymal stem cells suppress angiogenesis by down-regulating VEGF expression in breast cancer cells. PLoS ONE 2013, 8, e84256. [Google Scholar] [CrossRef] [PubMed]
- Fonsato, V.; Collino, F.; Herrera, M.B.; Cavallari, C.; Deregibus, M.C.; Cisterna, B.; Bruno, S.; Romagnoli, R.; Salizzoni, M.; Tetta, C.; et al. Human liver stem cell-derived microvesicles inhibit hepatoma growth in SCID mice by delivering antitumor microRNAs. Stem Cells 2012, 30, 1985–1998. [Google Scholar] [CrossRef] [PubMed]
- Kanlikilicer, P.; Rashed, M.H.; Bayraktar, R.; Mitra, R.; Ivan, C.; Aslan, B.; Zhang, X.; Filant, J.; Silva, A.M.; Rodriguez-Aguayo, C.; et al. Ubiquitous release of exosomal tumor suppressor miR-6126 from ovarian cancer cells. Cancer Res. 2016, 76. [Google Scholar] [CrossRef] [PubMed]
- Wahlgren, J.; De, L.K.T.; Brisslert, M.; Vaziri Sani, F.; Telemo, E.; Sunnerhagen, P.; Valadi, H. Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Res. 2012, 40, e130. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011, 29, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Iguchi, H.; Ochiya, T. Circulating microRNA in body fluid: A new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010, 101, 2087–2092. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; Hoon, D.S.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Savelyeva, A.V.; Kuligina, E.V.; Bariakin, D.N.; Kozlov, V.V.; Ryabchikova, E.I.; Richter, V.A.; Semenov, D.V. Variety of RNAs in peripheral blood cells, plasma, and plasma fractions. BioMed Res. Int. 2017, 2017, 7404912. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.K.; Yu, J.C. Circulating microRNAs and long non-coding RNAs in gastric cancer diagnosis: An update and review. World J. Gastroenterol. 2015, 21, 9863–9886. [Google Scholar] [CrossRef] [PubMed]
- Warnecke-Eberz, U.; Chon, S.H.; Holscher, A.H.; Drebber, U.; Bollschweiler, E. Exosomal onco-miRs from serum of patients with adenocarcinoma of the esophagus: Comparison of miRNA profiles of exosomes and matching tumor. Tumour Biol. 2015, 36, 4643–4653. [Google Scholar] [CrossRef] [PubMed]
- Manterola, L.; Guruceaga, E.; Gallego Perez-Larraya, J.; Gonzalez-Huarriz, M.; Jauregui, P.; Tejada, S.; Diez-Valle, R.; Segura, V.; Sampron, N.; Barrena, C.; et al. A small noncoding RNA signature found in exosomes of GBM patient serum as a diagnostic tool. Neuro Oncol. 2014, 16, 520–527. [Google Scholar] [CrossRef] [PubMed]
- Mihelich, B.L.; Dambal, S.; Lin, S.; Nonn, L. miR-182, of the miR-183 cluster family, is packaged in exosomes and is detected in human exosomes from serum, breast cells and prostate cells. Oncol. Lett. 2016, 12, 1197–1203. [Google Scholar] [CrossRef] [PubMed]
- Takeshita, N.; Hoshino, I.; Mori, M.; Akutsu, Y.; Hanari, N.; Yoneyama, Y.; Ikeda, N.; Isozaki, Y.; Maruyama, T.; Akanuma, N.; et al. Serum microRNA expression profile: miR-1246 as a novel diagnostic and prognostic biomarker for oesophageal squamous cell carcinoma. Br. J. Cancer 2013, 108, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Conley, A.; Minciacchi, V.R.; Lee, D.H.; Knudsen, B.S.; Karlan, B.Y.; Citrigno, L.; Viglietto, G.; Tewari, M.; Freeman, M.R.; Demichelis, F.; et al. High-throughput sequencing of two populations of extracellular vesicles provides an mRNA signature that can be detected in the circulation of breast cancer patients. RNA Biol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.D.; Gercel-Taylor, C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008, 110, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Joshi, G.K.; Deitz-McElyea, S.; Liyanage, T.; Lawrence, K.; Mali, S.; Sardar, R.; Korc, M. Label-free nanoplasmonic-based short noncoding RNA sensing at attomolar concentrations allows for quantitative and highly specific assay of microRNA-10b in biological fluids and circulating exosomes. ACS Nano 2015, 9, 11075–11089. [Google Scholar] [CrossRef] [PubMed]
- Mohankumar, S.; Patel, T. Extracellular vesicle long noncoding RNA as potential biomarkers of liver cancer. Brief. Funct. Genom. 2016, 15, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Shao, Y.; Zhang, X.; Zheng, T.; Miao, M.; Qin, L.; Wang, B.; Ye, G.; Xiao, B.; Guo, J. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumour Biol. 2015, 36, 2007–2012. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, X.; Gao, S.; Jing, F.; Yang, Y.; Du, L.; Zheng, G.; Li, P.; Li, C.; Wang, C. Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget 2016. [Google Scholar] [CrossRef] [PubMed]
- Balaj, L.; Lessard, R.; Dai, L.; Cho, Y.J.; Pomeroy, S.L.; Breakefield, X.O.; Skog, J. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2011, 2, 180. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Kamohara, H.; Kinoshita, K.; Kurashige, J.; Ishimoto, T.; Iwatsuki, M.; Watanabe, M.; Baba, H. Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer 2013, 119, 1159–1167. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Hou, L.; Li, A.; Duan, Y.; Gao, H.; Song, X. Expression of serum exosomal microRNA-21 in human hepatocellular carcinoma. BioMed Res. Int. 2014, 2014, 864894. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Sun, H.; Wang, X.; Yu, Q.; Li, S.; Yu, X.; Gong, W. Increased exosomal microRNA-21 and microRNA-146a levels in the cervicovaginal lavage specimens of patients with cervical cancer. Int. J. Mol. Sci. 2014, 15, 758–773. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, T.; Sugimachi, K.; Iinuma, H.; Takahashi, Y.; Kurashige, J.; Sawada, G.; Ueda, M.; Uchi, R.; Ueo, H.; Takano, Y.; et al. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br. J. Cancer 2015, 113, 275–281. [Google Scholar] [CrossRef]
- Shi, R.; Wang, P.Y.; Li, X.Y.; Chen, J.X.; Li, Y.; Zhang, X.Z.; Zhang, C.G.; Jiang, T.; Li, W.B.; Ding, W.; et al. Exosomal levels of miRNA-21 from cerebrospinal fluids associated with poor prognosis and tumor recurrence of glioma patients. Oncotarget 2015, 6, 26971–26981. [Google Scholar] [CrossRef] [PubMed]
- Sugimachi, K.; Matsumura, T.; Hirata, H.; Uchi, R.; Ueda, M.; Ueo, H.; Shinden, Y.; Iguchi, T.; Eguchi, H.; Shirabe, K.; et al. Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br. J. Cancer 2015, 112, 532–538. [Google Scholar] [CrossRef] [PubMed]
- Di Modica, M.; Regondi, V.; Sandri, M.; Iorio, M.V.; Zanetti, A.; Tagliabue, E.; Casalini, P.; Triulzi, T. Breast cancer-secreted miR-939 downregulates VE-cadherin and destroys the barrier function of endothelial monolayers. Cancer Lett. 2017, 384, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Soldevilla, B.; Rodriguez, M.; San Millan, C.; Garcia, V.; Fernandez-Perianez, R.; Gil-Calderon, B.; Martin, P.; Garcia-Grande, A.; Silva, J.; Bonilla, F.; et al. Tumor-derived exosomes are enriched in DeltaNp73, which promotes oncogenic potential in acceptor cells and correlates with patient survival. Hum. Mol. Genet. 2014, 23, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Stamatopoulos, B.; Van Damme, M.; Crompot, E.; Dessars, B.; Housni, H.E.; Mineur, P.; Meuleman, N.; Bron, D.; Lagneaux, L. Opposite prognostic significance of cellular and serum circulating microRNA-150 in patients with chronic lymphocytic leukemia. Mol. Med. 2015, 21, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.C.; Zhao, J.T.; Gundara, J.; Serpell, J.; Bach, L.A.; Sidhu, S. Papillary thyroid cancer-derived exosomes contain miRNA-146b and miRNA-222. J. Surg. Res. 2015, 196, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Fais, S.; O'Driscoll, L.; Borras, F.E.; Buzas, E.; Camussi, G.; Cappello, F.; Carvalho, J.; Cordeiro da Silva, A.; Del Portillo, H.; El Andaloussi, S.; et al. Evidence-based clinical use of nanoscale extracellular vesicles in nanomedicine. ACS Nano 2016, 10, 3886–3899. [Google Scholar] [CrossRef] [PubMed]
- Lener, T.; Gimona, M.; Aigner, L.; Borger, V.; Buzas, E.; Camussi, G.; Chaput, N.; Chatterjee, D.; Court, F.A.; Del Portillo, H.A.; et al. Applying extracellular vesicles based therapeutics in clinical trials—An ISEV position paper. J. Extracell. Vesicles 2015, 4, 30087. [Google Scholar] [CrossRef] [PubMed]
| microRNAs | Implications | Reference |
|---|---|---|
| TERRA | Genomic instability and transcriptional regulation of inflammatory cytokines | [72,73] |
| TERRA | DNA damage response and inflammatory responses | [73] |
| HOTAIR | Inflammatory responses and migration of active macrophages | [74] |
| HOTAIR | Urinary biomarker for bladder cancer diagnosis and prognosis | [75] |
| HOTAIR | Serum-based biomarker for laryngeal squamous cell carcinoma | [76] |
| lincRNA-p21, HOTAIR, ncRNA-CCND1, TUG1, GAS5, MALAT1 | Response to DNA damage | [78] |
| PARTICLE | Methylation, gene silencing and transcriptional repression | [44] |
| H19 and H19 antisense | Epstein-Barr virus induced expression in immortalized B cells | [81] |
| HN12 lncRNA | Inhibition of cell apoptosis and maintaining the function of mitochondria in Hirschsprung’s disease | [82] |
| linc-RoR | Modulation of chemosensitivity in human hepatocellular cancer | [97] |
| linc-RoR | Modulation of hypoxia-signaling pathways | [125] |
| UCA1 lncRNA | Enhanced tamoxifen resistance in breast cancer cells | [126] |
| TUC339 | Progression of hepatocellular carcinoma growth | [114] |
| H19 lncRNA | Modulation of endothelial cell phenotype and tumor angiogenesis | [115] |
| H19 lncRNA | Proliferation and anchor independent tumor growth of cervical cancer cells | [116] |
| BCAR4 | Serum-based diagnostic and prognostic markers for colorectal cancer | [118] |
| CRNDE-h | Serum-based biomarker for diagnosis and prognosis of colorectal cancer | [175] |
| microRNAs | Implications | Reference |
|---|---|---|
| miR-10b | Breast cancer cell invasion | [98] |
| miR-122 | Glucose metabolism in premetastatic niche and cancer metastasis | [99] |
| miR-1246 | Oral squamous cell carcinoma metastasis | [106] |
| miR-100-5p, miR-21-5p | Prostate cancer progression and metastasis | [107] |
| miR-7977 | Hematopoietic dysfunction and progression to myeloid neoplasms | [110] |
| miR-15a | Multiple myeloma progression | [111] |
| miR-146a | Multiple myeloma cell survival and migration | [112] |
| miR-221/222 | Enhanced drug resistance in breast cancer | [130] |
| miR-134 | Enhanced drug sensitivity and reduction in triple-negative breast cancer aggression | [131] |
| miR-34a | Response to chemotherapy in prostate cancer, prognostic biomarker | [134] |
| miR-1290, miR-375 | Prognostic markers in castration-resistant prostate cancer | [135] |
| miR-208a | Radio-resistance in human lung cancer cells | [137] |
| miR-29a, miR-150 | Prognostic markers against lung cancer radiotherapy | [138] |
| miR-122 | Hepatocellular carcinoma chemosensitivity and increased antitumor efficacy of chemotherapeutic agents | [140] |
| miR-222/223 | Dormancy in early stage breast cancer | [141] |
| miR-23b | Dormancy in metastatic breast cancer cells | [142] |
| miR-21-3p | Drug resistance in ovarian cancer | [144] |
| miR-512, miR-373 | Sensitivity against drug and lung cancer inhibition | [145] |
| miR-143 | Tumor inhibition | [146] |
| miR-375 | Tumor inhibition | [147] |
| miR-145 | Tumor inhibition | [148] |
| miR-29c | Tumor inhibition | [149] |
| miR-16 | Suppression of tumor angiogenesis | [156] |
| miR-451, miR-223, miR-24, miR-125b, miR-31, miR-122 | Inhibition of hepatoma growth | [157] |
| miR-6126 | Ovarian cancer metastasis | [158] |
| miR-1246 | Diagnostic and prognostic biomarker for esophageal squamous cell carcinoma | [169] |
| miR-21 | Diagnostic biomarker for esophageal squamous cell carcinoma, human hepatocellular carcinoma, cervical and ovarian cancer | [95,177,178,179] |
| miR-146b, miR-222 | Prognostic marker of recurrence in papillary thyroid cancer | [186] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fatima, F.; Nawaz, M. Vesiculated Long Non-Coding RNAs: Offshore Packages Deciphering Trans-Regulation between Cells, Cancer Progression and Resistance to Therapies. Non-Coding RNA 2017, 3, 10. https://doi.org/10.3390/ncrna3010010
Fatima F, Nawaz M. Vesiculated Long Non-Coding RNAs: Offshore Packages Deciphering Trans-Regulation between Cells, Cancer Progression and Resistance to Therapies. Non-Coding RNA. 2017; 3(1):10. https://doi.org/10.3390/ncrna3010010
Chicago/Turabian StyleFatima, Farah, and Muhammad Nawaz. 2017. "Vesiculated Long Non-Coding RNAs: Offshore Packages Deciphering Trans-Regulation between Cells, Cancer Progression and Resistance to Therapies" Non-Coding RNA 3, no. 1: 10. https://doi.org/10.3390/ncrna3010010
APA StyleFatima, F., & Nawaz, M. (2017). Vesiculated Long Non-Coding RNAs: Offshore Packages Deciphering Trans-Regulation between Cells, Cancer Progression and Resistance to Therapies. Non-Coding RNA, 3(1), 10. https://doi.org/10.3390/ncrna3010010







