High Percentage of Isomeric Human MicroRNA and Their Analytical Challenges
Abstract
:1. Introduction
2. Results and Discussion
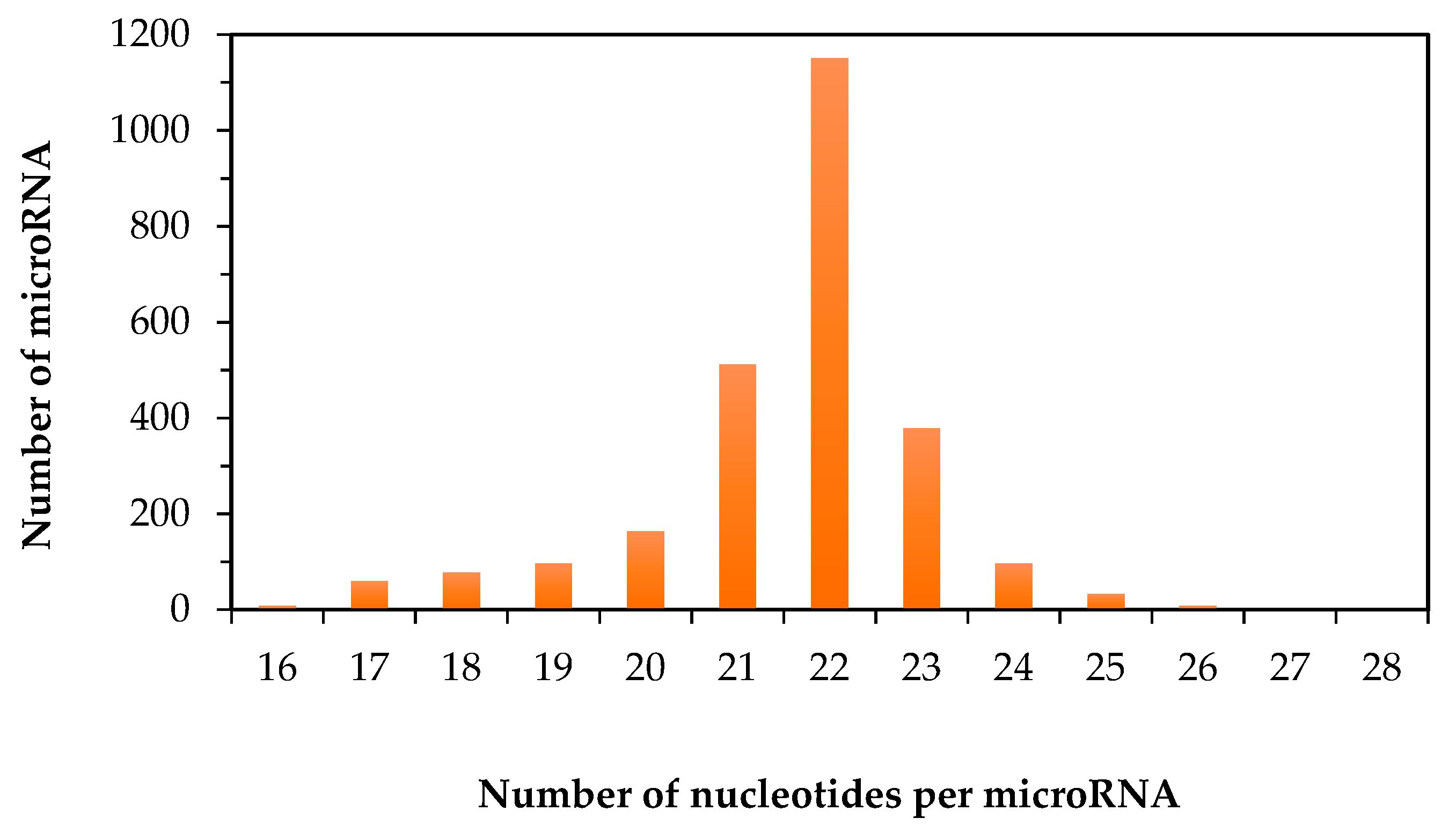
2.1. Size Distribution of Human Mature microRNA
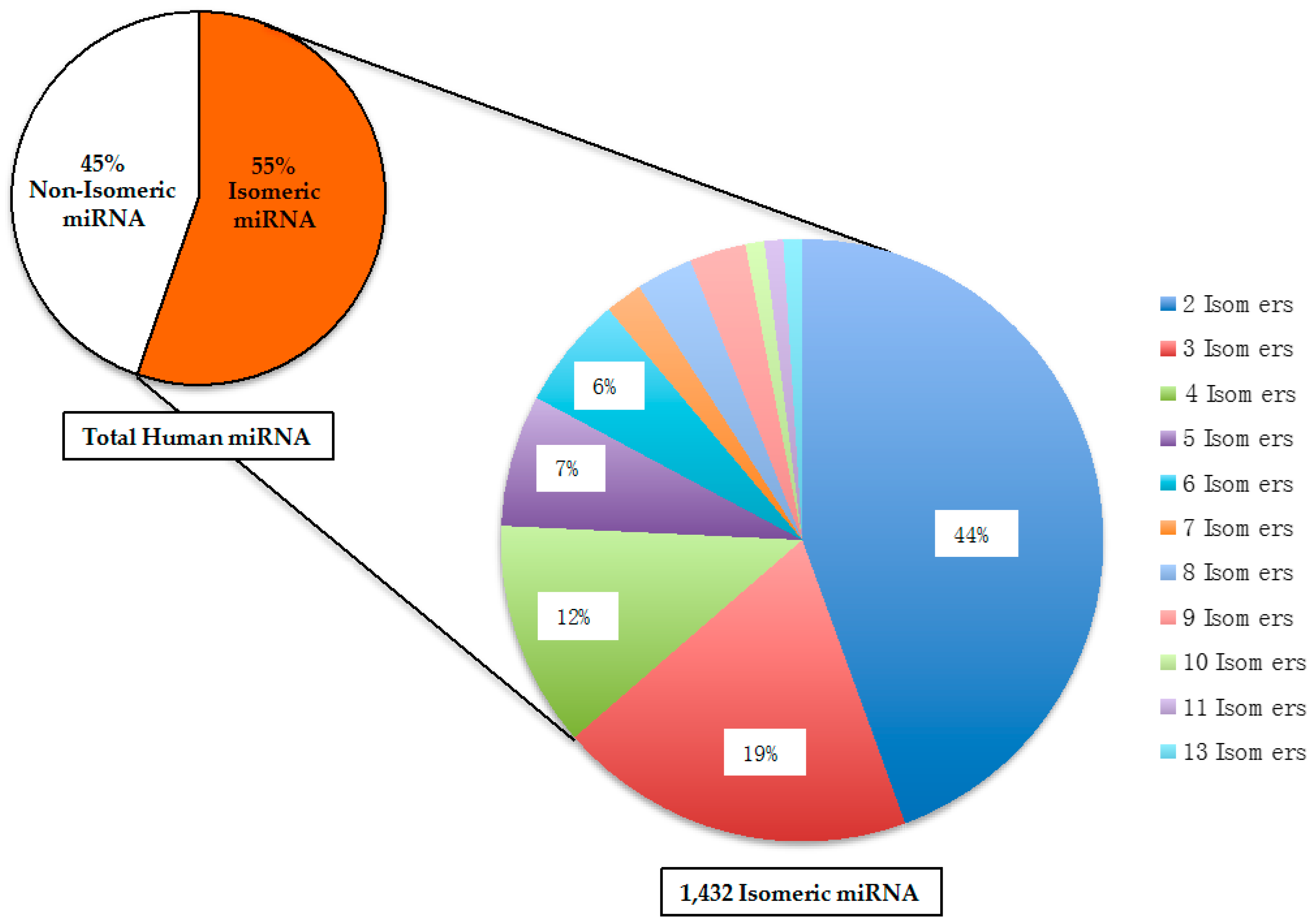
2.2. Isomeric microRNA
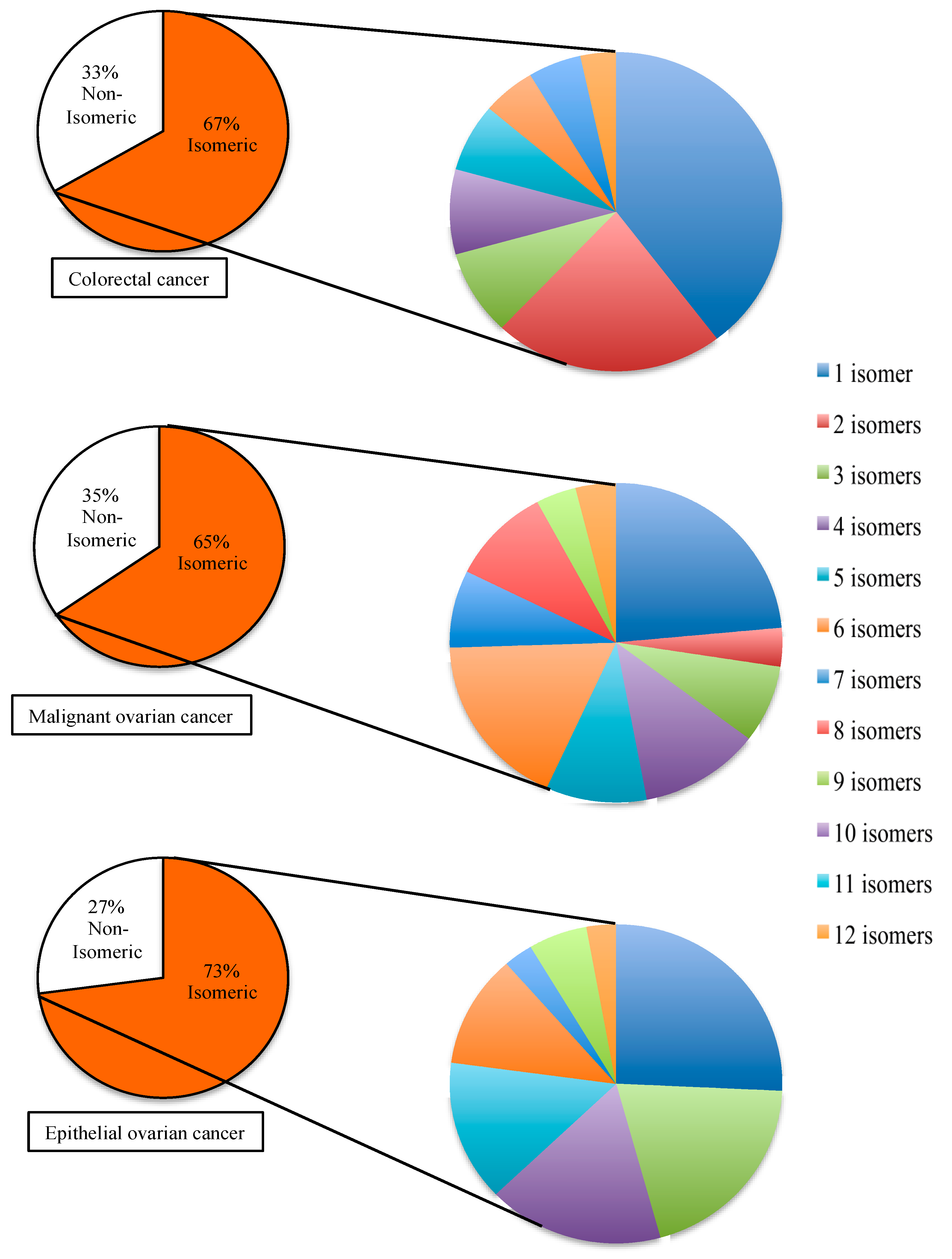
2.3. Disease-Associated Isomeric microRNA
2.4. Sequence Similarities among Isomeric microRNA
2.5. Analytical Challenges from Isomeric microRNA
3. Materials and Methods
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Watson, J.D.; Crick, F.H. Molecular Structure of Nucleic Acids. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef]
- Krueger, A.T.; Kool, E.T. Model systems for understanding DNA base pairing. Curr. Opin. Chem. Biol. 2007, 11, 588–594. [Google Scholar] [CrossRef] [PubMed]
- Helm, M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 2006, 34, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S.; Saini, H.K.; van Dongen, S.; Enright, A.J. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008, 36, D154–D158. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011, 39, D152–D157. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef] [PubMed]
- Asangani, I.A.; Rasheed, S.A.; Nikolova, D.A.; Leupold, J.H.; Colburn, N.H.; Post, S.; Allgayer, H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 2008, 27, 2128–2136. [Google Scholar] [CrossRef] [PubMed]
- Braconi, C.; Kogure, T.; Valeri, N.; Huang, N.; Nuovo, G.; Costinean, S.; Negrini, M.; Miotto, E.; Croce, C.M.; Patel, T. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene 2011, 30, 4750–4756. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Park, J.K.; Jiang, F.; Liu, Y.; McKearin, D.; Liu, Q. Dicer-1, but not Loquacious, is critical for assembly of miRNA-induced silencing complexes. RNA 2007, 13, 2324–2329. [Google Scholar] [CrossRef] [PubMed]
- Doerks, T.; Copley, R.R.; Schultz, J.; Ponting, C.P.; Bork, P. Systematic identification of novel protein domain families associated with nuclear functions. Genome Res. 2002, 12, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Mlcochova, H.; Hezova, R.; Stanik, M.; Slaby, O. Urine microRNAs as potential noninvasive biomarkers in urologic cancers. Urol. Oncol. 2014, 32, 41.e1–41.e9. [Google Scholar] [CrossRef] [PubMed]
- Scott, S.; Sun-Mi, P.; Amir, R.R.; Robert, S.; Emily, O.K.; David, J.; Christine, F.; Ernst, L.; Marcus, E.P. Let-7 expression defines two differentiation stages of cancer. Proc. Natl. Acad. Sci. USA 2007, 104, 11400–11405. [Google Scholar]
- Lu, L.; Katsaros, D.; de la Longrais, I.A.; Sochirca, O.; Yu, H. Hypermethylation of let-7a-3 in epithelial ovarian cancer is associated with low insulin-like growth factor-II expression and favorable prognosis. Cancer Res. 2007, 67, 10117–10122. [Google Scholar] [CrossRef] [PubMed]
- Heneghan, H.M.; Miller, N.; Lowery, A.J.; Sweeney, K.J.; Newell, J.; Kerin, M.J. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann. Surg. 2010, 251, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Xiao, L.; Li, J.; Kanwar, Y.S.; Liu, F.; Sun, L. Urine miRNAs: Potential biomarkers for monitoring progression of early stages of diabetic nephropathy. Med. Hypotheses 2013, 81, 274–278. [Google Scholar] [CrossRef] [PubMed]
- Peggy, E.; Tam, W.; Liping, S.; Amy, C.; Zongdong, L.; Mario, G.; Elsebet, L.; James, D. Accumaulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl. Acad. Sci. USA 2005, 102, 3627–3632. [Google Scholar]
- Bartels, C.L.; Tsongalis, G.J. MicroRNAs: Novel biomarkers for human cancer. Clin. Chem. 2009, 55, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Janssen, H.L.; Reesink, H.W.; Lawitz, E.J.; Zeuzem, S.; Rodriguez-Torres, M.; Patel, K.; van der Meer, A.J.; Patick, A.K.; Chen, A.; Zhou, Y.; et al. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013, 368, 1685–1694. [Google Scholar] [CrossRef] [PubMed]
- Piehl, F.; Jagodic, M. MicroRNAs as promising novel biomarkers and potential drug targets for inflammatory neurological diseases. J. Neurol. Sci. 2015, 356, 3–4. [Google Scholar] [CrossRef] [PubMed]
- Renfeng, W.F.Y.; Qiang, Z.; Tieying, S.; Guoliang, T.; Weiwei, C.; Yaxiao, Z.; Baolei, L.; Xiaojian, Z.; Jiabao, L. MicroRNA-148b is a potential prognostic biomarker and predictor of response to radiotherapy in non-small-cell lung cancer. Physiol. Biochem. 2016, 72, 337–343. [Google Scholar]
- Teplyuk, N.M.; Uhlmann, E.J.; Gabriely, G.; Volfovsky, N.; Wang, Y.; Teng, J.; Karmali, P.; Marcusson, E.; Peter, M.; Mohan, A.; et al. Therapeutic potential of targeting microRNA-10b in established intracranial glioblastoma: First steps toward the clinic. EMBO Mol. Med. 2016, 8, 268–287. [Google Scholar] [CrossRef] [PubMed]
- Jingsheng, S.; Yibing, W.; Jun, X.; Siqun, W.; Jianguo, W.; Feiyan, C.; Gangyong, H.; Jie, C. MicroRNAs are potential prognostic and therapeutic targets in diabetic osteoarthritis. J. Bone Miner. Metab. 2015, 33, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kalinowski, F.C.; Brown, R.A.; Ganda, C.; Giles, K.M.; Epis, M.R.; Horsham, J.; Leedman, P.J. microRNA-7: A tumor suppressor miRNA with therapeutic potential. Int. J. Biochem. Cell Biol. 2014, 54, 312–317. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, C.C.; Cheng, H.H.; Tewari, M. MicroRNA profiling: Approaches and considerations. Nat. Rev. Genet. 2012, 13, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Ibberson, D.; Benes, V.; Muckenthaler, M.U.; Castoldi, M. RNA degradation compromises the reliability of microRNA expression profiling. BMC Biotechnol. 2009, 9, 102. [Google Scholar] [CrossRef] [PubMed]
- Agnieszka, P.; Kaczkowski, B.; Thomas, L.; Marete, F.; Susanna, C. How the RNA isolation method can affect microRNA microarray results. Acta Biochem. Pol. 2011, 58, 535–540. [Google Scholar]
- Wang, W.X.; Wilfred, B.R.; Baldwin, D.A.; Isett, R.B.; Ren, N.; Stromberg, A.; Nelson, P.T. Focus on RNA isolation: Obtaining RNA for microRNA (miRNA) expression profiling analyses of neural tissue. Biochim. Biophys. Acta 1779, 1779, 749–757. [Google Scholar] [CrossRef] [PubMed]
- Ach, R.A.; Wang, H.; Curry, B. Measuring microRNAs: Comparisons of microarray and quantitative PCR measurements, and of different total RNA prep methods. BMC Biotechnol. 2008, 8, 69. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Gelfond, J.A.; McManus, L.M.; Shireman, P.K. Reproducibility of quantitative RT-PCR array in miRNA expression profiling and comparison with microarray analysis. BMC Genom. 2009, 10, 407. [Google Scholar] [CrossRef] [PubMed]
- Git, A.; Dvinge, H.; Salmon-Divon, M.; Osborne, M.; Kutter, C.; Hadfield, J.; Bertone, P.; Caldas, C. Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA 2010, 16, 991–1006. [Google Scholar] [CrossRef] [PubMed]
- Dame, G.; Lampe, J.; Hakenberg, S.; Urban, G. Development of a Fast miRNA Extraction System for Tumor Analysis Based on a Simple Lab on Chip Approach. Procedia Eng. 2015, 120, 158–162. [Google Scholar] [CrossRef]
- Wang, P.; Zhao, J.; Hu, B.; Cheng, Z.; Bai, Y.; Jin, Q.; Liu, H.; Mao, H.; Li, S.; Zhao, J. Chip-based visual detection of microRNA using DNA-functionalized gold nanoparticles. Sci. China Life Sci. 2016, 59, 510–515. [Google Scholar] [CrossRef] [PubMed]
- Stefan, S.; Bernal-Méndez, S.; Christian, L. Electrospray tandem mass spectrometry of mixed-sequence RNA:DNA oligonucleotides. J. Am. Soc. Mass Spectrom. 2002, 13, 936–945. [Google Scholar]
- Li, S.; Limbach, P.A. Identification of RNA sequence isomer by isotope labeling and LC-MS/MS. J. Mass Spectrom. 2014, 49, 1191–1198. [Google Scholar] [CrossRef] [PubMed]
- Hedegaard, J.; Thorsen, K.; Lund, M.K.; Hein, A.M.; Hamilton-Dutoit, S.J.; Vang, S.; Nordentoft, I.; Birkenkamp-Demtroder, K.; Kruhoffer, M.; Hager, H.; et al. Next-generation sequencing of RNA and DNA isolated from paired fresh-frozen and formalin-fixed paraffin-embedded samples of human cancer and normal tissue. PLoS ONE 2014, 9, e98187. [Google Scholar] [CrossRef] [PubMed]
- Wambua, D.M.; Tannous, B.A.; Chiu, N.H.L. Creating Mass signatures for Detection of microRNA. Anal. Methods 2012, 4, 3453–3459. [Google Scholar] [CrossRef]
- Wetzel, C.; Limbach, P.A. Mass spectrometry of modified RNAs: Recent developments. Analyst 2016, 141, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Gurman, P.C.; Jane, B.; Leigh, R.; Andrew, H. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res. 2007, 35, 60–69. [Google Scholar]
- Valoczi, A.; Hornyik, C.; Varga, N.; Burgyan, J.; Kauppinen, S.; Havelda, Z. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 2004, 32, e175. [Google Scholar] [CrossRef] [PubMed]
- Morin, R.D.; O’Connor, M.D.; Griffith, M.; Kuchenbauer, F.; Delaney, A.; Prabhu, A.L.; Zhao, Y.; McDonald, H.; Zeng, T.; Hirst, M.; et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008, 18, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Burroughs, A.M.; Ando, Y.; de Hoon, M.J.; Tomaru, Y.; Nishibu, T.; Ukekawa, R.; Funakoshi, T.; Kurokawa, T.; Suzuki, H.; Hayashizaki, Y.; et al. A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome Res. 2010, 20, 1398–1410. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.R.; Quinton, L.J.; Blahna, M.T.; Neilson, J.R.; Fu, S.; Ivanov, A.R.; Wolf, D.A.; Mizgerd, J.P. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat. Cell Biol. 2009, 11, 1157–1163. [Google Scholar] [CrossRef] [PubMed]
- Biba, M.; Jiang, E.; Mao, B.; Zewge, D.; Foley, J.P.; Welch, C.J. Factors influencing the separation of oligonucleotides using reversed-phase/ion-exchange mixed-mode high performance liquid chromatography columns. J. Chromatogr. A 1304, 1304, 69–77. [Google Scholar] [CrossRef] [PubMed]
- miRBase. Available online: http://mirbase.org/ (accessed on 29 November 2016).
- Rozenski, J.; Crain, P.F.; James, A.; McCloskey, J.A. The RNA Modification Database: 1999 update. Nucleic Acids Res. 1999, 27, 196–197. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Wang, Y.; Hao, Y.; Juan, L.; Teng, M.; Zhang, X.; Li, M.; Wang, G.; Liu, Y. miR2Disease: A manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009, 37, D98–D104. [Google Scholar] [CrossRef] [PubMed]
| miRNA | Sequence (5′–3′) | Chromosome | Location |
|---|---|---|---|
| miR-21-5p | UAGCUUAUCAGACUGAUGUUGA | 17 | NC_000017.11 (59841266..59841337) |
| miR-95-3p | UUCAACGGGUAUUUAUUGAGCA | 4 | NC_000004.12 (8005301..8005381) |
| miR-100-3p | CAAGCUUGUAUCUAUAGGUAUG | 11 | NC_000011.10 (122152229..122152308) |
| miR-513b-5p | UUCACAAGGAGGUGUCAUUUAU | X | NC_000023.11 (147199044..147199127) |
| miR-519a-3p | AAAGUGCAUCCUUUUAGAGUGU | 19 | NC_000019.10 (53752397..53752481) |
| miR-519b-3p | AAAGUGCAUCCUUUUAGAGGUU | 19 | NC_000019.10 (53695213..53695293) |
| miR-522-3p | AAAAUGGUUCCCUUUAGAGUGU | 19 | NC_000019.10 (53751211..53751297) |
| miR-548am-5p | AAAAGUAAUUGCGGUUUUUGCC | X | NC_000023.11 (16627012..16627085) |
| miR-548c-5p | AAAAGUAAUUGCGGUUUUUGCC | 12 | NC_000012.12 (64622509..64622605) |
| miR-548h-5p | AAAAGUAAUCGCGGUUUUUGUC | 6 | NC_000006.12 (131792172..131792231) |
| miR-548k | AAAAGUACUUGCGGAUUUUGCU | 11 | NC_000011.10 (70283955..70284070) |
| miR-548o-5p | AAAAGUAAUUGCGGUUUUUGCC | 20 | NC_000020.11 (38516563..38516632) |
| miR-4789-5p | GUAUACACCUGAUAUGUGUAUG | 3 | NC_000003.12 (175369540..175369621) |
| miRNA | Sequence (5′–3′) | Chromosome | Location |
|---|---|---|---|
| let-7a-2-3p | CUGUACAGCCUCCUAGCUUUCC | 11 | NC_000011.10 (122146522..12214659) |
| let-7e-3p | CUAUACGGCCUCCUAGCUUUCC | 19 | NC_000019.10 (51692786..51692864) |
| miR-301b-3p | CAGUGCAAUGAUAUUGUCAAAGC | 22 | NC_000022.11 (21652981..21653058) |
| miR-301a-3p | CAGUGCAAUAGUAUUGUCAAAGC | 17 | NC_000017.11 (59151136..59151221) |
| miR-378a-3p | ACUGGACUUGGAGUCAGAAGGC | 5 | NC_000005.10 (149732825..14973289) |
| miR-422a | ACUGGACUUAGGGUCAGAAGGC | 15 | NC_000015.10 (63870930..63871019) |
| miR-20b-5p | CAAAGUGCUCAUAGUGCAGGUAG | X | NC_000023.11 (134169809..13416987) |
| miR-17-5p | CAAAGUGCUUACAGUGCAGGUAG | 13 | NC_000013.11 (91350605..91350688) |
| miR-148a-3p | UCAGUGCACUACAGAACUUUGU | 7 | NC_000007.14 (25949919..25949986) |
| miR-148b-3p | UCAGUGCAUCACAGAACUUUGU | 12 | NC_000012.12 (54337216..54337314) |
| Analytical Method | Specificity for Detecting Specific microRNA | Sample Throughput | Costs | Differentiation of Isomeric microRNA |
|---|---|---|---|---|
| By Sequencing | ★★★ | Low to High | $$–$$$ | Yes |
| By Mass Measurement | ★★★ | Medium | $$ | Size- and Sequence-dependent |
| By Complementary Probe | ★★ to ★★★ | Low to High | $$ | Sequence-dependent |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mwangi, J.N.; Chiu, N.H.L. High Percentage of Isomeric Human MicroRNA and Their Analytical Challenges. Non-Coding RNA 2016, 2, 13. https://doi.org/10.3390/ncrna2040013
Mwangi JN, Chiu NHL. High Percentage of Isomeric Human MicroRNA and Their Analytical Challenges. Non-Coding RNA. 2016; 2(4):13. https://doi.org/10.3390/ncrna2040013
Chicago/Turabian StyleMwangi, Joseph N., and Norman H. L. Chiu. 2016. "High Percentage of Isomeric Human MicroRNA and Their Analytical Challenges" Non-Coding RNA 2, no. 4: 13. https://doi.org/10.3390/ncrna2040013
APA StyleMwangi, J. N., & Chiu, N. H. L. (2016). High Percentage of Isomeric Human MicroRNA and Their Analytical Challenges. Non-Coding RNA, 2(4), 13. https://doi.org/10.3390/ncrna2040013





