Current perspectives in Set7 mediated stem cell differentiation
Abstract
:1. Introduction
2. Non-Histone Methylation by Set7 and Genetic Set7 Knockout Mouse Studies
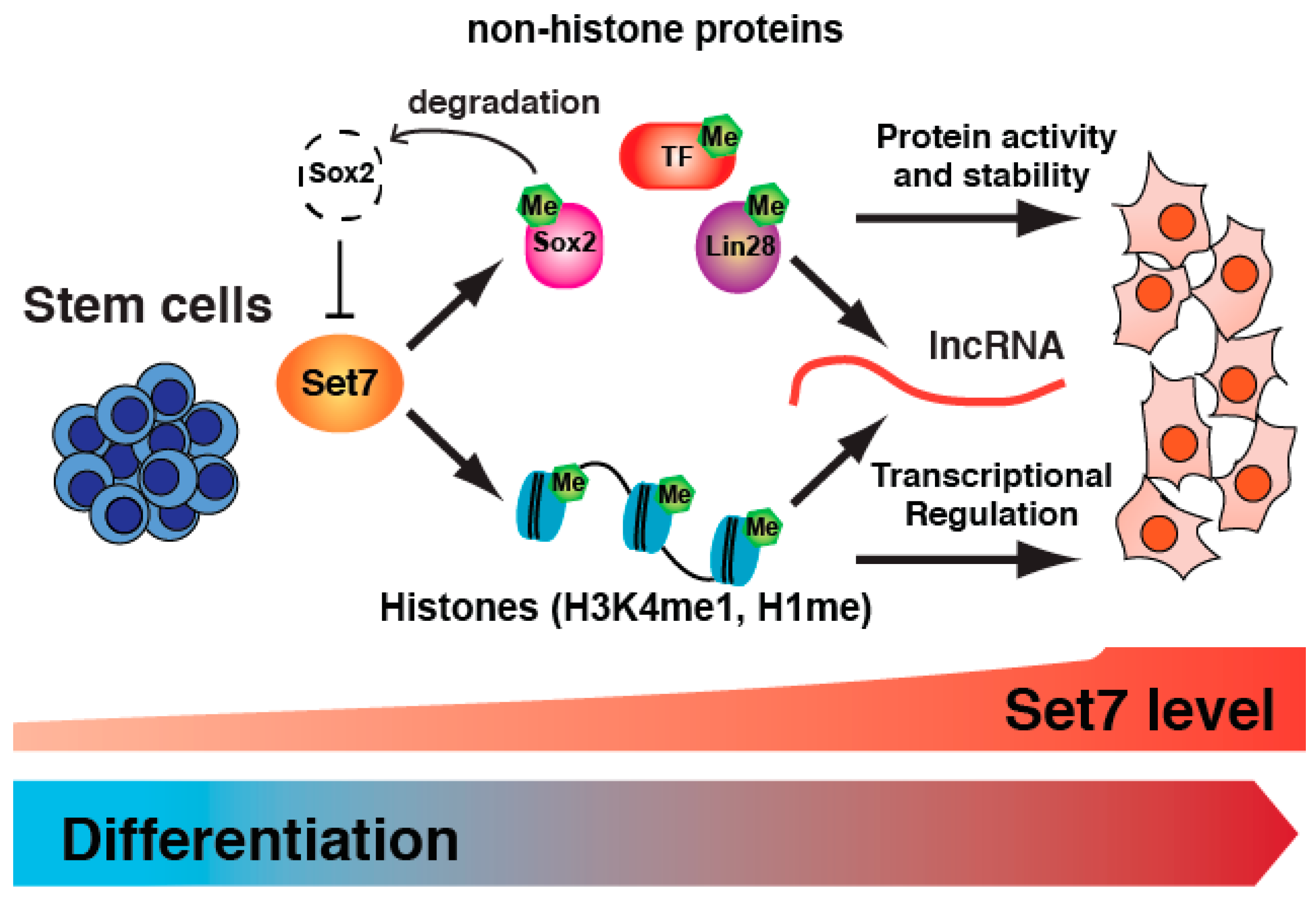
3. Set7 Regulates Stem Cell Differentiation
4. Is Set7 Restricted to SM-Associated Gene Regulation?
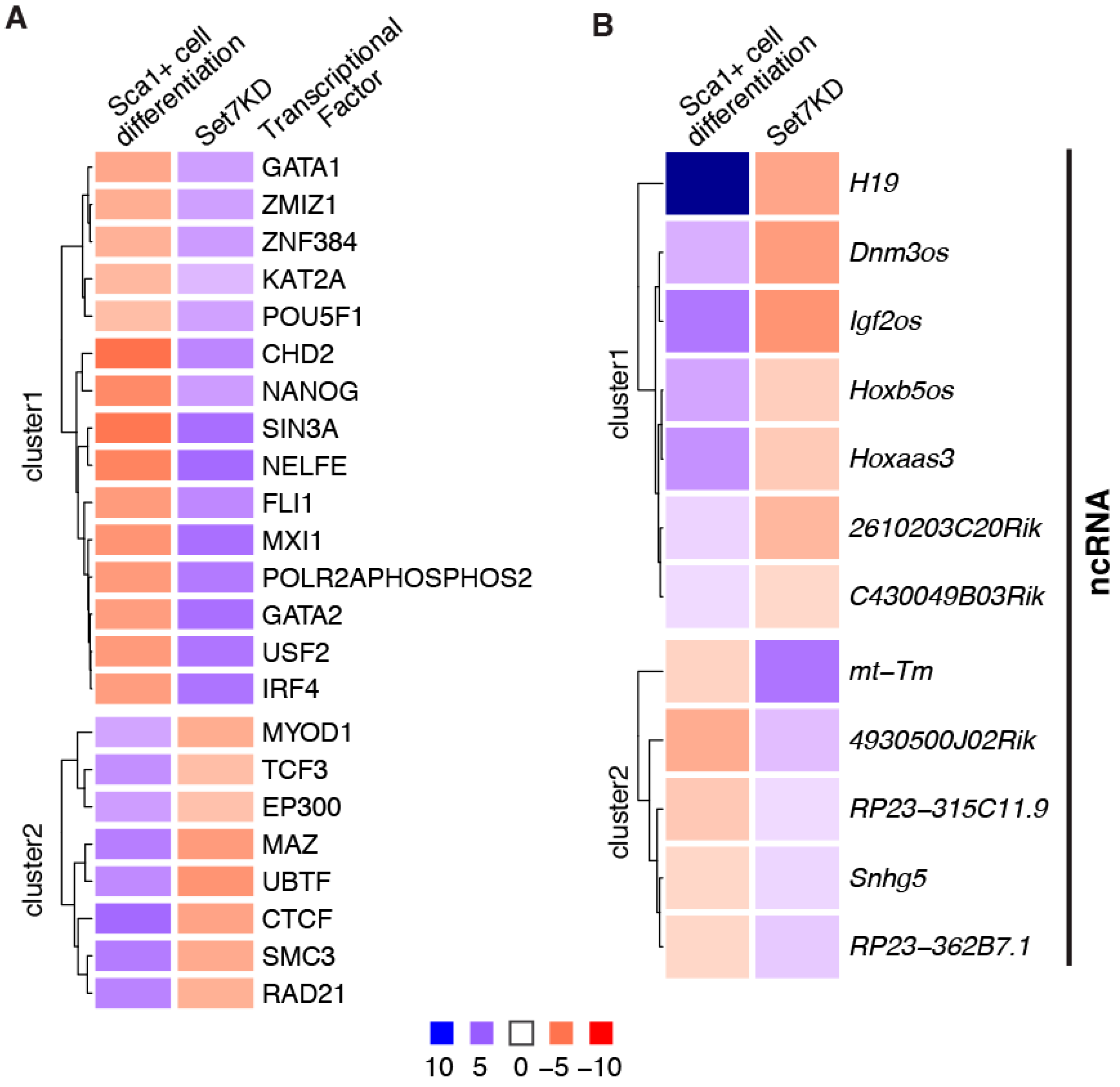
5. Set7 Regulates the Expression of Differentiation-Associated ncRNA
6. Conclusions and Future Prospects
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| Set7 | SET domain containing lysine methyltransferase 7 |
| H3K4me1 | Histone 3 lysine 4 monomethylation |
| TFs | Transcriptional Factors |
| ncRNA | non-coding RNA |
| ESC | Embryonic Stem Cell |
| KO | Knock Out |
| YAP | Yes-Associated Protein |
| TGFß | Transforming Growth Factor beta |
| Sox2 | Sry box-containing gene 2 |
| Oct4 | Octamer-binding transcriptional factor 4 |
| LIN28A | Lin-28 Homolog A |
| KD | Knock Down |
| RNA-Seq | RNA Sequencing |
| let-7 | Lethal-7 |
| MYOD | Myogenic differentiation protein |
| SM | Smooth Muscle |
| SRF | Serum Response Factor |
| HNF4 | Hepatocyte Nuclear Factor 4 |
| miRNA | microRNA |
| Sca1 | Stem cell antigen 1 |
| Igf2os | Insulin-like growth factor 2, opposite strand |
| Dnm3os | Dynamin 3, Opposite Strand |
| Hoxaas3 | HOXA Cluster Antisense RNA 3 |
References
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Cao, R.; Xia, L.; Erdjument-Bromage, H.; Borchers, C.; Tempst, P.; Zhang, Y. Purification and functional characterization of a histone h3-lysine 4-specific methyltransferase. Mol. Cell 2001, 8, 1207–1217. [Google Scholar] [CrossRef]
- Nishioka, K.; Chuikov, S.; Sarma, K.; Erdjument-Bromage, H.; Allis, C.D.; Tempst, P.; Reinberg, D. Set9, a novel histone h3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 2002, 16, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Keating, S.T.; Ziemann, M.; Okabe, J.; Khan, A.W.; Balcerczyk, A.; El-Osta, A. Deep sequencing reveals novel set7 networks. Cell. Mol. Life Sci. 2014, 71, 4471–4486. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, S.; Chin, H.G.; Esteve, P.O.; Jacobsen, S.E. Set7/9 mediated methylation of non-histone proteins in mammalian cells. Epigenetics 2009, 4, 383–387. [Google Scholar] [CrossRef] [PubMed]
- Keating, S.T.; El-Osta, A. Transcriptional regulation by the set7 lysine methyltransferase. Epigenetics 2013, 8, 361–372. [Google Scholar] [CrossRef] [PubMed]
- El-Osta, A.; Brasacchio, D.; Yao, D.; Pocai, A.; Jones, P.L.; Roeder, R.G.; Cooper, M.E.; Brownlee, M. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J. Exp. Med. 2008, 205, 2409–2417. [Google Scholar] [CrossRef] [PubMed]
- Okabe, J.; Orlowski, C.; Balcerczyk, A.; Tikellis, C.; Thomas, M.C.; Cooper, M.E.; El-Osta, A. Distinguishing hyperglycemic changes by set7 in vascular endothelial cells. Circ. Res. 2012, 110, 1067–1076. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Neppl, R.L.; Huang, Z.P.; Chen, J.; Tang, R.H.; Cao, R.; Zhang, Y.; Jin, S.W.; Wang, D.Z. The histone methyltransferase set7/9 promotes myoblast differentiation and myofibril assembly. J. Cell Biol. 2011, 194, 551–565. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Zhang, L.; Wei, W.; Jin, X.; Wang, P.; Tong, Y.; Li, J.; Du, J.X.; Wong, J. A methylation-phosphorylation switch determines sox2 stability and function in esc maintenance or differentiation. Mol. Cell 2014, 55, 537–551. [Google Scholar] [CrossRef] [PubMed]
- Castano, J.; Morera, C.; Sese, B.; Boue, S.; Bonet-Costa, C.; Marti, M.; Roque, A.; Jordan, A.; Barrero, M.J. Setd7 regulates the differentiation of human embryonic stem cells. PLoS ONE 2016, 11, e0149502. [Google Scholar] [CrossRef] [PubMed]
- Tuano, N.K.; Okabe, J.; Ziemann, M.; Cooper, M.E.; El-Osta, A. Set7 mediated interactions regulate transcriptional networks in embryonic stem cells. Nucleic Acids Res. 2016, 44, 9206–9217. [Google Scholar] [CrossRef] [PubMed]
- Keating, S.T.; El-Osta, A. Epigenetics and metabolism. Circ. Res. 2015, 116, 715–736. [Google Scholar] [CrossRef] [PubMed]
- Dhayalan, A.; Kudithipudi, S.; Rathert, P.; Jeltsch, A. Specificity analysis-based identification of new methylation targets of the set7/9 protein lysine methyltransferase. Chem. Biol. 2011, 18, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Chuikov, S.; Kurash, J.K.; Wilson, J.R.; Xiao, B.; Justin, N.; Ivanov, G.S.; McKinney, K.; Tempst, P.; Prives, C.; Gamblin, S.J.; et al. Regulation of p53 activity through lysine methylation. Nature 2004, 432, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Kurash, J.K.; Lei, H.; Shen, Q.; Marston, W.L.; Granda, B.W.; Fan, H.; Wall, D.; Li, E.; Gaudet, F. Methylation of p53 by set7/9 mediates p53 acetylation and activity in vivo. Mol. Cell 2008, 29, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Lehnertz, B.; Rogalski, J.C.; Schulze, F.M.; Yi, L.; Lin, S.; Kast, J.; Rossi, F.M. P53-dependent transcription and tumor suppression are not affected in set7/9-deficient mice. Mol. Cell 2011, 43, 673–680. [Google Scholar] [CrossRef] [PubMed]
- Campaner, S.; Spreafico, F.; Burgold, T.; Doni, M.; Rosato, U.; Amati, B.; Testa, G. The methyltransferase set7/9 (setd7) is dispensable for the p53-mediated DNA damage response in vivo. Mol. Cell 2011, 43, 681–688. [Google Scholar] [CrossRef] [PubMed]
- Oudhoff, M.J.; Freeman, S.A.; Couzens, A.L.; Antignano, F.; Kuznetsova, E.; Min, P.H.; Northrop, J.P.; Lehnertz, B.; Barsyte-Lovejoy, D.; Vedadi, M.; et al. Control of the hippo pathway by set7-dependent methylation of yap. Dev. Cell 2013, 26, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Elkouris, M.; Kontaki, H.; Stavropoulos, A.; Antonoglou, A.; Nikolaou, K.C.; Samiotaki, M.; Szantai, E.; Saviolaki, D.; Brown, P.J.; Sideras, P.; et al. Set9-mediated regulation of tgf-beta signaling links protein methylation to pulmonary fibrosis. Cell Rep. 2016, 15, 2733–2744. [Google Scholar] [CrossRef] [PubMed]
- Young, R.A. Control of the embryonic stem cell state. Cell 2011, 144, 940–954. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.K.; Lee, H.; Han, K.; Kim, S.C.; Choi, Y.; Park, S.W.; Bak, G.; Lee, Y.; Choi, J.K.; Kim, T.K.; et al. Set7/9 methylation of the pluripotency factor lin28a is a nucleolar localization mechanism that blocks let-7 biogenesis in human escs. Cell Stem Cell 2014, 15, 735–749. [Google Scholar] [CrossRef] [PubMed]
- Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar]
- Gabory, A.; Ripoche, M.A.; Yoshimizu, T.; Dandolo, L. The h19 gene: Regulation and function of a non-coding RNA. Cytogenet. Genome Res. 2006, 113, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Sato, T.; Amano, T.; Kawamura, Y.; Kawamura, N.; Kawaguchi, H.; Yamashita, N.; Kurihara, H.; Nakaoka, T. Dnm3os, a non-coding RNA, is required for normal growth and skeletal development in mice. Dev. Dyn. 2008, 237, 3738–3748. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.C.; Chang, Y.M.; Pan, C.T.; Chen, C.C.; Ling, L.; Tsao, K.C.; Yang, R.B.; Li, W.H. Functional evolution of cardiac micrornas in heart development and functions. Mol. Biol. Evol. 2014, 31, 2722–2734. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.X.; Yan, Y.W.; Chen, D.; Ai, C.Z.; Lu, X.; Xu, S.S.; Jiang, S.; Zhong, G.S.; Chen, D.B.; Jiang, Y.Z. Long non-coding RNA hoxa-as3 interacts with ezh2 to regulate lineage commitment of mesenchymal stem cells. Oncotarget 2016, in press. [Google Scholar] [CrossRef] [PubMed]
- Pagans, S.; Kauder, S.E.; Kaehlcke, K.; Sakane, N.; Schroeder, S.; Dormeyer, W.; Trievel, R.C.; Verdin, E.; Schnolzer, M.; Ott, M. The cellular lysine methyltransferase set7/9-kmt7 binds hiv-1 tar RNA, monomethylates the viral transactivator tat, and enhances hiv transcription. Cell. Host Microbe 2010, 7, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Wan, H.; Zhao, X.; Zhu, S.; Zhou, Q.; Ding, S. Brief report: Combined chemical treatment enables oct4-induced reprogramming from mouse embryonic fibroblasts. Stem Cells 2011, 29, 549–553. [Google Scholar] [CrossRef] [PubMed]
- Okabe, J.; Fernandez, A.Z.; Ziemann, M.; Keating, S.T.; Balcerczyk, A.; El-Osta, A. Endothelial transcriptome in response to pharmacological methyltransferase inhibition. ChemMedChem 2014, 9, 1755–1762. [Google Scholar] [CrossRef] [PubMed]
- Barsyte-Lovejoy, D.; Li, F.; Oudhoff, M.J.; Tatlock, J.H.; Dong, A.; Zeng, H.; Wu, H.; Freeman, S.A.; Schapira, M.; Senisterra, G.A.; et al. (r)-pfi-2 is a potent and selective inhibitor of setd7 methyltransferase activity in cells. Proc. Natl. Acad. Sci. USA 2014, 111, 12853–12858. [Google Scholar] [CrossRef] [PubMed]
| Publication | Year | Knockout Type | Deletion Site | Knockout Strategy | Survival | Other Knockout Phenotype |
|---|---|---|---|---|---|---|
| Kurash et al. [16] | 2008 | Constitutive | Exon 2 | Insertion of promoter less LacZ-Neo-poly-A cassette | Half of Set7 -/- mice died during embryogenesis. | Set7KO mice survived to adulthood appeared grossly normal. Set7KO could not induce p53 downstream targets upon DNA damage. |
| Lehnertz et al. [17] | 2011 | Conditional | Exon 4-8 | Crossing to an actin-Flp deleter strain | Viable with no gross abnormality | Normal ability to p53 mediated cell cycle arrest and apoptosis following genotoxic stimuli in Set7KO mice |
| Campaner et al. [18] | 2011 | Constitutive | Exon 2 | Red/ET-based recombineering | Viable with normal life span No increased predisposition to tumorigenesis | No effect on p53 dependent cell cycle arrest and apoptosis following DNA damage |
| Oudhoff et al. [19] | 2013 | Conditional | Exon 2 | Intestinal epithelial cells (IECs) specific deletion | No overt phenotype | Shorter and wider intestinal crypts. Increase expression of YAP target genes in IECs |
| Elkouris et al. [20] | 2016 | Constitutive | Exon 4 | Crossing to a CMV-Cre strain | Normal development and fertile | Set7KO has a protective effect against pulmonary fibrosis |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karimnia, N.; Rafehi, H.; Tuano, N.K.; Ziemann, M.; K.N, H.; Okabe, J.; El-Osta, A. Current perspectives in Set7 mediated stem cell differentiation. Non-Coding RNA 2016, 2, 14. https://doi.org/10.3390/ncrna2040014
Karimnia N, Rafehi H, Tuano NK, Ziemann M, K.N H, Okabe J, El-Osta A. Current perspectives in Set7 mediated stem cell differentiation. Non-Coding RNA. 2016; 2(4):14. https://doi.org/10.3390/ncrna2040014
Chicago/Turabian StyleKarimnia, Nazanin, Haloom Rafehi, Natasha K Tuano, Mark Ziemann, Harikrishnan K.N, Jun Okabe, and Assam El-Osta. 2016. "Current perspectives in Set7 mediated stem cell differentiation" Non-Coding RNA 2, no. 4: 14. https://doi.org/10.3390/ncrna2040014
APA StyleKarimnia, N., Rafehi, H., Tuano, N. K., Ziemann, M., K.N, H., Okabe, J., & El-Osta, A. (2016). Current perspectives in Set7 mediated stem cell differentiation. Non-Coding RNA, 2(4), 14. https://doi.org/10.3390/ncrna2040014






