High Sensitivity and Specificity Platform to Validate MicroRNA Biomarkers in Cancer and Human Diseases
Abstract
1. Introduction
2. Results and Discussion
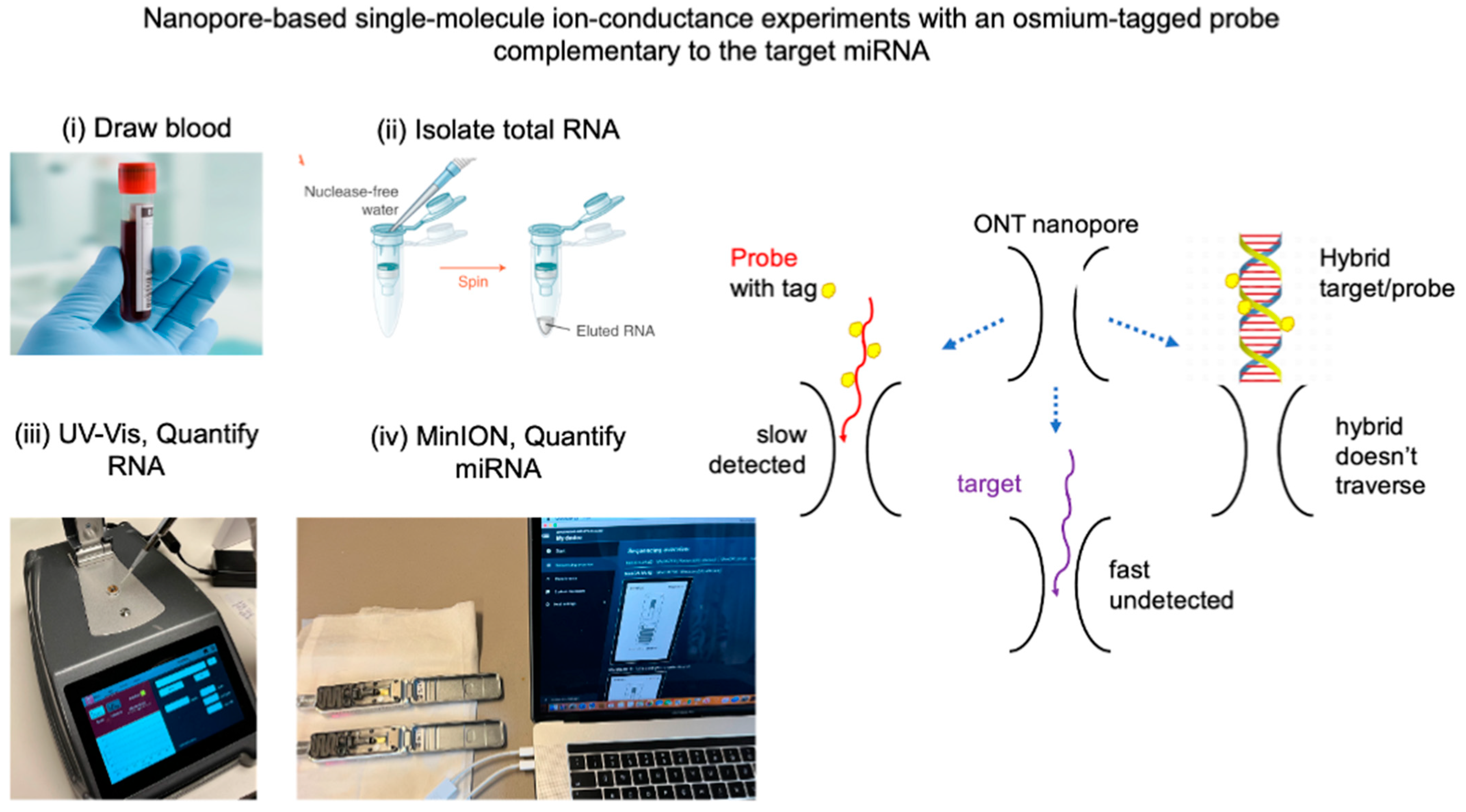
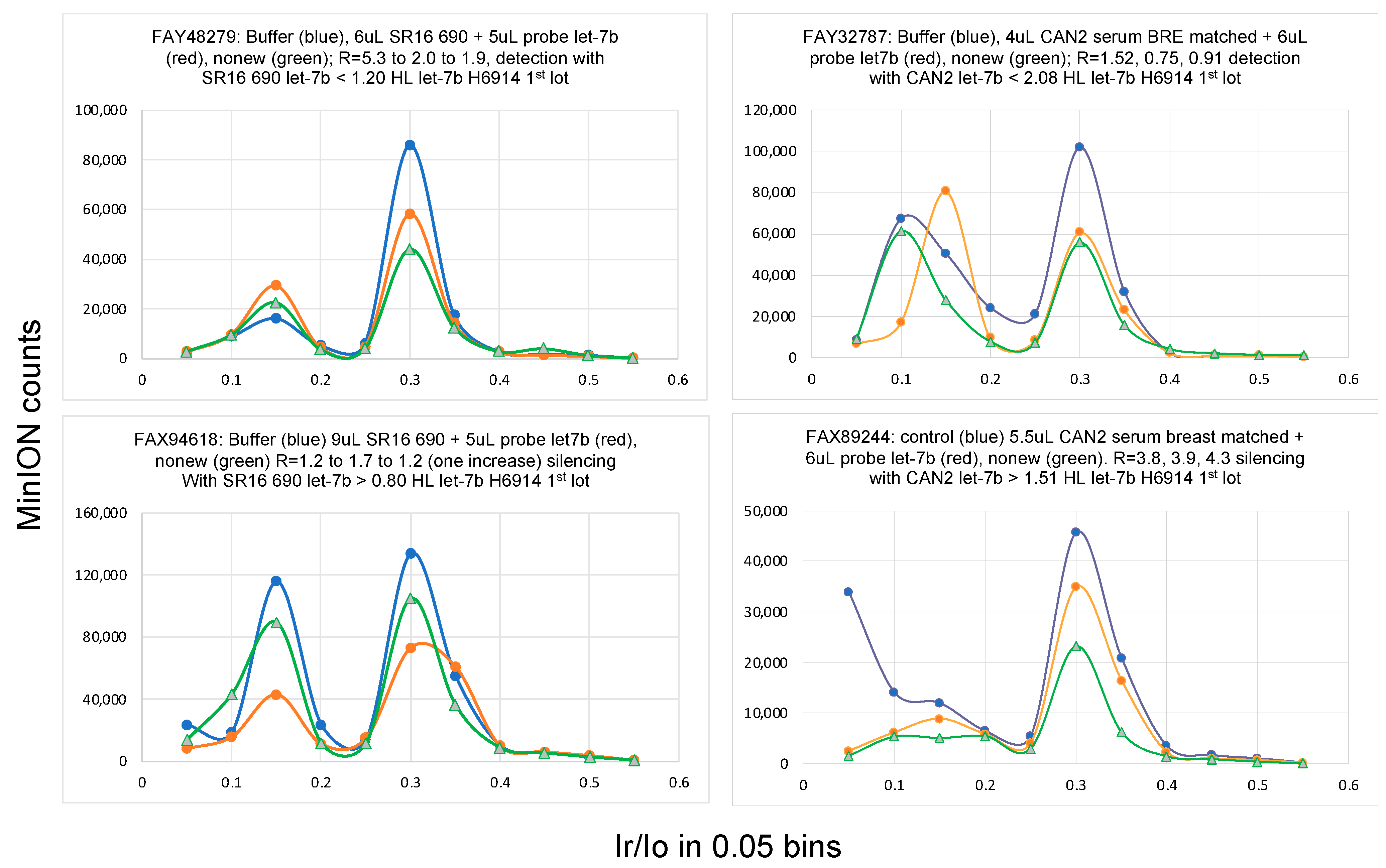
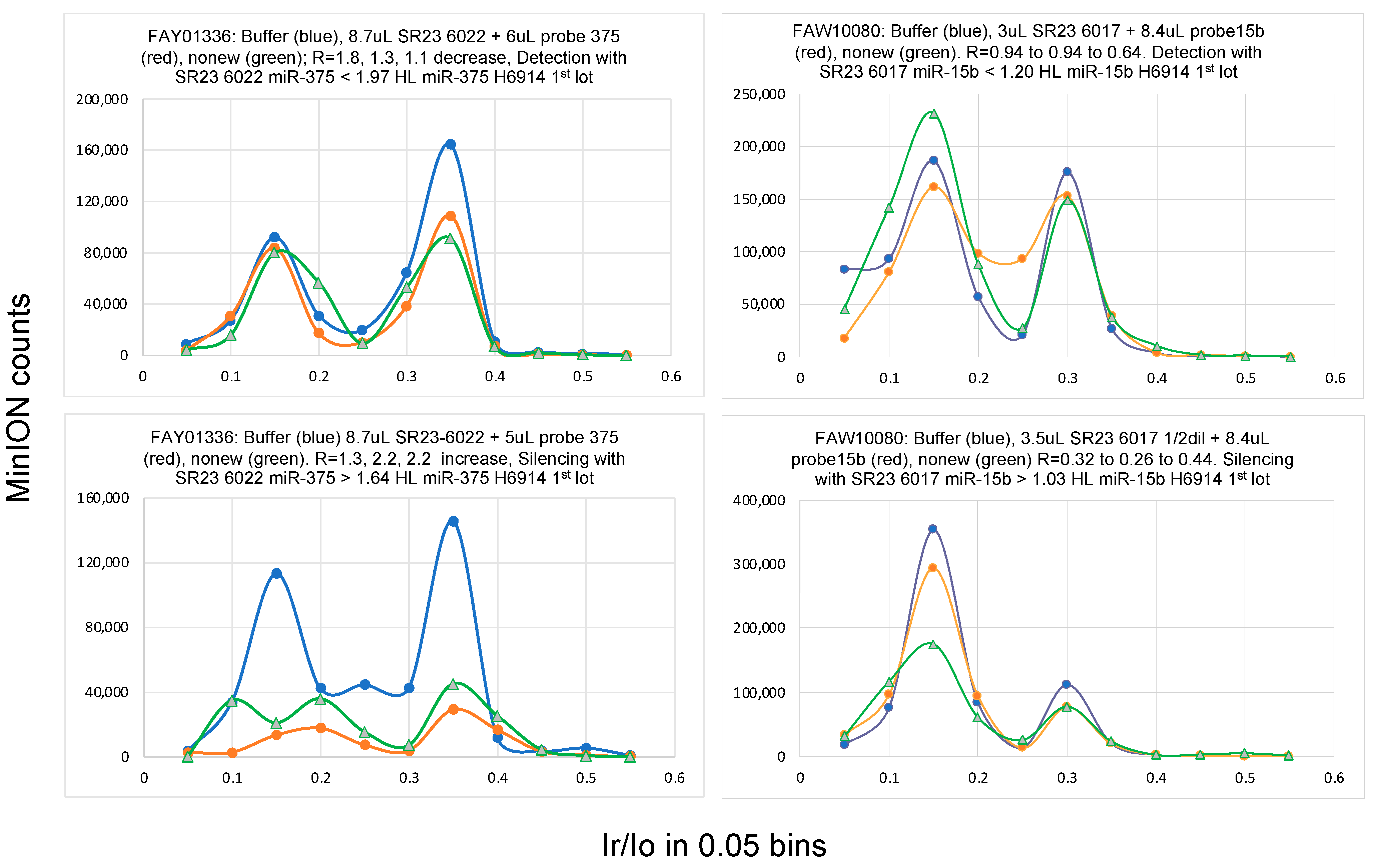
2.1. Single-Molecule Ion Conductance Experiments Using the MinION from ONT
| Biospecimen | H6914 (1) 1st Lot (HL) (2) | H6914 (1) 2nd Lot | H6914 (1) 3rd Lot | H6914 (1) 4th Lot | Serum H2 (3) | Urine1 H2 (3) | Urine2 H2 (3,4) |
|---|---|---|---|---|---|---|---|
| Total RNA, ng/mL | 16.0 | 16.5 | 15.9 | 14.3 | 20.7 | 16.8 | 27.4 (4) |
| miRNA | Copies (+/−%) | Copies (+/−%) | |||||
| miR-16 | 210,250 | ||||||
| miR-15b | 17,710 | 16,716 | 17,687 | 21,852 | 15,517 (7) | ||
| let-7b | 12,150 | 8668 (5) | 19,853 (37) | ||||
| miR-21-5p | 10,494 | 10,514 | >2.0 × HL | 9855 (14) | 21,352 (22) | ||
| miR-375-3p | 9240 | 9636 | 8292 | 1.5 to 2.0 × HL | |||
| miR-141-3p | 6096 | 5341 | 4919 (5) | 1.5 to 2.0 × HL | 5313 (12) |
| Subject ID (1) | Biospecimen | Condition (2) | Isolated Total RNA (ng/μL) | RNA (μL) (3) | Probe Let-7b 30 fM (μL) (3) | Probe Copies per 1 μL of RNA | Normalized Probe Copies (4) | Normalized Probe Copies/12,150 (5) | Ratio of Late to Early (Ir/Io)max (6) | Result (7) | Let-7b/ Let-7b HL (8) | Let-7b/ Let-7b HL (+/−) (8) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H1 | urine | healthy | 18.8 | 9.5 | 6.0 | 11,368 | 9675 | 0.80 | R = 3.5, 4.8, 9.0 (increase ×2) | SIL | >0.80 * | |
| “ | “ | “ | “ | 6.3 | 6.0 | 17,143 | 14,590 | 1.20 | R = 2.4, 1.2, 1.2 (decrease ×2) | DET | <1.20 * | 1.00 (0.20) |
| H2 1/3 dilution | “ | “ | 27.4 | 6.0 | 6.0 | 18,000 | 10,499 | 0.86 | R = 2.1, 3.9, 1.1 (increase ×1, decrease ×1) | Note1 | ||
| “ | “ | “ | “ | 4.0 | 6.0 | 27,000 | 15,749 | 1.30 | R = 6.7, 4.3, 1.9 (decrease ×2) | DET | <1.30 * | |
| “ | “ | “ | “ | 4.0 | 6.0 | 27,000 | 15,749 | 1.30 | R = 1.6, 1.5, 1.1 (decrease ×1) | DET | <1.30 * | |
| “ | “ | “ | “ | 8.5 | 6.0 | 12,706 | 7411 | 0.61 | R = 4.1, 3.7, 5.4 (increase ×1) | SIL | >0.61 * | 1.07 (0.37) |
| H6914 4th lot | serum comb. men | “ | 14.3 | 10.5 | 5.0 | 8571 | 9590 | 0.79 | R = 2.2, 1.5, 2.3 (decrease ×1) | DET | <0.79 * | |
| “ | “ | “ | “ | 6.8 | 5.0 | 13,235 | 14,809 | 1.22 | R = 3.5, 3.8, 2.4 (decrease ×1) | DET | <1.22 | |
| “ | “ | “ | “ | 13.0 | 5.0 | 6923 | 7746 | 0.64 | R = 1.0, 1.5, 1.0 (increase ×1) | SIL | >0.64 * | 0.72 (0.08) |
| SR16-690 | serum | PAN cancer | 16.4 | 9.0 | 5.0 | 10,000 | 9756 | 0.80 | R = 1.2, 1.7, 1.2 (increase ×1) | SIL | >0.80 * | |
| “ | “ | “ | “ | 6.0 | 5.0 | 15,000 | 14,634 | 1.20 | R = 5.3, 2.0, 1.9 (decrease ×2) | DET | <1.20 * | |
| “ B | “ | “ | 14.3 | 6.5 | 5.0 | 13,846 | 15,492 | 1.28 | R = 3.6, 1.9, 1.6 (decrease ×2) | DET | <1.28 | |
| “ B | “ | “ | “ | 13.0 | 5.0 | 6923 | 7746 | 0.64 | R = 1.0, 1.6, 0.8 (increase ×1) | SIL | >0.64 | 1.00 (0.20) |
| SR17-248 B | “ | “ | 21.0 | 4.5 | 5.0 | 20,000 | 15,238 | 1.25 | R = 1.6, 0.9, 1.6 (decrease ×1) | DET | <1.25 | |
| “ | “ | “ | “ | 7.5 | 5.0 | 12,000 | 9143 | 0.75 | R = 1.1, 0.6, 0.7 (decrease ×2) | DET | <0.75 * | |
| “ | “ | “ | “ | 9.5 | 5.0 | 9474 | 7218 | 0.59 | R = 1.2, 2.3, 2.2 (increase ×2) | SIL | >0.59 * | 0.67 (0.08) |
| SR23-6022 | urine | PRO cancer | 14.5 | 4.0 | 5.0 | 22,500 | 24,828 | 2.04 | R = 3.7, 3.0, 1.0 (decrease ×2) | DET | <2.04 * | |
| “ | “ | “ | “ | 5.0 | 4.0 | 14,400 | 15,890 | 1.31 | severely reduced events | SIL | >1.31 | |
| “ | “ | “ | “ | 8.0 | 4.0 | 9000 | 9931 | 0.82 | “ | SIL | >0.82 | |
| “ | “ | “ | “ | 4.0 | 4.0 | 18,000 | 19,862 | 1.63 | “ | SIL | >1.63 * | 1.82 (0.20) |
| “ | “ | “ | “ | 5.3 | 5.0 | 16,981 | 18,738 | 1.54 | R = 1.6, 1.8, 1.3 (comparable) | SIL | Note1 | |
| SR23-6028 | “ | “ | 12.4 | 10.0 | 4.0 | 7200 | 9290 | 0.76 | R = 0.7, 0.9, 1.7 (increase ×1) | SIL | >0.76 | |
| “ | “ | “ | “ | 6.0 | 4.0 | 12,000 | 15,484 | 1.27 | R = 1.9, 1.5, 2.7 (increase ×1) | SIL | >1.27 * | |
| “ | “ | “ | “ | 4.7 | 4.0 | 15,319 | 19,767 | 1.63 | R = 2.7, 1.7, 2.0 (decrease ×2) | DET | <1.63 * | 1.45 (0.18) |
| SR23-6016 1/4 dilution | “ | BRE cancer | 43.7 | 4.0 | 6.0 | 27,000 | 9886 | 0.81 | R = 1.5, 2.9, 2.3 (increase ×2) | SIL | >0.81 | |
| “ | “ | “ | “ | 2.7 | 6.0 | 40,000 | 14,645 | 1.21 | R = 2.0, 2.3, 2.4 | Note1 | ||
| 1/8 dilution | “ | “ | 21.8 | 4.0 | 7.5 | 33,750 | 24,771 | 2.04 | R = 6.6, 2.2, 3.6 decrease ×2) | DET | <2.04 * | |
| “ | “ | “ | “ | 4.0 | 6.0 | 27,000 | 19,817 | 1.63 | R = 1.7, -, 3.2 (increase ×1) | SIL | >1.63 * | 1.77 (0.28) |
| “ | “ | “ | “ | 5.5 | 7.5 | 24,545 | 18,015 | 1.48 | R = 1.1, 1.9, 3.2 (increase ×2) | SIL | >1.48 | |
| 101499 | serum match | “ | 17.1 | 5.5 | 6.0 | 19,636 | 18,373 | 1.51 | R = 3.8, 3.9, 4.3 (increase ×1) | SIL | >1.51 * | |
| “ | “ | “ | “ | 4.0 | 6.0 | 27,000 | 25,263 | 2.08 | R = 1.5, 0.8, 0.9 (decrease ×2) | DET | <2.08 * | 1.80 (0.28) |
| ID (1) | Indication | Isolated Total RNA, ng/μL (2) | miRNA Targets in HL Units (3) | ||||
|---|---|---|---|---|---|---|---|
| miR-15b | miR-21 | miR-375 | miR-141 | miR-375 + miR-141 | |||
| Cancer serum | |||||||
| CAN7 | breast | 6.9 | 0.79 | 1.60 | |||
| CAN9 | “ | 9.1 | 0.89 | 1.79 | 1.80 | ||
| CAN4 | prostate | 12.0 | 0.88 | 1.79 | 1.81 | ||
| CAN6 | “ | 8.0 | 0.90 | 1.87 | 1.84 | ||
| SR16-690 | pancreatic | 16.4 | 1.01 | 1.63 | |||
| 1.88 | |||||||
| SR17-248 | pancreatic | 14.4 | 1.00 | 1.88 | |||
| Cancer urine | |||||||
| SR23 6016 | breast | 174.6 | 1.34 | 1.75 | 1.76 | 1.69 | |
| SR23 6017 | “ | 88.8 | 1.12 | 2.13 | 1.73 | 1.66 | |
| 2.20 | |||||||
| SR23 6018 | breast | 16.1 | 1.72 | ||||
| 2.25 | |||||||
| SR23 6022 | prostate | 15.3 | 1.81 | 1.80 | |||
| SR23 6028 | “ | 13.3 | 1.81 | 1.82 | |||
| SR23 6023 | “ | 18.5 | 1.82 | 1.63 | |||
| 2.00 | |||||||
| SR23 6033 | pancreatic | 13.4 | 1.82 | 1.82 | |||
| Healthy urine | |||||||
| 7.5, 22.9 | 0.97 | 1.04 | |||||
| 21.7 | 0.97 | ||||||
| 9.5 | 1.00 | ||||||
| H2 | 16.8, 82.3 | 0.84 | 0.90 | 0.83 | 0.84 | ||
| 1.19 | |||||||
| 8.7 | 1.01 | ||||||
| 16.5 | 0.89 | ||||||
| 12.2 | 1.00 | ||||||
| 57.4 | 0.98 | 0.80 | |||||
| 163.0 | 0.84 | 1.02 | |||||
| 11.7 | 0.96 | ||||||
| 25.6 | 1.03 | 0.87 | |||||
| 14.5 | 0.92 | ||||||
| 16.4 | 1.17 | 1.16 | |||||
| 13.8 | 0.79 | 1.17 | 1.06 | ||||
| 12.4,15.7 | 1.02 | 1.42 | 1.30 | ||||
| 14.7 | 1.30 | 1.31 | |||||
| Biobank | ID | Age Group | Gender | Cancer | T Stage | N Stage | Specimen |
|---|---|---|---|---|---|---|---|
| Tissue for Research, UK | 101499 | 56–60 | F | breast | - | - | serum matched |
| SR23 6016 | 51–55 | F | “ | pT1b | pN0 | urine | |
| SR23 6017 | 66–70 | F | “ | pT1b | “ | “ | |
| SR23 6018 | 51–55 | F | “ | pT1a | “ | “ | |
| SR23 6022 | 71–75 | M | prostate | pT2 | “ | “ | |
| SR23 6023 | 66–70 | M | “ | “ | “ | “ | |
| SR23 6028 | 51–55 | M | “ | “ | “ | “ | |
| SR23 6033 | 66–70 | F | pancreatic | “ | “ | “ | |
| SR16 690 | 51–55 | M | “ | pT2 | “ | serum | |
| SR17 248 | 51–55 | M | “ | pT1 | “ | “ | |
| Discovery Life Sciences, US | CAN4 | 66–70 | M | prostate | newly diagnosed, pretreatment | serum | |
| CAN6 | 56–60 | M | “ | “ | |||
| CAN7 | 51–55 | F | breast | “ | |||
| CAN9 | 56–60 | F | “ | “ | |||
| ID: DNA Oligo Sequence Used for Probe | In Sequence mU is 2′-OMeU and dU is 2′-deoxyU | Concen, fM (1) | # OsBp Average (2) |
|---|---|---|---|
| Probe 375T5 | (A)5dUCACGCGAGCCGAACGAACAAAC(T)5C(A)5 | 42.0 | 5.1 |
| Probe m21T5 | (A)5mUCAACAmUCAGmUCmUGAmUAAGCmUA(T)5C(A)6 | 27.1 | 4.4 |
| Probe m141T5 | (A)4CCAmUC(mU)3ACCAGACAGmUG(mU)2A(T)5(A)5 | 33.5 | 4.7 |
| Probe 15bT5 | (A)6dUGdUAAACCAdUGAdUGdUGCdUGCdUAT5A6 | 35.0 | 5.9 |
| Probe let7bT5 | (A)6CCACACAACCmUACmUACCmUCA(T)5(A)5 | 30.0 | 5.5 |
2.2. Can Urine Replace Blood as a Biospecimen for miRNA Determination?
2.3. Biospecimen Used in This Study
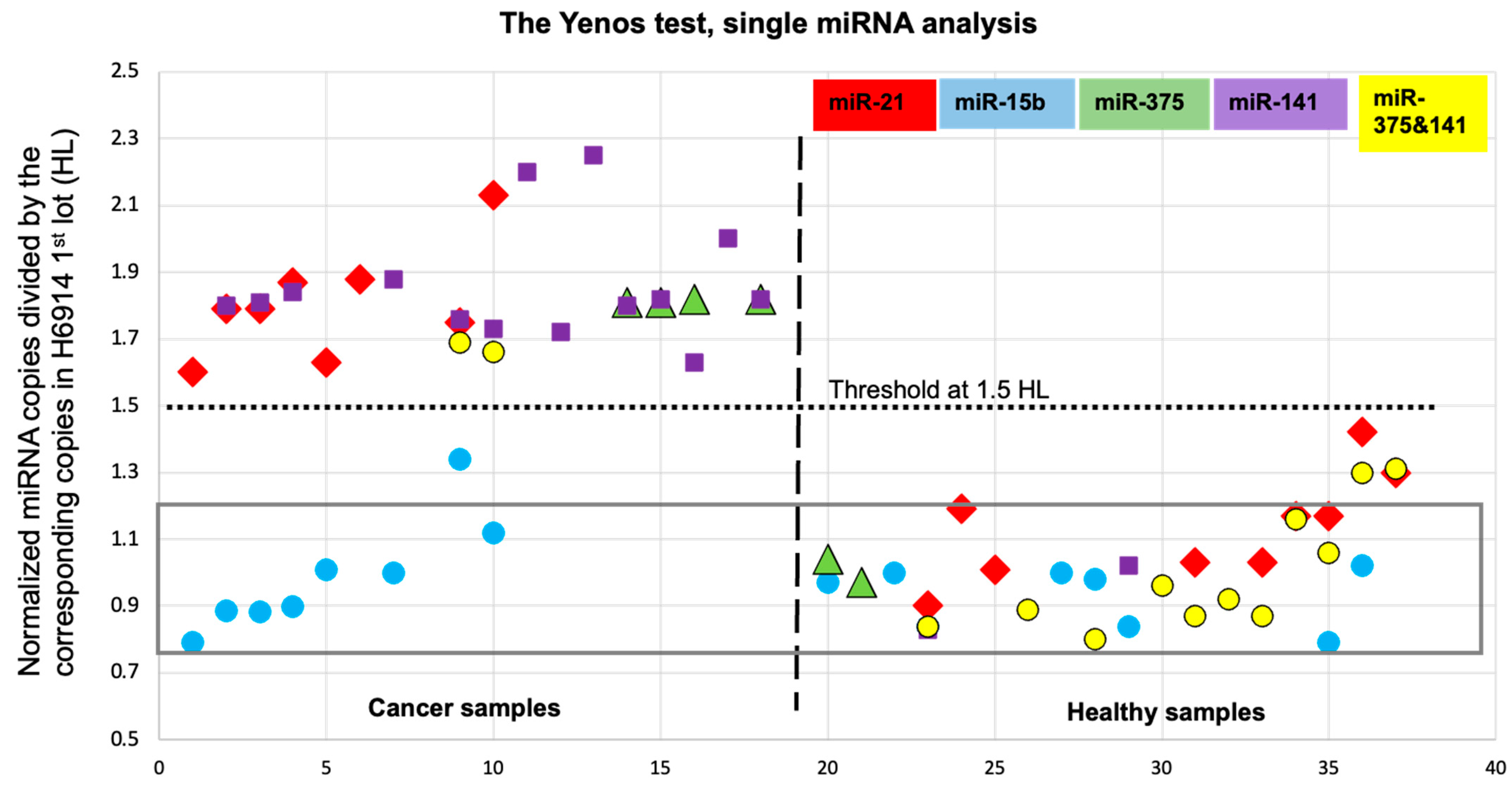
2.4. Validation Strategy for an miRNA Cancer Biomarker
2.5. The miRNA Copy Number Is Proportional to the Total RNA Isolated from the Biospecimen
2.6. Bracketing the miRNA Copy Number Leads to High Accuracy
2.7. Protein-Based vs. Solid-State Nanopores
2.8. Potential, Limitations, Biomarker Multiplexing and Implementation of a Multi-Cancer Screening Test
3. Materials and Methods
3.1. Human Samples
3.2. Oligos, Probes and Other Reagents
3.3. Osmylation of Nucleic Acids
3.4. The Development, Optimization and Validation of Probes
3.5. Single-Molecule Ion-Channel Conductance Experiments on the MinION (MinION Mk1B Platform)
3.6. Data Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics. CA A Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- World Health Organization. Cancer. Available online: http://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 21 July 2024).
- National Cancer Institute. Age and Cancer Risk. Available online: https://www.cancer.gov/about-cancer/causes-prevention/risk/age (accessed on 21 July 2024).
- Accuracy of Mammograms. Available online: https://www.komen.org/breast-cancer/screening/mammography/accuracy/ (accessed on 21 July 2024).
- Women’s Health Network Mammograms. Available online: https://www.womenshealthnetwork.com/breast-health/mammograms/ (accessed on 21 July 2024).
- National Health Service England, Prostate. Available online: https://www.nhs.uk/conditions/prostate-cancer/should-i-have-psa-test/ (accessed on 21 July 2024).
- Borbiev, T.; Kohaar, I.; Petrovics, G. Clinical Biofluid Assays for Prostate Cancer. Cancers 2023, 16, 165. [Google Scholar] [CrossRef]
- Kane, L.E.; Mellotte, G.S.; Mylod, E.; O’Brien, R.M.; O’Connell, F.; Buckley, C.E.; Arlow, J.; Nguyen, K.; Mockler, D.; Meade, A.D.; et al. Diagnostic Accuracy of Blood-based Biomarkers for Pancreatic Cancer: A Systematic Review and Meta-analysis. Cancer Res. Commun. 2022, 2, 1229–1243. [Google Scholar] [CrossRef]
- Stosic, K.; Senar, O.A.; Tarfouss, J.; Bouchart, C.; Navez, J.; Van Laethem, J.-L.; Arsenijevic, T. A Comprehensive Review of the Potential Role of Liquid Biopsy as a Diagnostic, Prognostic, and Predictive Biomarker in Pancreatic Ductal Adenocarcinoma. Cells 2023, 13, 3. [Google Scholar] [CrossRef]
- Connal, S.; Cameron, J.M.; Sala, A.; Brennan, P.M.; Palmer, D.S.; Palmer, J.D.; Perlow, H.; Baker, M.J. Liquid biopsies: The future of cancer early detection. J. Transl. Med. 2023, 21, 118. [Google Scholar] [CrossRef] [PubMed]
- Armakolas, A.; Kotsari, M.; Koskinas, J. Liquid Biopsies, Novel Approaches and Future Directions. Cancers 2023, 15, 1579. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef]
- Beer, T.M. Novel blood-based early cancer detection: Diagnostics in development. Am. J. Manag. Care 2020, 26, S292–S299. [Google Scholar] [CrossRef]
- Duffy, M.J.; Crown, J. Circulating tumor DNA (ctDNA): Can it be used as a pan-cancer early detection test? Crit. Rev. Clin. Lab. Sci. 2024, 61, 241–253. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, B.D.; Oke, J.; Virdee, P.S.; Harris, D.A.; O’Doherty, C.; Park, J.E.; Hamady, Z.; Sehgal, V.; Millar, A.; Medley, L.; et al. Multi-cancer early detection test in symptomatic patients referred for cancer investigation in England and Wales (SYMPLIFY): A large-scale, observational cohort study. Lancet Oncol. 2023, 24, 733–743. [Google Scholar] [CrossRef]
- Ambros, V. microRNAs. Cell 2001, 107, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Alles, J.; Fehlmann, T.; Fischer, U.; Backes, C.; Galata, V.; Minet, M.; Hart, M.; Abu-Halima, M.; Grässer, F.A.; Lenhof, H.-P.; et al. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019, 47, 3353–3364. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- Valihrach, L.; Androvic, P.; Kubista, M. Circulating miRNA analysis for cancer diagnostics and therapy. Mol. Asp. Med. 2019, 72, 100825. [Google Scholar] [CrossRef] [PubMed]
- Ban, E.; Lim, H.J.; Kwon, H.; Song, E.J. Practical magnetic bead-based capillary electrophoresis with laser-induced fluorescence for detecting endogenous miRNA in plasma. Heliyon 2023, 9, e22809. [Google Scholar] [CrossRef] [PubMed]
- Porzycki, P.; Ciszkowicz, E.; Semik, M.; Tyrka, M. Combination of three miRNA (miR-141, miR-21, and miR-375) as potential diagnostic tool for prostate cancer recognition. Int. Urol. Nephrol. 2018, 50, 1619–1626. [Google Scholar] [CrossRef] [PubMed]
- Zografos, E.; Zagouri, F.; Kalapanida, D.; Zakopoulou, R.; Kyriazoglou, A.; Apostolidou, K.; Gazouli, M.; Dimopoulos, M.-A. Prognostic role of microRNAs in breast cancer: A systematic review. Oncotarget 2019, 10, 7156–7178. [Google Scholar] [CrossRef]
- Jenike, A.E.; Halushka, M.K. miR-21: A non-specific biomarker of all maladies. Biomark. Res. 2021, 9, 18. [Google Scholar] [CrossRef]
- Filella, X.; Foj, L. miRNAs as novel biomarkers in the management of prostate cancer. Clin. Chem. Lab. Med. 2017, 55, 715–736. [Google Scholar] [CrossRef]
- Sequeira, J.P.; Barros-Silva, D.; Ferreira-Torre, P.; Salta, S.; Braga, I.; Carvalho, J.; Freitas, R.; Henrique, R.; Jerónimo, C. OncoUroMiR: Circulating miRNAs for Detection and Discrimination of the Main Urological Cancers Using a ddPCR-Based Approach. Int. J. Mol. Sci. 2023, 24, 13890. [Google Scholar] [CrossRef] [PubMed]
- Gahlawat, A.W.; Witte, T.; Sinn, P.; Schott, S. Circulating cf-miRNA as a more appropriate surrogate liquid biopsy marker than cfDNA for ovarian cancer. Sci. Rep. 2023, 13, 5503. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Feng, B.; Han, S.; Zhang, K.; Chen, J.; Li, C.; Wang, R.; Chen, L. The Roles of MicroRNA-141 in Human Cancers: From Diagnosis to Treatment. Cell. Physiol. Biochem. 2016, 38, 427–448. [Google Scholar] [CrossRef] [PubMed]
- Alahdal, M.; Perera, R.A.; Moschovas, M.C.; Patel, V.; Perera, R.J. Current advances of liquid biopsies in prostate cancer: Molecular biomarkers. Mol. Ther.—Oncolytics 2023, 30, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Godoy, P.M.; Barczak, A.J.; DeHoff, P.; Srinivasan, S.; Etheridge, A.; Galas, D.; Das, S.; Erle, D.J.; Laurent, L.C. Comparison of Reproducibility, Accuracy, Sensitivity, and Specificity of miRNA Quantification Platforms. Cell Rep. 2019, 29, 4212–4222.e5. [Google Scholar] [CrossRef] [PubMed]
- Hindson, C.M.; Chevillet, J.R.; Briggs, H.A.; Gallichotte, E.N.; Ruf, I.K.; Hindson, B.J.; Vessella, R.L.; Tewari, M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 2013, 10, 1003–1005. [Google Scholar] [CrossRef] [PubMed]
- Jet, T.; Gines, G.; Rondelez, Y.; Taly, V. Advances in multiplexed techniques for the detection and quantification of microRNAs. Chem. Soc. Rev. 2021, 50, 4141–4161. [Google Scholar] [CrossRef]
- Mestdagh, P.; Hartmann, N.; Baeriswyl, L.; Andreasen, D.; Bernard, N.; Chen, C.; Cheo, D.; D’Andrade, P.; DeMayo, M.; Dennis, L.; et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat. Methods 2014, 11, 809–815. [Google Scholar] [CrossRef]
- Seyhan, A.A. Circulating microRNAs as Potential Biomarkers in Pancreatic Cancer—Advances and Challenges. Int. J. Mol. Sci. 2023, 24, 13340. [Google Scholar] [CrossRef]
- Kang, A.S.W.; Bernasconi, J.G.; Jack, W.; Kanavarioti, A. Ready-to-use nanopore platform for the detection of any DNA/RNA oligo at attomole range using an Osmium tagged complementary probe. Sci. Rep. 2020, 10, 19790. [Google Scholar] [CrossRef] [PubMed]
- Kanavarioti, A. Femtomolar-level PCR-free quantification of microRNA cancer biomarkers in serum. BioRxiv 2023. [Google Scholar] [CrossRef]
- Kasianowicz, J.J.; Brandin, E.; Branton, D.; Deamer, D.W. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl. Acad. Sci. USA 1996, 93, 13770–13773. [Google Scholar] [CrossRef]
- Butler, T.Z.; Gundlach, J.H.; Troll, M. Ionic current blockades from DNA and RNA molecules in the alpha-hemolysin nanopore. Biophys. J. 2007, 93, 3229–3240. [Google Scholar] [CrossRef] [PubMed]
- Maglia, G.; Heron, A.J.; Stoddart, D.; Japrung, D.; Bayley, H. Analysis of single nucleic acid molecules with protein nanopores. Methods Enzymol. 2010, 475, 591–623. [Google Scholar] [CrossRef] [PubMed]
- Haque, F.; Li, J.; Wu, H.-C.; Liang, X.-J.; Guo, P. Solid-state and biological nanopore for real-time sensing of single chemical and sequencing of DNA. Nano Today 2013, 8, 56–74. [Google Scholar] [CrossRef] [PubMed]
- Xi, D.; Shang, J.; Fan, E.; You, J.; Zhang, S.; Wang, H. Nanopore-based selective discrimination of microRNAs with single- nucleotide difference using locked nucleic acid-modified probes. Anal. Chem. 2016, 88, 10540–10546. [Google Scholar] [CrossRef]
- Zahid, O.K.; Wang, F.; Ruzicka, J.A.; Taylor, E.W.; Hall, A.R. Sequence-specific recognition of microRNAs and other short nucleic acids with solid-state nanopores. Nano Lett. 2016, 16, 2033–2039. [Google Scholar] [CrossRef]
- Henley, R.Y.; Vazquez-Pagan, A.G.; Johnson, M.; Kanavarioti, A.; Wanunu, M. Osmium-based on pyrimidine contrast tags for enhanced nanopore-based DNA base discrimination. PLoS ONE 2015, 10, e0142155. [Google Scholar] [CrossRef]
- Ding, Y.; Kanavarioti, A. Single pyrimidine discrimination during voltage-driven translocation of osmylated oligodeoxynucleotides via the α-hemolysin nanopore. Beilstein J. Nanotechnol. 2016, 7, 91–101. [Google Scholar] [CrossRef]
- Sultan, M.; Kanavarioti, A. Nanopore device-based fingerprinting of RNA oligos and microRNAs enhanced with an Osmium tag. Sci. Rep. 2019, 9, 14180. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Pataillot-Meakin, T.; Shibakawa, A.; Ren, R.; Bevan, C.L.; Ladame, S.; Ivanov, A.P.; Edel, J.B. Single-molecule amplification-free multiplexed detection of circulating microRNA cancer biomarkers from serum. Nat. Commun. 2021, 12, 3515. [Google Scholar] [CrossRef] [PubMed]
- Koch, C.; Reilly-O’donnell, B.; Gutierrez, R.; Lucarelli, C.; Ng, F.S.; Gorelik, J.; Ivanov, A.P.; Edel, J.B. Nanopore sequencing of DNA-barcoded probes for highly multiplexed detection of microRNA, proteins and small biomarkers. Nat. Nanotechnol. 2023, 18, 1483–1491. [Google Scholar] [CrossRef] [PubMed]
- Maguire, S.; Guan, S. Rolling circle reverse transcription enables high fidelity nanopore sequencing of small RNA. PLoS ONE 2022, 17, e0275471. [Google Scholar] [CrossRef] [PubMed]
- Quint, I.; Simantzik, J.; Kaiser, L.; Laufer, S.; Csuk, R.; Smith, D.; Kohl, M.; Deigner, H.-P. Ready-to-use nanopore platform for label-free small molecule quantification: Ethanolamine as first example. Nanomed. Nanotechnol. Biol. Med. 2024, 55, 102724. [Google Scholar] [CrossRef] [PubMed]
- Kanavarioti, A.; Kang, A.S.W. See RNA(OsBp) Event Detection Python Package in a Public Repository. Available online: https://github.com/kangaroo96/osbp_detect/blob/master/instructions.md (accessed on 21 July 2024).
- Chomczynski, P.; Wilfinger, W.W.; Eghbalnia, H.R.; Kennedy, A.; Rymaszewski, M.; Mackey, K. Inter-Individual Differences in RNA Levels in Human Peripheral Blood. PLoS ONE 2016, 11, e0148260. [Google Scholar] [CrossRef]
- Mall, C.; Rocke, D.M.; Durbin-Johnson, B.; Weiss, R.H. Stability of miRNA in Human Urine Supports its Biomarker Potential. Biomarkers Med. 2013, 7, 623–631. [Google Scholar] [CrossRef]
- Benke, M.; Zeöld, A.; Kittel, Á.; Khamari, D.; Hritz, I.; Horváth, M.; Keczer, B.; Borka, K.; Szücs, Á.; Wiener, Z. MiR-200b categorizes patients into pancreas cystic lesion subgroups with different malignant potential. Sci. Rep. 2023, 13, 19280. [Google Scholar] [CrossRef] [PubMed]
- Kanavarioti, A. HPLC methods for purity evaluation of man-made single-stranded RNAs. Sci. Rep. 2019, 9, 1019. [Google Scholar] [CrossRef]
- Chang, C.-H.; Beer, M.; Marzilli, L.G. Osmium-labeled polynucleotides. The reaction of osmium tetroxide with deoxyribonucleic acid and synthetic polynucleotides in the presence of tertiary nitrogen donor ligands. Biochemistry 1977, 16, 33–38. [Google Scholar] [CrossRef]
- Palecek, E. Probing DNA structure with osmium tetroxide complexes in vitro. Methods Enzymol. 1992, 212, 139–155. [Google Scholar] [PubMed]
- Reske, T.; Surkus, A.-E.; Duwensee, H.; Flechsig, G.-U. Kinetics of the labeling reactions of thymine, cytosine and uracil with osmium tetroxide bipyridine. Microchim. Acta 2009, 166, 197–201. [Google Scholar] [CrossRef]
- Debnath, T.K.; Okamoto, A. Osmium Tag for Post-transcriptionally Modified RNA. ChemBioChem 2018, 19, 1653–1656. [Google Scholar] [CrossRef] [PubMed]
- Kanavarioti, A.; Greenman, K.L.; Hamalainen, M.; Jain, A.; Johns, A.M.; Melville, C.R.; Kemmish, K.; Andregg, W. Capillary electrophoretic separation-based approach to determine the labeling kinetics of oligodeoxynucleotides. Electrophoresis 2012, 33, 3529–3543. [Google Scholar] [CrossRef]
- Kanavarioti, A. False positives and false negatives measure less than 0.001% in labeling ssDNA with osmium tetroxide 2,2’-bipyridine. Beilstein J. Nanotechnol. 2016, 7, 1434–1446. [Google Scholar] [CrossRef]
- MSDS and User Information. Available online: https://www.ehs.harvard.edu/sites/default/files/lab_safety_guideline_osmium_tetroxide.pdf (accessed on 21 July 2024).
| (A) | |||||
| Accuracy of Measurement (+/−) | Normalized Control, Range | Average Control | Normalized Disease Overexpressed, Range | Average Disease | miRNA in Disease/Control, x-Fold |
| 15% | 0.85 to 1.15 | 1.0 | 1.15 to 1.55 | 1.35 | >1.35 |
| 20% | 0.8 to 1.2 | 1.0 | 1.2 to 1.8 | 1.5 | >1.5 |
| 30% | 0.7 to 1.3 | 1.0 | 1.3 to 2.4 | 1.85 | >1.85 |
| 40% | 0.6 to 1.4 | 1.0 | 1.4 to 3.2 | 2.3 | >2.3 |
| 50% | 0.5 to 1.5 | 1.0 | 1.5 to 4.5 | 3.0 | >3.0 |
| 75% | 0.25 to 1.75 | 1.0 | 1.75 to 12.25 | 7.0 | >7.0 |
| (B) | |||||
| Accuracy of Measurement (+/−) | Normalized Control, Range | Average Control | Normalized Disease Underexpressed, Range | Average Disease | miRNA in Control/Disease, x-Fold |
| 15% | 0.85 to 1.15 | 1.0 | 0.63 to 0.85 | 0.74 | >1.35 |
| 20% | 0.8 to 1.2 | 1.0 | 0.54 to 0.8 | 0.67 | >1.5 |
| 30% | 0.7 to 1.3 | 1.0 | 0.4 to 0.7 | 0.55 | >1.8 |
| 40% | 0.6 to 1.4 | 1.0 | 0.3 to 0.6 | 0.45 | >2.2 |
| 50% | 0.5 to 1.5 | 1.0 | 0.18 to 0.5 | 0.34 | >2.9 |
| 75% | 0.25 to 1.75 | 1.0 | 0.036 to 0.25 | 0.14 | > 7.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kanavarioti, A.; Rehman, M.H.; Qureshi, S.; Rafiq, A.; Sultan, M. High Sensitivity and Specificity Platform to Validate MicroRNA Biomarkers in Cancer and Human Diseases. Non-Coding RNA 2024, 10, 42. https://doi.org/10.3390/ncrna10040042
Kanavarioti A, Rehman MH, Qureshi S, Rafiq A, Sultan M. High Sensitivity and Specificity Platform to Validate MicroRNA Biomarkers in Cancer and Human Diseases. Non-Coding RNA. 2024; 10(4):42. https://doi.org/10.3390/ncrna10040042
Chicago/Turabian StyleKanavarioti, Anastassia, M. Hassaan Rehman, Salma Qureshi, Aleena Rafiq, and Madiha Sultan. 2024. "High Sensitivity and Specificity Platform to Validate MicroRNA Biomarkers in Cancer and Human Diseases" Non-Coding RNA 10, no. 4: 42. https://doi.org/10.3390/ncrna10040042
APA StyleKanavarioti, A., Rehman, M. H., Qureshi, S., Rafiq, A., & Sultan, M. (2024). High Sensitivity and Specificity Platform to Validate MicroRNA Biomarkers in Cancer and Human Diseases. Non-Coding RNA, 10(4), 42. https://doi.org/10.3390/ncrna10040042









