An Emphasis on the Role of Long Non-Coding RNAs in Viral Gene Expression, Pathogenesis, and Innate Immunity in Viral Chicken Diseases
Abstract
1. Introduction
2. Long Non-Coding RNAs (lncRNAs)
2.1. Classification of Long Non-Coding RNAs (lncRNAs)
2.2. Biogenesis of lncRNAs
2.3. Mechanism of lncRNA Action
2.4. Role of lncRNAs in Gene Expression
2.4.1. Chromatin Structure Regulation
2.4.2. Regulation of Transcription by lncRNA
2.4.3. Role of lncRNA in Post-Transcriptional Regulation
2.5. Role of lncRNAs in the Innate Antiviral Response
2.6. Role of lncRNAs in Virus Pathogenesis
2.6.1. lncRNAs in Viral Gene Expression
2.6.2. lncRNAs in Viral Replication
2.6.3. lncRNAs in Viral Assembly and Release
2.7. Methods of Detecting lncRNAs
2.7.1. Full-Length cDNA Sequencing
2.7.2. Chromatin State Maps
2.7.3. RNA Sequencing
2.7.4. Chromatin Isolation by RNA Purification (ChIRP)
2.7.5. Modified Crosslinking and Immunoprecipitation (M-CLIP) Assay
2.7.6. Single-Cell RNA Sequencing
3. Role of lncRNAs in Specific Viral Chicken Diseases
3.1. Avian Leukosis
3.2. Marek’s Disease
3.3. Infectious Bursal Disease (Gumboro Disease)
3.4. Avian Influenza
3.5. Infectious Bronchitis
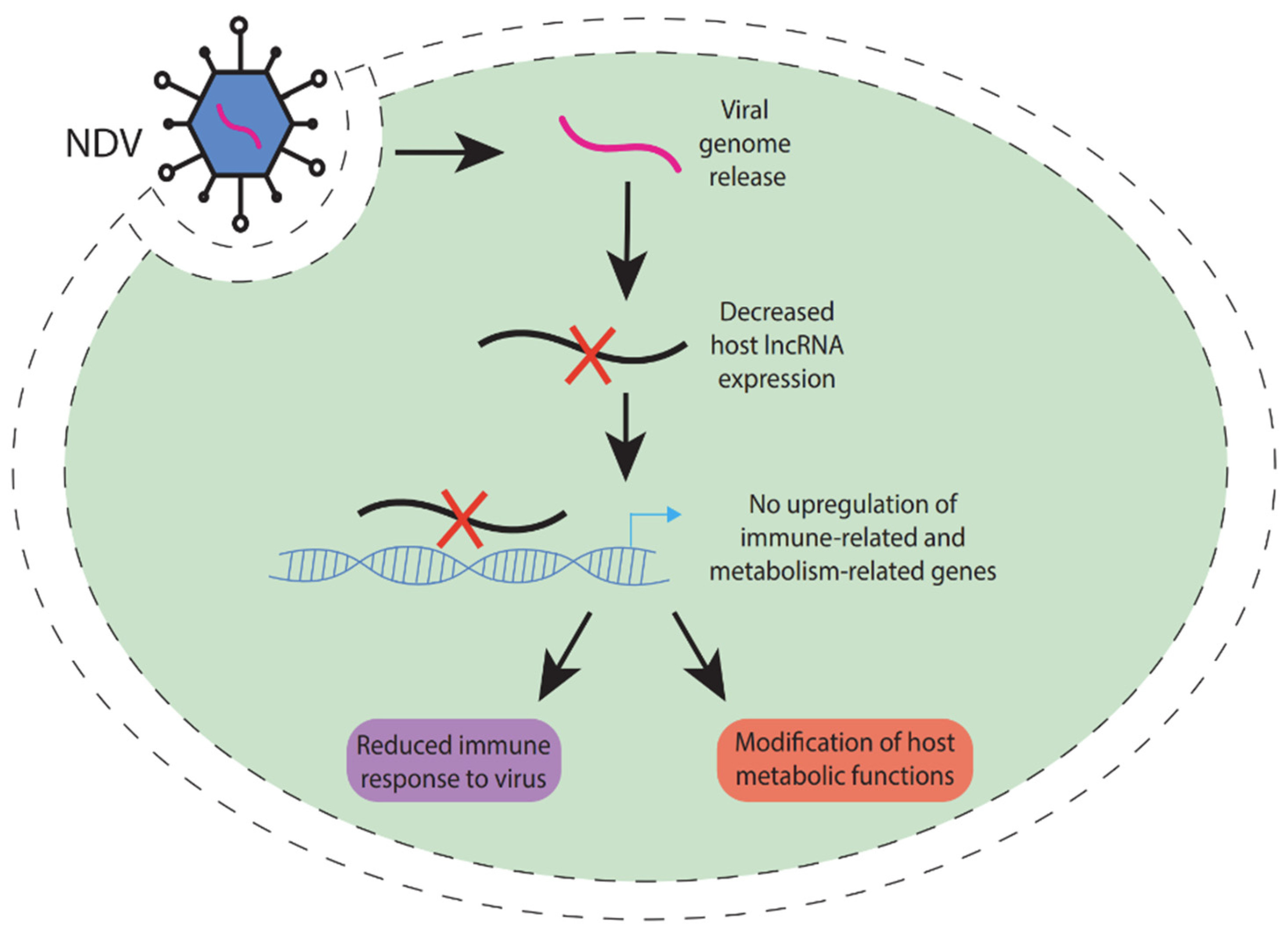
3.6. Newcastle Disease
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bacon, L.D. The National Registry of Genetically Unique Animal Populations: USDA-ADOL Chicken Genetic Lines; National Animal Germplasm Program: East Lansing, MI, USA, 2002. [Google Scholar]
- Mohandas, S.S.; Thakur, M.; Gangil, R. A Mini-Review on Immunosuppressive Viral Diseases and their Containment Methods at Commercial Poultry Farms. Vet. Immunol. Biotechnol. 2018, 1, 31. [Google Scholar]
- Balamurugan, V.; Kataria, J.M. Economically Important Non-Oncogenic Immunosuppressive Viral Diseases of Chicken—Current Status. Vet. Res. Commun. 2006, 30, 541–566. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ouyang, H.; Zheng, M.; Cai, B.; Han, P.; Abdalla, B.A.; Nie, Q.; Zhang, X. Integrated analysis of long non-coding RNAs (LncRNAs) and mRNA expression profiles reveals the potential role of LncRNAs in skeletal muscle development of the chicken. Front. Physiol. 2017, 7, 687. [Google Scholar] [CrossRef]
- Peng, Y.; Chang, L.; Wang, Y.; Wang, R.; Hu, L.; Zhao, Z.; Geng, L.; Liu, Z.; Gong, Y.; Li, J.; et al. Genome-wide differential expression of long noncoding RNAs and mRNAs in ovarian follicles of two different chicken breeds. Genomics 2018, 111, 1395–1403. [Google Scholar] [CrossRef]
- Wower, I.K.; Brandebourg, T.D.; Wower, J. New insights on the mobility of viral and host non-coding RNAs reveal extracellular vesicles as intriguing candidate antiviral targets. Pathogens 2020, 9, 876. [Google Scholar] [CrossRef]
- Ahmad, P.; Bensaoud, C.; Mekki, I.; Rehman, M.U.; Kotsyfakis, M. Long Non-Coding RNAs and their potential roles in the vector–host–pathogen triad. Life 2021, 11, 56. [Google Scholar] [CrossRef]
- Narayanan, A.; Iordanskiy, S.; Das, R.; Van Duyne, R.; Santos, S.; Jaworski, E.; Guendel, I.; Sampey, G.; Dalby, E.; Iglesias-Ussel, M.; et al. Exosomes Derived from HIV-1-infected Cells Contain Trans-activationResponse Element RNA. J. Biol. Chem. 2013, 288, 20014–20033. [Google Scholar] [CrossRef]
- Zhou, W.; Woodson, M.; Neupane, B.; Bai, F.; Sherman, M.B.; Choi, K.H.; Neelakanta, G.; Sultana, H. Exosomes serve as novel modes of tick-borne flavivirus transmission from arthropod to human cellsandfacilitates dissemination of viral RNA and proteins to the vertebrate neuronal cells. PLoS Pathog. 2018, 14, e1006764. [Google Scholar] [CrossRef]
- Samir, M. Host response to veterinary infectious diseases: Role of coding and non-coding RNAs as biomarkers and disease modulators. Front. Vet. Sci. 2023, 10, 1275169. [Google Scholar] [CrossRef]
- Kosinska-Selbi, B.; Mielczarek, M.; Szyda, J. Long non-coding RNA in livestock. Animal 2020, 14, 2003–2013. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Cen, S. Roles of lncRNAs in influenza virus infection. Emerg. Microbes Infect. 2020, 9, 1407–1414. [Google Scholar] [CrossRef] [PubMed]
- Braconi, T.; Kogure, N.; Valeri, N.; Huang, G.; Nuovo, S.; Costinean, M.; Negrini, E.; Miotto, C.; Croce, M.; Patel, T. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene 2011, 30, 4750–4756. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 925–933. [Google Scholar] [CrossRef]
- Zhang, Y.; Cao, X. Long noncoding RNAs in innate immunity. Cell Mol. Immunol. 2016, 13, 138–147. [Google Scholar] [CrossRef]
- Beermann, J.; Piccoli, M.T.; Viereck, J.; Thum, T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol. Rev. 2016, 96, 1297–1325. [Google Scholar] [CrossRef]
- Guttman, M.; Russell, P.; Ingolia, N.T.; Weissman, J.S.; Lander, E.S. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 2013, 154, 240–251. [Google Scholar] [CrossRef]
- Tahira, A.C.; Kubrusly, M.S.; Faria, M.F.; Dazzani, B.; Fonseca, R.S.; Maracaja-Coutinho, V.; Verjovski-Almeida, S.; Machado, M.C.; Reis, E.M. Long noncoding intronic RNAs are differentially expressed in primary and metastatic pancreatic cancer. Mol. Cancer 2011, 10, 141. [Google Scholar] [CrossRef]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef]
- Guttman, M.; Garber, M.; Levin, J.Z.; Donaghey, J.; Robinson, J.; Adiconis, X.; Fan, L.; Koziol, M.J.; Gnirke, A.; Nusbaum, C.; et al. Ab initio reconstruction of cell type–specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol. 2010, 28, 503–510. [Google Scholar] [CrossRef]
- Khalil, A.M.; Guttman, M.; Huarte, M.; Garber, M.; Raj, A.; Rivea Morales, D.; Thomas, K.; Presser, A.; Bernstein, B.E.; van Oudenaarden, A.; et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 11667–11672. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.J.; Ma, X.K.; Xing, Y.H.; Zheng, C.C.; Xu, Y.F.; Shan, L.; Zhang, J.; Wang, S.; Wang, Y.; Carmichael, G.G.; et al. Distinct processing of lncRNAs contributes to non-conserved functions in stem cells. Cell 2020, 181, 621–636. [Google Scholar] [CrossRef]
- Melé, M.; Mattioli, K.; Mallard, W.; Shechner, D.M.; Gerhardinger, C.; Rinn, J.L. Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs. Genome Res. 2017, 27, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Shukla, C.J.; McCorkindale, A.L.; Gerhardinger, C.; Korthauer, K.D.; Cabili, M.N.; Shechner, D.M.; Irizarry, R.A.; Maass, P.G.; Rinn, J.L. High-throughput identification of RNA nuclear enrichment sequences. EMBO J. 2018, 37, 98452. [Google Scholar] [CrossRef] [PubMed]
- Carlevaro-Fita, J.; Rahim, A.; Guigó, R.; Vardy, L.A.; Johnson, R. Cytoplasmic long noncoding RNAs are frequently bound to and degraded at ribosomes in human cells. RNA 2016, 22, 867–882. [Google Scholar] [CrossRef]
- Brown, C.J.; Hendrich, B.; Rupert, J.L.; Lafrenière, R.G.; Xing, Y.; Lawrence, J.; Willard, H.F. The human XIST gene: Analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 1992, 71, 527–542. [Google Scholar] [CrossRef]
- Chaumeil, J.; Le Baccon, P.; Wutz, A.; Heard, E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006, 20, 2223–2237. [Google Scholar] [CrossRef]
- Gebert, C.; Kunkel, D.; Grinberg, A.; Pfeifer, K. H19 Imprinting Control Region Methylation Requires an Imprinted Environment Only in the Male Germ Line. Mol. Cell Biol. 2009, 30, 1108–1115. [Google Scholar] [CrossRef]
- Lee, J.T. Epigenetic Regulation by Long Noncoding RNAs. Science 2012, 338, 1435–1439. [Google Scholar] [CrossRef]
- Martens, J.A.; Laprade, L.; Winston, F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 2004, 429, 571–574. [Google Scholar] [CrossRef]
- Redon, S.; Reichenbach, P.; Lingner, J. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 2010, 38, 5797–5806. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular Mechanisms of Long Noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed]
- van Werven, F.J.; Neuert, G.; Hendrick, N.; Lardenois, A.; Buratowski, S.; Van Oudenaarden, A.; Primig, M.; Amon, A. Transcription of Two Long Noncoding RNAs Mediates Mating-Type Control of Gametogenesis in Budding Yeast. Cell 2012, 150, 1170–1181. [Google Scholar] [CrossRef] [PubMed]
- Baniushin, B.F. Methylation of adenine residues in DNA of eukaryotes. Mol. Biol. 2005, 39, 557–566. [Google Scholar]
- Bao, X.; Wu, H.; Zhu, X.; Guo, X.; Hutchins, A.P.; Luo, Z.; Song, H.; Chen, Y.; Lai, K.; Yin, M.; et al. The p53- 1040 induced lincRNA-p21 derails somatic cell reprogramming by sustaining H3K9me3 and CpG methylation at pluripotency gene promoters. Cell Res. 2014, 25, 80–92. [Google Scholar] [CrossRef]
- O’Leary, V.B.; Ovsepian, S.V.; Carrascosa, L.G.; Buske, F.A.; Radulovi´c, V.; Niyazi, M.; Mörtl, S.; Trau, M.; Atkinson, M.J.; Anastasov, N. PARTICLE, a triplex-forming long ncRNA, regulates locus-specific methylation in response to low-dose Irradiation. Cell Rep. 2015, 11, 474–485. [Google Scholar] [CrossRef]
- Wang, J.; Gong, C.; Maquat, L.E. Control of myogenesis by rodent SINE-containing lncRNAs. Genes Dev. 2013, 27, 793–804. [Google Scholar] [CrossRef]
- Wilusz, J.E.; Sunwoo, H.; Spector, D. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev. 2009, 23, 1494–1504. [Google Scholar] [CrossRef]
- Zeng, W.; Chu, Q.; Xu, T. The long noncoding RNA NARL regulates immune responses via microRNA-mediated NOD1 downregulation in teleost fish. J. Biol. Chem. 2021, 296, 100414. [Google Scholar]
- Beltrán, M.; Puig, I.; Peña, C.; García, J.M.; Lvarez, A.B.Á.; Peña, R.; Bonilla, F.; De Herreros, A.G. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008, 22, 756–769. [Google Scholar] [CrossRef]
- Latos, P.A.; Pauler, F.M.; Koerner, M.V.; Senergin, H.B.; Hudson, Q.; Stocsits, R.R.; Allhoff, W.; Stricker, S.H.; Klement, R.M.; Warczok, K.E.; et al. Airn Transcriptional Overlap, But Not Its lncRNA Products, Induces Imprinted Igf2r Silencing. Science 2012, 338, 1469–1472. [Google Scholar] [CrossRef] [PubMed]
- Jackson, R.; Kroehling, L.; Khitun, A.; Bailis, W.; Jarret, A.; York, A.G.; Khan, O.; Brewer, J.R.; Skadow, M.H.; Duizer, C.; et al. The Translation of Non-Canonical Open Reading Frames Controls Mucosal Immunity. Nature 2018, 564, 434–438. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, A.M.; Chang, H.Y. Long noncoding RNAs in cancer pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef]
- Wang, K.; Liu, F.; Liu, C.Y.; An, T.; Zhang, J.; Zhou, L.Y.; Wang, M.; Dong, Y.H.; Li, N.; Gao, J.N.; et al. The long noncoding RNA NRF regulates programmed necrosis and myocardial injury during ischemia and reperfusion by targeting miR-873. Cell Death Differ. 2016, 23, 1394–1405. [Google Scholar] [CrossRef]
- Wang, A.; Bao, Y.; Wu, Z.; Zhao, T.; Wang, D.; Shi, J.; Liu, B.; Sun, S.; Yang, F.; Wang, L.; et al. Long noncoding RNA EGFR-AS1 promotes cell growth and metastasis via affecting HuR mediated mRNA stability of EGFR in renal cancer. Cell Death Dis. 2019, 10, 154. [Google Scholar] [CrossRef]
- Zhao, C.; Li, X.; Han, B.; Qu, L.; Liu, C.; Song, J.; Lian, L.; Yang, N. Gga-miR-130b- 3p inhibits MSB1 cell proliferation, migration, invasion, and its downregulation in MD tumor is attributed to hypermethylation. Oncotarget 2018, 9, 24187–24198. [Google Scholar] [CrossRef][Green Version]
- Kotake, Y.; Nakagawa, T.; Kitagawa, K.; Suzuki, S.; Liu, N.; Kitagawa, M.; Xiong, Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15INK4B tumor suppressor gene. Oncogene 2011, 30, 1956–1962. [Google Scholar] [CrossRef]
- Chu, C.; Qu, K.; Zhong, F.L.; Artandi, S.E.; Chang, H.Y. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 2011, 44, 667–678. [Google Scholar] [CrossRef]
- Vance, K.W.; Ponting, C.P. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014, 30, 348–355. [Google Scholar] [CrossRef]
- Yang, L.; Froberg, J.E.; Lee, J.T. Long noncoding RNAs: Fresh perspectives into the RNA world. Trends Biochem. Sci. 2014, 39, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Zhang, R.; Sun, X. Enhancer lncrnas influence chromatin interactions in different ways. Front. Genet. 2019, 10, 936. [Google Scholar] [CrossRef] [PubMed]
- Hezroni, H.; Koppstein, D.; Schwartz, M.G.; Avrutin, A.; Bartel, D.P.; Ulitsky, I. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep. 2015, 11, 1110–1122. [Google Scholar] [CrossRef] [PubMed]
- Tilgner, H.; Knowles, D.G.; Johnson, R.; Davis, C.A.; Chakrabortty, S.; Djebali, S.; Curado, J.; Snyder, M.; Gingeras, T.R.; Guigó, R. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res. 2012, 22, 1616–1625. [Google Scholar] [CrossRef]
- Zuckerman, B.; Ulitsky, I. Predictive models of subcellular localization of long RNAs. RNA 2019, 25, 557–572. [Google Scholar] [CrossRef]
- Bonetti, A.; Agostini, F.; Suzuki, A.M.; Hashimoto, K.; Pascarella, G.; Gimenez, J.; Roos, L.; Nash, A.J.; Ghilotti, M.; Cameron, C.J.; et al. RADICL-seq identifies general and cell type-specific principles of genome-wide RNA–chromatin interactions. Nat. Commun. 2020, 11, 1018. [Google Scholar] [CrossRef]
- Li, X.; Zhou, B.; Chen, L.; Gou, L.T.; Li, H.; Fu, X.D. GRID-seq reveals the global RNA–chromatin interactome. Nat. Biotechnol. 2017, 35, 940–950. [Google Scholar] [CrossRef]
- Saldaña-Meyer, R.; Rodriguez-Hernaez, J.; Escobar, T.; Nishana, M.; Jácome-López, K.; Nora, E.P.; Bruneau, B.G.; Tsirigos, A.; Furlan-Magaril, M.; Skok, J.; et al. RNA interactions are essential for CTCF-mediated genome organization. Mol. Cell 2019, 76, 412–422. [Google Scholar] [CrossRef]
- Tan-Wong, S.M.; Dhir, S.; Proudfoot, N.J. R-loops promote antisense transcription across the mammalian genome. Mol. Cell 2016, 76, 600–616. [Google Scholar] [CrossRef]
- Niehrs, C.; Luke, B. Regulatory R-loops as facilitators of gene expression and genome stability. Nat. Rev. Mol. Cell Biol. 2020, 21, 167–178. [Google Scholar] [CrossRef]
- Grote, P.; Wittler, L.; Hendrix, D.; Koch, F.; Währisch, S.; Beisaw, A.; Macura, K.; Bläss, G.; Kellis, M.; Werber, M.; et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell 2013, 24, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Martianov, I.; Ramadass, A.; Serra Barros, A.; Chow, N.; Akoulitchev, A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 2007, 445, 666–670. [Google Scholar] [CrossRef] [PubMed]
- Mondal, T.; Subhash, S.; Vaid, R.; Enroth, S.; Uday, S.; Reinius, B.; Mitra, S.; Mohammed, A.; James, A.R.; Hoberg, E.; et al. MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA–DNA triplex structures. Nat. Commun. 2015, 6, 7743. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, K.M.; Mayer, C.; Postepska, A.; Grummt, I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010, 24, 2264–2269. [Google Scholar] [CrossRef]
- Seila, A.C.; Calabrese, J.M.; Levine, S.S.; Yeo, G.W.; Rahl, P.B.; Flynn, R.A.; Young, R.A.; Sharp, P.A. Divergent transcription from active promoters. Science 2008, 322, 1849–1851. [Google Scholar] [CrossRef]
- Luo, S.; Lu, J.Y.; Liu, L.; Yin, Y.; Chen, C.; Han, X.; Wu, B.; Xu, R.; Liu, W.; Yan, P.; et al. Divergent lncRNAs regulate gene expression and lineage differentiation in pluripotent cells. Cell Stem Cell 2016, 18, 637–652. [Google Scholar] [CrossRef]
- Stojic, L.; Niemczyk, M.; Orjalo, A.; Ito, Y.; Ruijter, A.E.M.; Uribe-Lewis, S.; Joseph, N.; Weston, S.; Menon, S.; Odom, D.T.; et al. Transcriptional silencing of long noncoding RNA GNG12-AS1 uncouples its transcriptional and product-related functions. Nat. Commun. 2016, 7, 10406. [Google Scholar] [CrossRef]
- Thebault, P.; Boutin, G.; Bhat, W.; Rufiange, A.; Martens, J.; Nourani, A. Transcription regulation by the noncoding RNA SRG1 requires Spt2-dependent chromatin deposition in the wake of RNA polymerase II. Mol. Cell. Biol. 2011, 31, 1288–1300. [Google Scholar] [CrossRef]
- Rom, A.; Melamed, L.; Gil, N.; Goldrich, M.J.; Kadir, R.; Golan, M.; Biton, I.; Perry, R.B.T.; Ulitsky, I. Regulation of CHD2 expression by the Chaserr long noncoding RNA gene is essential for viability. Nat. Commun. 2019, 10, 5092. [Google Scholar] [CrossRef]
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285–298. [Google Scholar] [CrossRef]
- Hartford, C.C.R.; Lal, A. When long noncoding becomes protein coding. Mol. Cell. Biol. 2020, 40, e00528-19. [Google Scholar] [CrossRef] [PubMed]
- Yap, K.; Mukhina, S.; Zhang, G.; Tan, J.S.; Ong, H.S.; Makeyev, E.V. A short tandem repeat-enriched RNA assembles a nuclear compartment to control alternative splicing and promote cell survival. Mol. Cell 2013, 72, 525–540. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kopp, F.; Chang, T.C.; Sataluri, A.; Chen, B.; Sivakumar, S.; Yu, H.; Xie, Y.; Mendell, J.T. Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell 2016, 164, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Tichon, A.; Perry, R.B.; Stojic, L.; Ulitsky, I. SAM68 is required for regulation of Pumilio by the NORAD long noncoding RNA. Genes Dev. 2018, 32, 70–78. [Google Scholar] [CrossRef]
- Wang, X.; McLachlan, J.; Zamore, P.D.; Hall, T.M. Modular recognition of RNA by a human Pumilio homology domain. Cell 2002, 110, 501–512. [Google Scholar] [CrossRef]
- Altfeld, M.; Gale, M., Jr. Innate immunity against HIV-1 infection. Nat. Immunol. 2015, 16, 554–562. [Google Scholar] [CrossRef]
- Takeuchi, O.; Akira, S. Innate immunity to virus infection. Immunol. Rev. 2009, 227, 75–86. [Google Scholar] [CrossRef]
- Coccia, E.M.; Battistini, A. Early IFN type I response: Learning from microbial evasion strategies. Semin. Immunol. 2015, 27, 85–101. [Google Scholar] [CrossRef]
- Xu, Y.; Zhong, J. Innate immunity against hepatitis C virus. Curr. Opin. Immunol. 2016, 42, 98–104. [Google Scholar] [CrossRef]
- Carnero, E.; Barriocanal, M.; Prior, C.; Pablo Unfried, J.; Segura, V.; Guruceaga, E.; Enguita, M.; Smerdou, C.; Gastaminza, P.; Fortes, P. Long noncoding RNA EGOT negatively affects the antiviral response and favours HCV replication. EMBO Rep. 2016, 17, 1013–1028. [Google Scholar] [CrossRef]
- Schultheiss, M.; Thimme, R. Toll like receptor 7 and hepatitis C virus infection. J. Hepatol. 2007, 47, 165–167. [Google Scholar] [CrossRef] [PubMed]
- Velloso, L.A.; Folli, F.; Saad, M.J. TLR4 at the crossroads of nutrients, gut microbiota, and metabolic inflammation. Endocr. Rev. 2015, 36, 245–271. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.Y.; Jouanguy, E.; Sancho-Shimizu, V.; Von Bernuth, H.; Yang, K.; Abel, L.; Picard, C.; Puel, A.; Casanova, J.L. Human Toll-like receptor-dependent induction of interferons in protective immunity to viruses. Immunol. Rev. 2007, 220, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Doyle, T.; Goujon, C.; Malim, M.H. HIV-1 and interferons: Who’s interfering with whom? Nat. Rev. Microbiol. 2015, 13, 403–413. [Google Scholar] [CrossRef]
- Kotzin, J.J.; Iseka, F.; Wright, J.; Basavappa, M.G.; Clark, M.L.; Ali, M.A.; Abdel-Hakeem, M.S.; Robertson, T.F.; Mowel, W.K.; Joannas, L.; et al. The long noncoding RNA Morrbid regulates CD8 T cells in response to viral infection. Proc. Natl. Acad. Sci. USA 2019, 116, 11916–11925. [Google Scholar] [CrossRef]
- Li, Z.; Chao, T.C.; Chang, K.Y.; Lin, N.; Patil, V.S.; Shimizu, C.; Head, S.R.; Burns, J.C.; Rana, T.M. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc. Natl. Acad. Sci. USA 2014, 111, 1002–1007. [Google Scholar] [CrossRef]
- Ouyang, J.; Hu, J.; Chen, J.L. lncRNAs regulate the innate immune response to viral infection. Wiley Interdiscip. Rev. RNA 2016, 7, 129–143. [Google Scholar] [CrossRef]
- Zhou, B.; Qi, F.; Wu, F.; Nie, H.; Song, Y.; Shao, L.; Han, J.; Wu, Z.; Saiyin, H.; Wei, G.; et al. Endogenous retrovirus-derived long noncoding RNA enhances innate immune responses via derepressing RELA expression. mBio 2019, 10, e00937-19. [Google Scholar] [CrossRef]
- Liu, W.; Ding, C. Roles of LncRNAs in viral infections. Front. Cell Infect. Microbiol. 2017, 7, 205. [Google Scholar] [CrossRef]
- Ma, H.; Han, P.; Ye, W.; Chen, H.; Zheng, X.; Cheng, L.; Zhang, L.; Yu, L.; Wu, X.A.; Xu, Z.; et al. The long noncoding RNA NEAT1 exerts anti-hantaviral effects by acting as positive feedback for RIG-I signalling. J. Virol. 2017, 91, 10–1128. [Google Scholar] [CrossRef]
- Cadena, C.; Hur, S. Antiviral immunity and circular RNA: No end in sight. Mol. Cell 2017, 67, 163–164. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.G.; Kim, M.V.; Chen, X.; Batista, P.J.; Aoyama, S.; Wilusz, J.E.; Iwasaki, A.; Chang, H.Y. Sensing self and foreign circular RNAs by intron identity. Mol. Cell 2017, 67, 228–238. [Google Scholar] [CrossRef] [PubMed]
- Tam, W.; Ben-Yehuda, D.; Hayward, W.S. bic, A Novel Gene Activated by Proviral Insertions in Avian Leukosis Virus Induced Lymphomas, Is Likely to Function Through Its Noncoding RNA. Mol. Cell Biol. 1997, 17, 1490–1502. [Google Scholar] [CrossRef] [PubMed]
- Sonkoly, E.; Bata-Csorgo, Z.; Pivarcsi, A.; Polyanka, H.; Kenderessy-Szabo, A.; Molnar, G.; Szentpali, K.; Bari, L.; Megyeri, K.; Mandi, Y.; et al. Identification and characterization of a novel, psoriasis susceptibility-related noncoding RNA gene, PRINS. J. Biol. Chem. 2005, 280, 24159–24167. [Google Scholar] [CrossRef]
- Ahanda, M.L.; Ruby, T.; Wittzell, H.; Bed’Hom, B.; Chausse, A.M.; Morin, V.; Oudin, A.; Chevalier, C.; Young, J.R.; Zoorob, R. Non-coding RNAs revealed during identification of genes involved in chicken immune responses. Immunogenetics 2009, 61, 55–70. [Google Scholar] [CrossRef]
- Zhang, Q.; Chen, C.Y.; Yedavalli, V.S.; Jeang, K.T. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. mBio 2013, 4, e00596-12. [Google Scholar] [CrossRef]
- Peng, X.; Gralinski, L.; Armour, C.D.; Ferris, M.T.; Thomas, M.J.; Proll, S.; Bradel-Tretheway, B.G.; Korth, M.J.; Castle, J.C.; Biery, M.C.; et al. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signalling. mBio 2010, 1, e00206-10. [Google Scholar] [CrossRef]
- Yao, R.W.; Wang, Y.; Chen, L.L. Cellular functions of long noncoding RNAs. Nat. Cell Biol. 2019, 21, 542–551. [Google Scholar] [CrossRef]
- Flores, R.; Delgado, S.; Gas, M.E.; Carbonell, A.; Molina, D.; Gago, S.; De la Pena, M. Viroids: The minimal non-coding RNAs with autonomous replication. FEBS Lett. 2004, 567, 42–48. [Google Scholar]
- Moon, S.L.; Anderson, J.R.; Kumagai, Y.; Wilusz, C.J.; Akira, S.; Khromykh, A.A.; Wilusz, J. A noncoding RNA produced by arthropod-borne flaviviruses inhibits the cellular exoribonuclease XRN1 and alters host mRNA stability. RNA 2012, 18, 2029–2040. [Google Scholar] [CrossRef]
- Rossetto, C.C.; Tarrant-Elorza, M.; Verma, S.; Purushothaman, P.; Pari, G.S. Regulation of viral and cellular gene expression by Kaposi’s sarcoma-associated herpesvirus polyadenylated nuclear RNA. J. Virol. 2013, 87, 5540–5553. [Google Scholar] [CrossRef]
- Juranic, V.; Babic Cac, M.; Lisnic, B.; Trsan, T.; Mefferd, A.; Mukhopadhyay, C.D.; Trgovcich, J. Dual analysis of the murine cytomegalovirus and host cell transcriptomes reveal new aspects of the virus-host cell interface. PLoS Pathog. 2013, 9, e1003611. [Google Scholar] [CrossRef]
- Funk, A.; Truong, K.; Nagasaki, T.; Torres, S.; Floden, N.; Balmori Melian, E.; Edmonds, J.; Dong, H.; Shi, P.Y.; Khromykh, A.A. RNA structures required for production of subgenomic flavivirus RNA. J. Virol. 2010, 84, 11407–11417. [Google Scholar] [CrossRef]
- Pijlman, G.P.; Funk, A.; Kondratieva, N.; Leung, J.; Torres, S.; van der Aa, L.; Liu, W.J.; Palmenberg, A.C.; Shi, P.Y.; Hall, R.A.; et al. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe 2008, 4, 579–591. [Google Scholar] [CrossRef]
- Wang, Z.; Deng, X.; Zou, W.; Engelhardt, J.F.; Yan, Z.; Qiu, J. Human Bocavirus 1 Is a Novel Helper for Adeno-associated Virus Replication. J. Virol. 2017, 91, e00710-17. [Google Scholar] [CrossRef]
- Doerr, A. Predicting PPIs. Nat. Methods 2012, 9, 1139. [Google Scholar] [CrossRef]
- Ilott, N.E.; Ponting, C.P. Predicting long non-coding RNAs using RNA sequencing. Methods 2013, 63, 50–59. [Google Scholar] [CrossRef]
- Carninci, P.; Kasukawa, T.; Katayama, S.; Gough, J.; Frith, M.C.; Maeda, N.; Oyama, R.; Ravasi, T.; Lenhard, B.; Wells, C.; et al. The transcriptional landscape of the mammalian genome. Science 2005, 309, 1559–1563. [Google Scholar] [CrossRef]
- Bernstein, B.E.; Kamal, M.; Lindblad-Toh, K.; Bekiranov, S.; Bailey, D.K.; Huebert, D.J.; McMahon, S.; Karlsson, E.K.; Kulbokas, E.J.; Gingeras, T.R.; et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 2005, 120, 169–181. [Google Scholar] [CrossRef]
- Heintzman, N.D.; Stuart, R.K.; Hon, G.; Fu, Y.; Ching, C.W.; Hawkins, R.D.; Barrera, L.O.; Van Calcar, S.; Qu, C.; Ching, K.A.; et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007, 39, 311–318. [Google Scholar] [CrossRef]
- Wang, P.; Xu, J.; Wang, Y.; Cao, X. An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism. Science 2017, 358, 1051–1055. [Google Scholar] [CrossRef]
- Pauli, A.; Valen, E.; Lin, M.F.; Garber, M.; Vastenhouw, N.L.; Levin, J.Z.; Schier, A.F. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012, 22, 577–591. [Google Scholar] [CrossRef]
- Tian, C.; Hu, G. Chromatin isolation by RNA purification (ChIRP) and its applications. In Epigenetics Methods; Academic Press: Cambridge, MA, USA, 2020; pp. 507–521. [Google Scholar]
- Hu, T.; Pi, W.; Zhu, X.; Yu, M.; Ha, H.; Shi, H.; Choi, J.H.; Tuan, D. Long non-coding RNAs transcribed by ERV-9 LTR retrotransposon act in cis to modulate long-range LTR enhancer function. Nucleic Acids Res. 2017, 45, 4479–4492. [Google Scholar] [CrossRef]
- Nadhan, R.; Gomathinayagam, R.; Radhakrishnan, R.; Ha, J.H.; Jayaraman, M.; Dhanasekaran, D.N. Optimizing lncRNA-miRNA interaction analysis: Modified crosslinking and immunoprecipitation (M-CLIP) assay. MethodsX 2024, 13, 103028. [Google Scholar] [CrossRef]
- Cagnin, S.; Alessio, E.; Bonadio, R.S.; Sales, G. Single-cell RNAseq analysis of lncRNAs. In Long Non-Coding RNAs in Cancer; Springer: New York, NY, USA, 2021; pp. 71–90. [Google Scholar]
- Qu, X.; Li, X.; Li, Z.; Liao, M.; Dai, M. Chicken peripheral blood mononuclear cells response to Avian Leukosis virus subgroup J infection assessed by single-cell RNA sequencing. Front. Microbiol. 2022, 13, 800618. [Google Scholar] [CrossRef]
- Adil, A.; Kumar, V.; Jan, A.T.; Asger, M. Single-cell transcriptomics: Current methods and challenges in data acquisition and analysis. Front. Neurosci. 2021, 15, 591122. [Google Scholar] [CrossRef]
- Lan, X.; Wang, Y.; Tian, K.; Ye, F.; Yin, H.; Zhao, X.; Xu, H.; Huang, Y.; Liu, H.; Hsieh, J.C.; et al. Integrated host and viral transcriptome analyses reveal pathology and inflammatory response mechanisms to ALV-J injection in SPF chickens. Sci. Rep. 2017, 7, 46156. [Google Scholar] [CrossRef]
- Qiu, L.; Chang, G.; Li, Z.; Bi, Y.; Liu, X.; Chen, G. Comprehensive Transcriptome Analysis Reveals Competing Endogenous RNA Networks During Avian Leukosis Virus, Subgroup J-Induced Tumorigenesis in Chickens. Front. Physiol. 2018, 9, 996. [Google Scholar] [CrossRef]
- Hu, X.; Chen, S.; Jia, C.; Xue, S.; Dou, C.; Dai, Z.; Xu, H.; Sun, Z.; Geng, T.; Cui, H. Gene expression profile and long non-coding RNA analysis, using RNA-Seq, in chicken embryonic fibroblast cells infected by avian leukosis virus. J. Arch. Virol. 2018, 163, 639–647. [Google Scholar] [CrossRef]
- Dai, M.; Feng, M.; Xie, T.; Zhang, X. Long non-coding RNA and MicroRNA profiling provides comprehensive insight into non-coding RNA involved host immune responses in ALV-J-infected chicken primary macrophage. Dev. Comp. Immunol. 2019, 100, 103414. [Google Scholar] [CrossRef]
- Davison, A.J.; Eberle, R.; Ehlers, B.; Hayward, G.S.; McGeoch, D.J.; Minson, A.C.; Pellett, P.E.; Roizman, B.; Studdert, M.J.; Thiry, E. The order Herpesvirales. Arch. Virol. 2009, 154, 171–177. [Google Scholar] [CrossRef]
- Figueroa, T.; Boumart, I.; Coupeau, D.; Rasschaert, D. Hyperediting by ADAR1 of a new herpesvirus lncRNA during the lytic phase of the oncogenic Marek’s disease virus. J. Gen. Virol. 2016, 97, 2973–2988. [Google Scholar] [CrossRef]
- Fenner, J.E.; Starr, R.; Cornish, A.L.; Zhang, J.G.; Metcalf, D.; Schreiber, R.D.; Sheehan, K.; Hilton, D.J.; Alexander, W.S.; Hertzog, P.J. Suppressor of cytokine signaling 1 regulates the immune response to infection by a unique inhibition of type I interferon activity. Nat. Immunol. 2006, 7, 33–39. [Google Scholar] [CrossRef]
- Burnside, J.; Ouyang, M.; Anderson, A.; Bernberg, E.; Lu, C.; Meyers, B.C.; Green, P.J.; Markis, M.; Isaacs, G.; Huang, E.; et al. Deep sequencing of chicken microRNAs. BMC Genom. 2008, 9, 185. [Google Scholar] [CrossRef]
- Zhao, P.; Li, X.J.; Teng, M.; Dang, L.; Yu, Z.H.; Chi, J.Q.; Su, J.W.; Zhang, G.P.; Luo, J. In vivo expression patterns of microRNAs of Gallid herpesvirus 2 (GaHV-2) during the virus life cycle and development of Marek’s disease lymphomas. Virus Genes 2015, 50, 245–252. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, S.; Wang, G.; Feng, S.; Han, K.; Han, L.; Han, L. Role of microRNA and long non-coding RNA in Marek’s disease tumorigenesis in chicken. Res. Vet. Sci. 2021, 135, 134–142. [Google Scholar] [CrossRef]
- Han, B.; He, Y.; Zhang, L.; Ding, Y.; Lian, L.; Zhao, C.; Song, J.; Yang, N. Long intergenic non-coding RNA GALMD3 in chicken Marek’s disease. Sci. Rep. 2017, 7, 10294. [Google Scholar] [CrossRef]
- Yao, Y.; Zhao, Y.; Smith, L.P.; Lawrie, C.H.; Saunders, N.J.; Watson, M.; Nair, V. Differential expression of microRNAs in Marek’s disease virus-transformed T-lymphoma cell lines. J. Gen. Virol. 2009, 90, 1551–1559. [Google Scholar] [CrossRef]
- He, Y.; Ding, Y.; Zhan, F.; Zhang, H.; Han, B.; Hu, G.; Zhao, K.; Yang, N.; Yu, Y.; Mao, L.; et al. The conservation and signatures of lincRNAs in Marek’s disease of chicken. Sci. Rep. 2015, 5, 15184. [Google Scholar] [CrossRef]
- He, Y.; Han, B.; Ding, Y.; Zhang, H.; Chang, S.; Zhang, L.; Zhao, C.; Yang, N.; Song, J. Linc-GALMD1 regulates Viral Gene Expression in the Chicken. Front. Genet. 2019, 10, 1122. [Google Scholar] [CrossRef]
- Sharma, J.M.; Kim, I.; Routenshlein, S.; Yeh, H.Y. Infectious bursal disease virus of chickens: Pathogenesis and immunosuppression. Dev. Comp. Immunol. 2000, 24, 223–235. [Google Scholar] [CrossRef]
- Banchereau, J.; Steinman, R.M. Dendritic cells and the control of immunity. Nature 1998, 392, 245–252. [Google Scholar] [CrossRef]
- Lin, J.; Xia, J.; Zhang, K.; Yang, Q. Genome-wide profiling of chicken dendritic cell response to infectious bursal disease. BMC Genom. 2016, 17, 878. [Google Scholar] [CrossRef]
- Huang, X.; Xu, Y.; Lin, Q.; Guo, W.; Zhao, D.; Wang, C.; Tang, L. Determination of antiviral action of long non-coding RNA loc107051710 during infectious bursal disease virus infection due to enhancement of interferon production. Virulence 2020, 11, 68–79. [Google Scholar] [CrossRef]
- Lu, C.; Xing, Y.; Cai, H.; Shi, Y.; Liu, J.; Huang, Y. Identification and analysis of long non-coding RNAs in response to H5N1 influenza viruses in duck (Anas platyrhynchos). BMC Genom. 2019, 20, 36. [Google Scholar] [CrossRef]
- Ma, Y.; Ouyang, J.; Wei, J.; Maarouf, M.; Chen, J.L. Involvement of host non-coding RNAs in the pathogenesis of the influenza virus. Int. J. Mol. Sci. 2016, 18, 39. [Google Scholar] [CrossRef]
- Ouyang, J.; Zhu, X.; Chen, Y.; Wei, H.; Chen, Q.; Chi, X.; Qi, B.; Zhang, L.; Zhao, Y.; Gao, G.F.; et al. NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell Host Microbe 2014, 16, 616–626. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, D.; Feng, W.; Guo, Y.; Sun, X.; Zhang, M.; Guan, Z.; Duan, M. Long noncoding RNA TSPOAP1 antisense RNA 1 negatively modulates type I IFN signaling to facilitate influenza A virus replication. J. Med. Virol. 2022, 94, 557–566. [Google Scholar] [CrossRef]
- Bamunuarachchi, G.; Pushparaj, S.; Liu, L. Interplay between host non-coding RNAs and influenza viruses. RNA Biol. 2021, 18, 767–784. [Google Scholar] [CrossRef]
- Jiang, M.; Zhang, S.; Yang, Z.; Lin, H.; Zhu, J.; Liu, L.; Wang, W.; Liu, S.; Liu, W.; Ma, Y.; et al. Self-Recognition of an Inducible Host lncRNA by RIG-I Feedback Restricts Innate Immune Response. Cell 2018, 173, 906–919.e13. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Y.; Li, Q.; Zhao, J.; Yi, D.; Ding, J.; Zhao, F.; Hu, S.; Zhou, J.; Deng, T.; et al. Influenza virus exploits an interferon-independent lncRNA to preserve viral RNA synthesis through stabilizing viral RNA polymerase PB1. Cell Rep. 2019, 27, 3295–3304. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, Y.; Zhou, R.; Zhao, J.; Zhang, Y.; Yi, D.; Li, Q.; Zhou, J.; Guo, F.; Liang, C.; et al. Host long noncoding RNA lncRNA-PAAN regulates the replication of influenza A virus. Viruses 2018, 10, 330. [Google Scholar] [CrossRef] [PubMed]
- Winterling, C.; Koch, M.; Koeppel, M.; Garcia-Alcalde, F.; Karlas, A.; Meyer, T.F. Evidence for a crucial role of a host non-coding RNA in influenza A virus replication. RNA Biol. 2014, 11, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Pushparaj, S.; Zhu, Z.; Huang, C.; More, S.; Liang, Y.; Lin, K.; Vaddadi, K.; Liu, L. Regulation of influenza A virus infection by Lnc-PINK1-2:5. J. Cell. Mol. Med. 2022, 26, 2285–2298. [Google Scholar] [CrossRef]
- Wang, K.; Gong, M.; Zhao, S.; Lai, C.; Zhao, L.; Cheng, S.; Xia, M.; Li, Y.; Wang, K.; Sun, H.; et al. A novel lncRNA DFRV plays a dual function in influenza A virus infection. Front. Microbiol. 2023, 14, 1171423. [Google Scholar] [CrossRef]
- An, H.; Cai, Z.; Yang, Y.; Wang, Z.; Liu, D.X.; Fang, S. Identification and formation mechanism of a novel noncoding RNA produced by avian infectious bronchitis virus. Virology 2019, 528, 176–180. [Google Scholar] [CrossRef]
- Lin, H.; Jiang, M.; Liu, L.; Yang, Z.; Ma, Z.; Liu, S.; Ma, Y.; Zhang, L.; Cao, X. long noncoding RNA Lnczc3h7a promotes a TRIM25-mediated RIG-I antiviral innate immune response. Nat. Immunol. 2019, 27, 812–823. [Google Scholar] [CrossRef]
- Vanamamalai, V.K.; Priyanka, E.; Kannaki, T.R.; Sharma, S. Integrated analysis of genes and long non-coding RNAs in trachea transcriptome to decipher the host response during Newcastle disease challenge in different breeds of chicken. Int. J. Biol. Macromol. 2023, 253, 127183. [Google Scholar] [CrossRef]
- Sha, Y.; Liu, X.; Yan, W.; Wang, M.; Li, H.; Jiang, S.; Yin, R. Long Non-Coding RNA Analysis: Severe Pathogenicity in Chicken Embryonic Visceral Tissues Infected with Highly Virulent Newcastle Disease Virus—A Comparison to the Avirulent Vaccine Virus. Microorganisms 2024, 12, 971. [Google Scholar] [CrossRef]
- Ju, M.; Kim, D.; Son, G.; Han, J. Circular RNAs in and out of Cells: Therapeutic Usages of Circular RNAs. Mol. Cells 2023, 46, 33–40. [Google Scholar] [CrossRef]


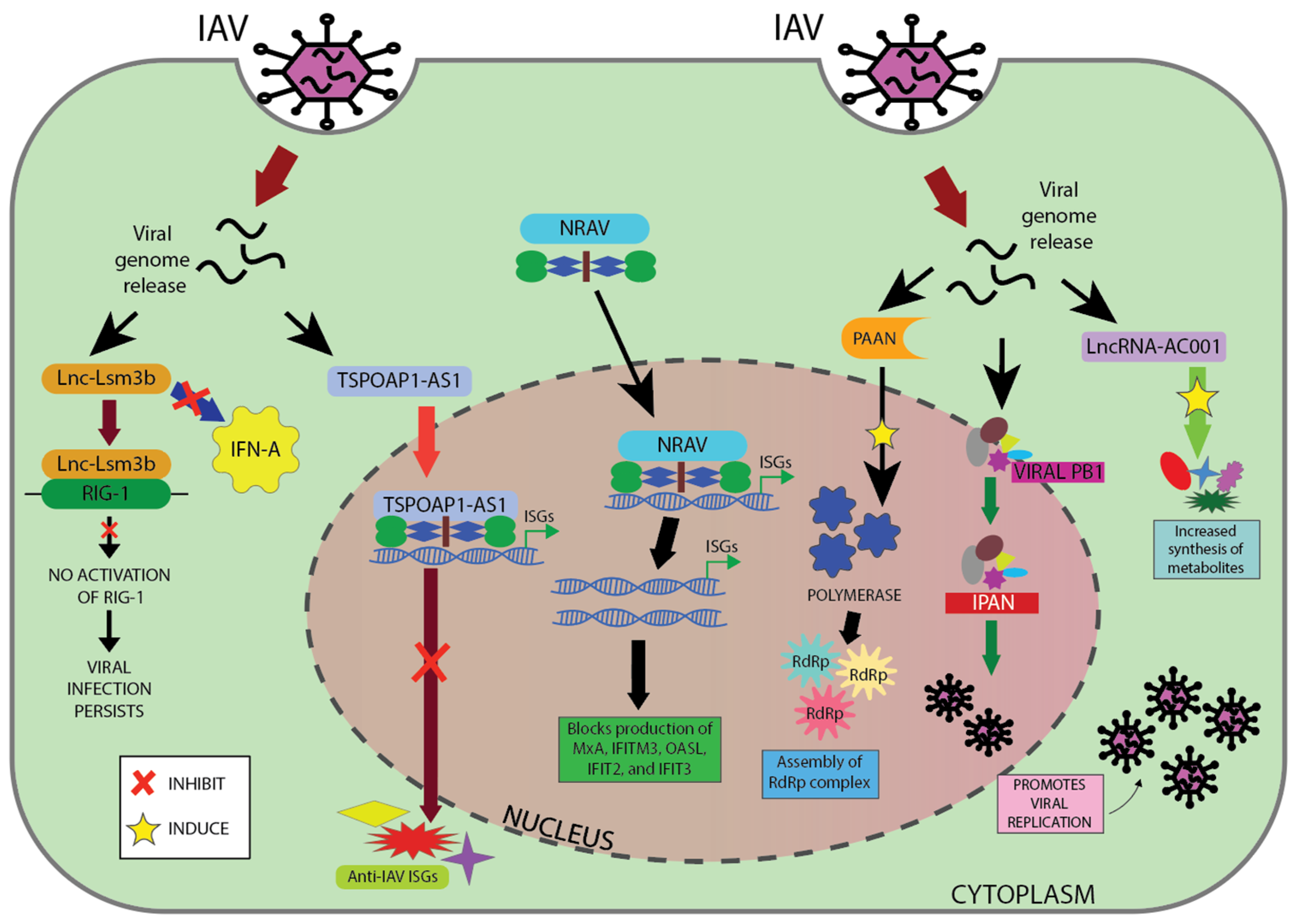
| lncRNA | Influenza Strain | Mechanism | Subcellular Localization | Potential Application | References |
|---|---|---|---|---|---|
| NRAV | A/WSN/1933 (H1N1) | Suppresses the initial transcription of a number of important ISGs, including MxA, IFITM3, OASL, IFIT2, and IFIT3. | Nucleus | Potential antiviral target | Ouyang et al., 2014 [140] |
| TSPOAP1- AS1 | A/Puerto Rico/8/1934 (H1N1) | OASL, ISG20, IFIT1, IFITM1, and other anti-IAV ISGs are negatively regulated, which suppresses IAV-triggered type I IFN signaling. | Nucleus | Potential antiviral target | Wang Q et al., 2022 [141] |
| lnc-Lsm3b | A/Puerto Rico/8/1934 (H1N1) | Blocks overproduction of type Ά IFNs and inhibits RIG-I activation by competing with viral RNAs for the binding of RIG-I monomers. | Cytoplasm | Potential antiviral target | Jiang et al., 2018 [143] |
| IPAN | A/WSN/1933 (H1N1) | Enhances the stability of viral PB1 by forming an association that promotes IAV transcription and replication. | Cytoplasm/Nucleus | Potential antiviral target | Wang et al., 2019 [144] |
| lncRNA-PAAN | A/WSN/1933 (H1N1) | Increases viral RNA polymerase activity by facilitating assembly of the RdRp complex. | Nucleus | Potential antiviral target | Wang J et al., 2018 [145] |
| lncRNA-ACOD1 | A/Puerto Rico/8/1934 (H1N1) | Increases the synthesis of metabolites and the catalytic activity of GOT2. | Cytoplasm | Potential antiviral target | Wang P et al., 2017 [112] |
| VIN | A/WSN/1933 (H1N1) | Restricts IAV replication and viral protein synthesis. | Nucleus | Increase expression using small molecule agonist | Winterling et al., 2014 [146] |
| lnc-PINK1-2 | A/Puerto Rico/8/1934 (H1N1) | Upregulates TXNIP and reduces IAV replication. | Nucleus | Increase expression using small molecule agonist | Pushparaj et al., 2022 [147] |
| lncRNA DFRV | A/Beijing/501/2009 (BJ501, H1N1) | Positively regulates IL-1β and TNF-α and inhibits viral replication. | Nucleus | Potential antiviral target | Wang et al., 2023 [148] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarma, A.; Suri, P.; Justice, M.; Angamuthu, R.; Pushparaj, S. An Emphasis on the Role of Long Non-Coding RNAs in Viral Gene Expression, Pathogenesis, and Innate Immunity in Viral Chicken Diseases. Non-Coding RNA 2025, 11, 42. https://doi.org/10.3390/ncrna11030042
Sarma A, Suri P, Justice M, Angamuthu R, Pushparaj S. An Emphasis on the Role of Long Non-Coding RNAs in Viral Gene Expression, Pathogenesis, and Innate Immunity in Viral Chicken Diseases. Non-Coding RNA. 2025; 11(3):42. https://doi.org/10.3390/ncrna11030042
Chicago/Turabian StyleSarma, Anindita, Parul Suri, Megan Justice, Raja Angamuthu, and Samuel Pushparaj. 2025. "An Emphasis on the Role of Long Non-Coding RNAs in Viral Gene Expression, Pathogenesis, and Innate Immunity in Viral Chicken Diseases" Non-Coding RNA 11, no. 3: 42. https://doi.org/10.3390/ncrna11030042
APA StyleSarma, A., Suri, P., Justice, M., Angamuthu, R., & Pushparaj, S. (2025). An Emphasis on the Role of Long Non-Coding RNAs in Viral Gene Expression, Pathogenesis, and Innate Immunity in Viral Chicken Diseases. Non-Coding RNA, 11(3), 42. https://doi.org/10.3390/ncrna11030042







