Differential Expression of lncRNAs in HIV Patients with TB and HIV-TB with Anti-Retroviral Treatment
Abstract
1. Introduction
2. Results
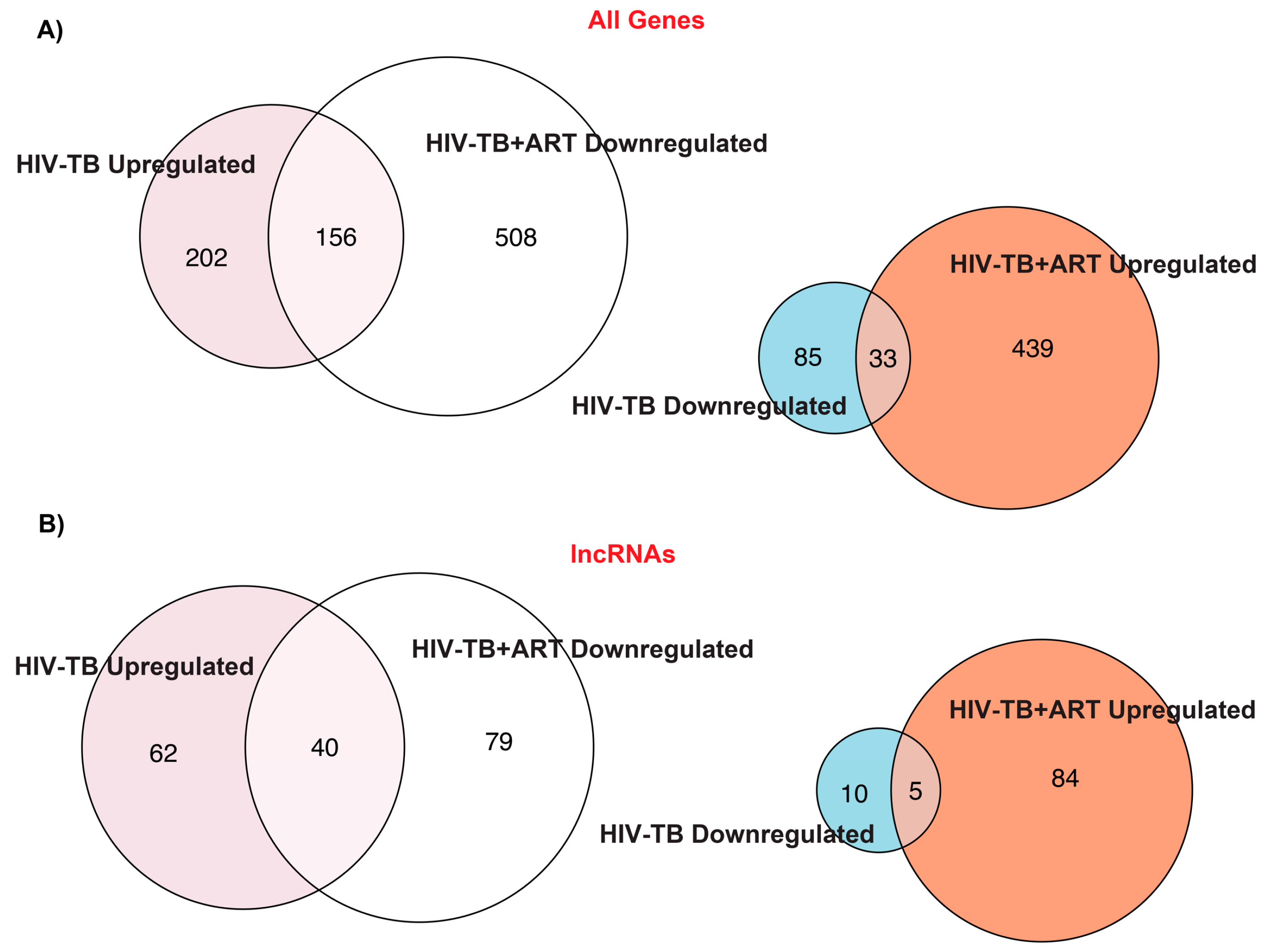
2.1. TB and ART Modulate Gene Expression Profiles in HIV Patients, including Non-Coding Transcripts
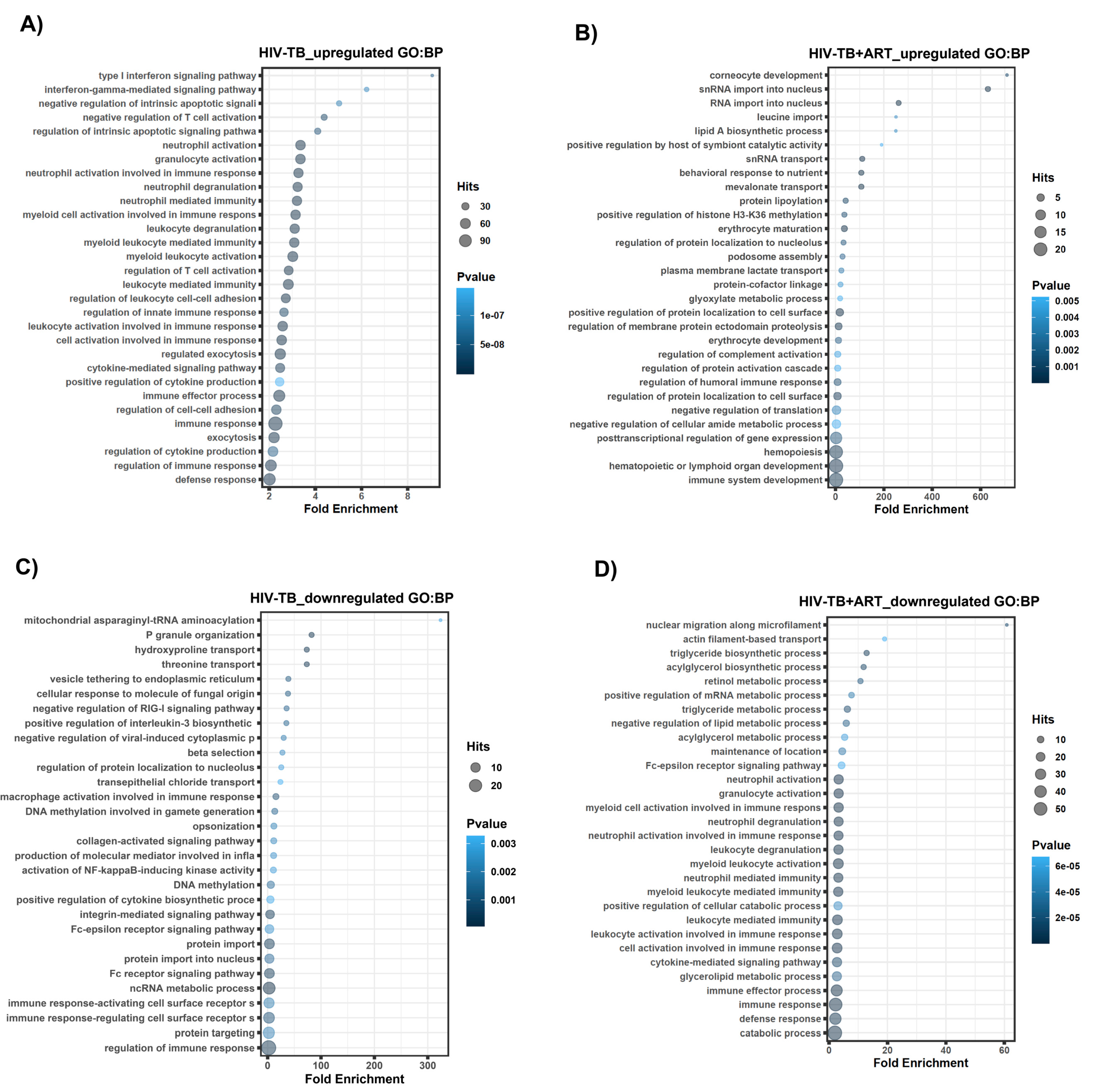
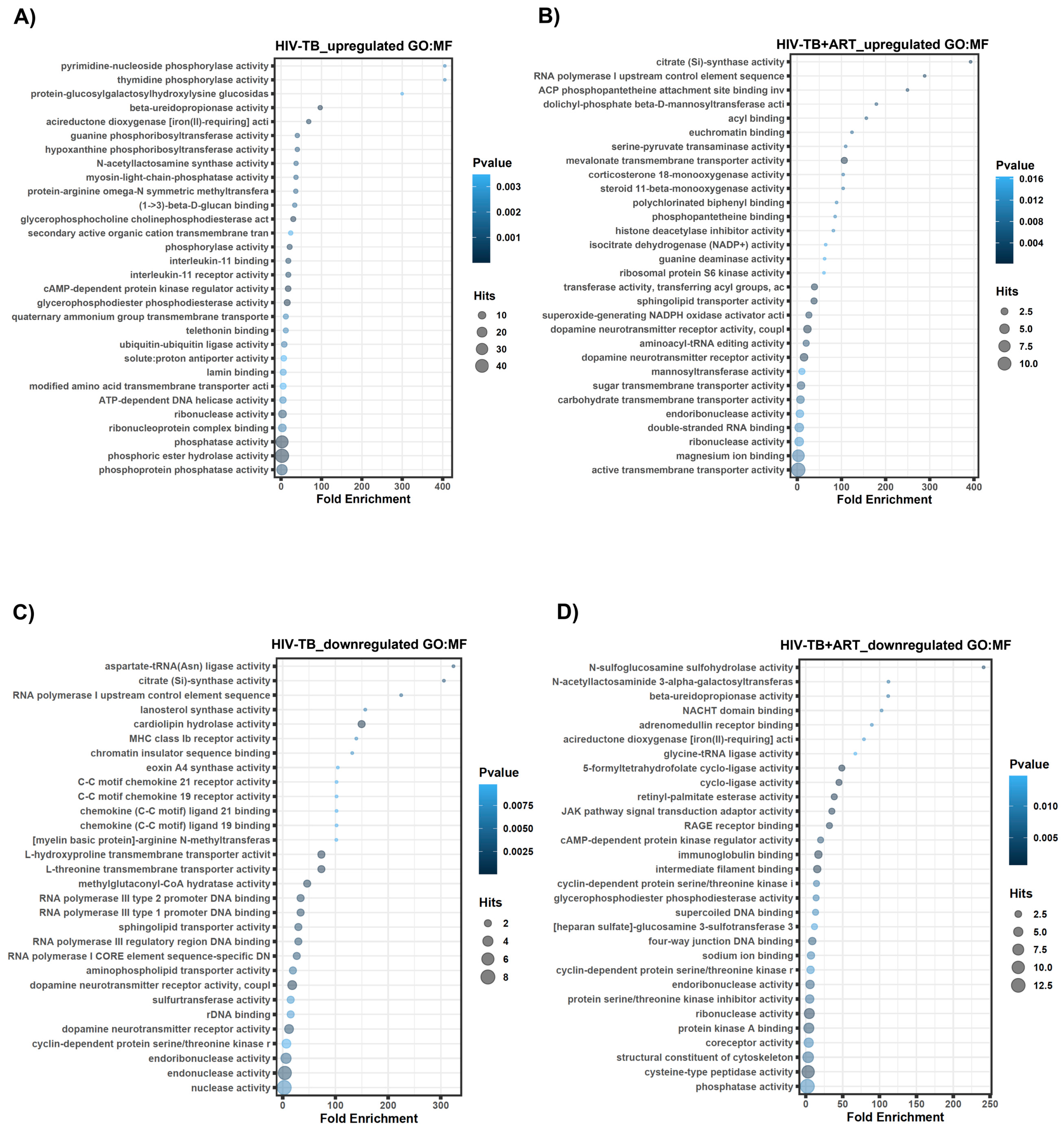
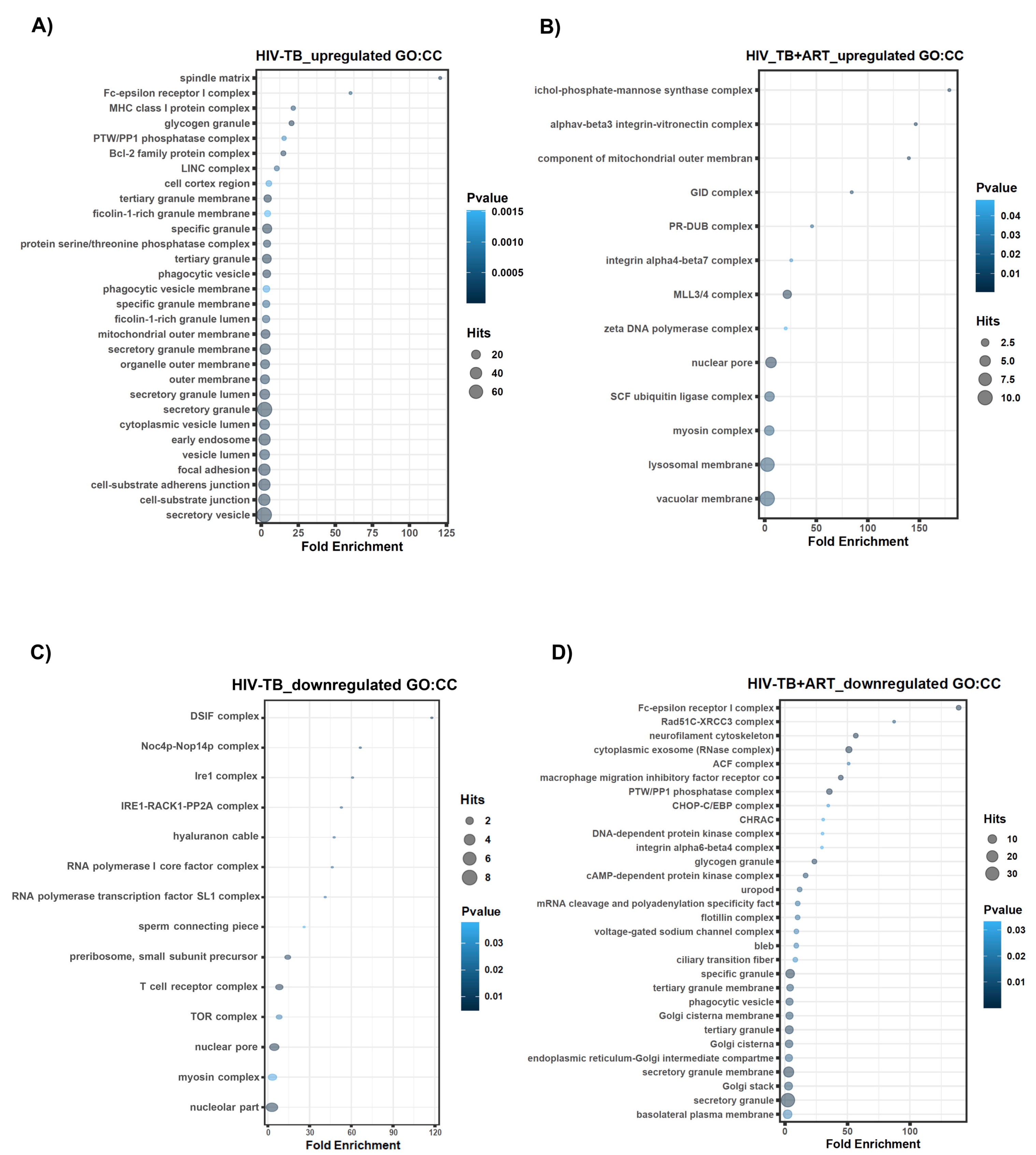
2.2. Assignment of Biological Meaning to lncRNA Genes and Predicting Their Potential Role in Various Cellular Functions
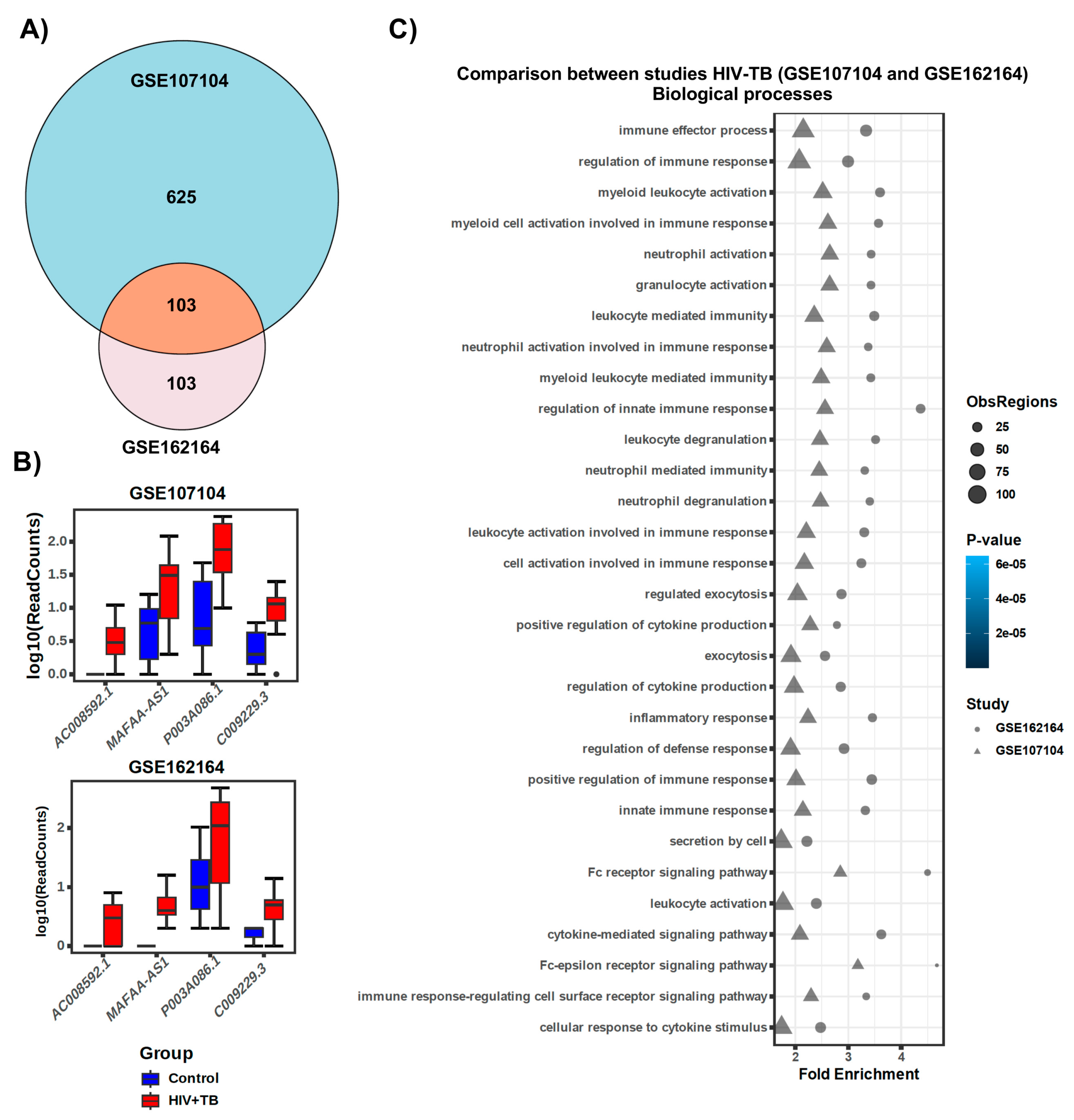
2.3. Comparison of lncRNA Gene Expression Analyses among HIV-TB Cohorts between Datasets
3. Discussion
4. Materials and Methods
4.1. Transcriptomic Data
4.2. Transcriptome Assembly and Differential Expression
4.3. Read Count Normalization from Genomic Datasets
4.4. Annotation and Composition of Non-Coding Genes
4.5. Genomic Regions Enrichment of Annotations Tool (GREAT) Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Global Tuberculosis Report 2023; WHO: Geneva, Switzerland, 2023.
- Houben, R.M.; Dodd, P.J. The Global Burden of Latent Tuberculosis Infection: A Re-estimation Using Mathematical Modelling. PLoS Med. 2016, 13, e1002152. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, L. Revisiting the role of the granuloma in tuberculosis. Nat. Rev. Immunol. 2012, 12, 352–366. [Google Scholar] [CrossRef]
- Cronan, M.R. In the Thick of It: Formation of the Tuberculous Granuloma and Its Effects on Host and Therapeutic Responses. Front. Immunol. 2022, 13, 820134. [Google Scholar] [CrossRef] [PubMed]
- Ehlers, S.; Schaible, U.E. The granuloma in tuberculosis: Dynamics of a host-pathogen collusion. Front. Immunol. 2012, 3, 411. [Google Scholar] [CrossRef] [PubMed]
- Keane, J.; Gershon, S.; Wise, R.P.; Mirabile-Levens, E.; Kasznica, J.; Schwieterman, W.D.; Siegel, J.N.; Braun, M.M. Tuberculosis associated with infliximab, a tumor necrosis factor alpha-neutralizing agent. N. Engl. J. Med. 2001, 345, 1098–1104. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yang, S.; Sun, G.; Tang, X.; Lu, S.; Neyrolles, O.; Gao, Q. Comparative miRNA expression profiles in individuals with latent and active tuberculosis. PLoS ONE 2011, 6, e25832. [Google Scholar] [CrossRef] [PubMed]
- Ai, J.W.; Ruan, Q.L.; Liu, Q.H.; Zhang, W.H. Updates on the risk factors for latent tuberculosis reactivation and their managements. Emerg. Microbes Infect. 2016, 5, e10. [Google Scholar] [CrossRef]
- Landry, J.; Menzies, D. Preventive chemotherapy. Where has it got us? Where to go next? Int. J. Tuberc. Lung Dis. 2008, 12, 1352–1364. [Google Scholar] [PubMed]
- Horsburgh, C.R., Jr.; Rubin, E.J. Clinical practice. Latent tuberculosis infection in the United States. N. Engl. J. Med. 2011, 364, 1441–1448. [Google Scholar] [CrossRef]
- WHO. HIV Data and Statistics; WHO: Geneva, Switzerland, 2023.
- Saharia, K.K.; Koup, R.A. T cell susceptibility to HIV influences outcome of opportunistic infections. Cell 2013, 155, 505–514. [Google Scholar] [CrossRef][Green Version]
- Pawlowski, A.; Jansson, M.; Skold, M.; Rottenberg, M.E.; Kallenius, G. Tuberculosis and HIV co-infection. PLoS Pathog. 2012, 8, e1002464. [Google Scholar] [CrossRef] [PubMed]
- Bell, L.C.K.; Noursadeghi, M. Pathogenesis of HIV-1 and Mycobacterium tuberculosis co-infection. Nat. Rev. Microbiol. 2018, 16, 80–90. [Google Scholar] [CrossRef] [PubMed]
- North, R.J.; Jung, Y.J. Immunity to tuberculosis. Annu. Rev. Immunol. 2004, 22, 599–623. [Google Scholar] [CrossRef] [PubMed]
- Cooper, A.M. Cell-mediated immune responses in tuberculosis. Annu. Rev. Immunol. 2009, 27, 393–422. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, S.H. Tuberculosis vaccines: Time to think about the next generation. Semin. Immunol. 2013, 25, 172–181. [Google Scholar] [CrossRef]
- Diedrich, C.R.; Flynn, J.L. HIV-1/mycobacterium tuberculosis coinfection immunology: How does HIV-1 exacerbate tuberculosis? Infect. Immun. 2011, 79, 1407–1417. [Google Scholar] [CrossRef] [PubMed]
- Collins, K.R.; Quinones-Mateu, M.E.; Toossi, Z.; Arts, E.J. Impact of tuberculosis on HIV-1 replication, diversity, and disease progression. AIDS Rev. 2002, 4, 165–176. [Google Scholar] [PubMed]
- Getahun, H.; Gunneberg, C.; Granich, R.; Nunn, P. HIV infection-associated tuberculosis: The epidemiology and the response. Clin. Infect. Dis. 2010, 50 (Suppl. 3), S201–S207. [Google Scholar] [CrossRef] [PubMed]
- Agliano, F.; Rathinam, V.A.; Medvedev, A.E.; Vanaja, S.K.; Vella, A.T. Long Noncoding RNAs in Host-Pathogen Interactions. Trends Immunol. 2019, 40, 492–510. [Google Scholar] [CrossRef]
- Wen, Y.; Chen, H.; Luo, F.; Zhou, H.; Li, Z. Roles of long noncoding RNAs in bacterial infection. Life Sci. 2020, 263, 118579. [Google Scholar] [CrossRef]
- Schmerer, N.; Schulte, L.N. Long noncoding RNAs in bacterial infection. Wiley Interdiscip. Rev. RNA 2021, 12, e1664. [Google Scholar] [CrossRef]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Huang, F.; Fu, J.; Dou, B.; Xu, B.; Miao, L.; Liu, W.; Yang, X.; Tan, C.; Chen, H.; et al. Differential transcription profiles of long non-coding RNAs in primary human brain microvascular endothelial cells in response to meningitic Escherichia coli. Sci. Rep. 2016, 6, 38903. [Google Scholar] [CrossRef] [PubMed]
- Imamura, K.; Takaya, A.; Ishida, Y.I.; Fukuoka, Y.; Taya, T.; Nakaki, R.; Kakeda, M.; Imamachi, N.; Sato, A.; Yamada, T.; et al. Diminished nuclear RNA decay upon Salmonella infection upregulates antibacterial noncoding RNAs. EMBO J. 2018, 37. [Google Scholar] [CrossRef] [PubMed]
- Yi, Z.; Li, J.; Gao, K.; Fu, Y. Identifcation of differentially expressed long non-coding RNAs in CD4+ T cells response to latent tuberculosis infection. J. Infect. 2014, 69, 558–568. [Google Scholar] [CrossRef]
- Menard, K.L.; Haskins, B.E.; Colombo, A.P.; Denkers, E.Y. Toxoplasma gondii Manipulates Expression of Host Long Noncoding RNA during Intracellular Infection. Sci. Rep. 2018, 8, 15017. [Google Scholar] [CrossRef] [PubMed]
- Riege, K.; Holzer, M.; Klassert, T.E.; Barth, E.; Brauer, J.; Collatz, M.; Hufsky, F.; Mostajo, N.; Stock, M.; Vogel, B.; et al. Massive Effect on LncRNAs in Human Monocytes During Fungal and Bacterial Infections and in Response to Vitamins A and D. Sci. Rep. 2017, 7, 40598. [Google Scholar] [CrossRef] [PubMed]
- Ji, S.; Zhu, M.; Zhang, J.; Cai, Y.; Zhai, X.; Wang, D.; Li, G.; Su, S.; Zhou, J. Microarray analysis of lncRNA expression in rabies virus infected human neuroblastoma cells. Infect. Genet. Evol. 2019, 67, 88–100. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, C.; Qin, L.; Li, D.; Zhou, G.; Dang, D.; Chen, S.; Sun, T.; Zhang, R.; Wu, W.; et al. Characterization of Critical Functions of Long Non-Coding RNAs and mRNAs in Rhabdomyosarcoma Cells and Mouse Skeletal Muscle Infected by Enterovirus 71 Using RNA-Seq. Viruses 2018, 10, 556. [Google Scholar] [CrossRef]
- Liu, X.; Duan, X.; Holmes, J.A.; Li, W.; Lee, S.H.; Tu, Z.; Zhu, C.; Salloum, S.; Lidofsky, A.; Schaefer, E.A.; et al. A Long Noncoding RNA Regulates Hepatitis C Virus Infection Through Interferon Alpha-Inducible Protein 6. Hepatology 2019, 69, 1004–1019. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Ou, Q.; Liu, C.; Shi, L.; Zhao, C.; Xu, Y.; Kong, S.K.; Loo, J.F.C.; Li, B.; Gu, D. Differential expression of long non-coding RNAs in patients with tuberculosis infection. Tuberculosis (Edinb) 2017, 107, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.L.; Wei, L.L.; Shi, L.Y.; Li, M.; Jiang, T.T.; Chen, J.; Liu, C.M.; Yang, S.; Tu, H.H.; Hu, Y.T.; et al. Screening and identification of lncRNAs as potential biomarkers for pulmonary tuberculosis. Sci. Rep. 2017, 7, 16751. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.; Haleyurgirisetty, M.; Ragupathy, V.; Wang, X.; Lee, S.; Hewlett, I.; Devadas, K. Differentially expressed host long intergenic noncoding RNA and mRNA in HIV-1 and HIV-2 infection. Sci. Rep. 2018, 8, 2546. [Google Scholar] [CrossRef] [PubMed]
- Trypsteen, W.; Mohammadi, P.; Van Hecke, C.; Mestdagh, P.; Lefever, S.; Saeys, Y.; De Bleser, P.; Vandesompele, J.; Ciuffi, A.; Vandekerckhove, L.; et al. Differential expression of lncRNAs during the HIV replication cycle: An underestimated layer in the HIV-host interplay. Sci. Rep. 2016, 6, 36111. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.; Du, P.; Nakanjako, D.; Hermans, S.; Briggs, J.; Nakiyingi, L.; Ellner, J.J.; Manabe, Y.C.; Salgame, P. Tuberculosis in advanced HIV infection is associated with increased expression of IFNgamma and its downstream targets. BMC Infect. Dis. 2018, 18, 220. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, V.; Queiroz, A.T.L.; Sangle, S.; Kagal, A.; Salvi, S.; Gupta, A.; Ellner, J.; Kadam, D.; Rolla, V.C.; Andrade, B.B.; et al. A Two-Gene Signature for Tuberculosis Diagnosis in Persons with Advanced HIV. Front. Immunol. 2021, 12, 631165. [Google Scholar] [CrossRef] [PubMed]
- Kacprzak, A.; Oniszh, K.; Podlasin, R.; Marczak, M.; Cielniak, I.; Augustynowicz-Kopec, E.; Tomkowski, W.; Szturmowicz, M. Atypical Pulmonary Tuberculosis as the First Manifestation of Advanced HIV Disease-Diagnostic Difficulties. Diagnostics 2022, 12, 1886. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, Q.; Lin, S.; Lai, J.; Lin, J.; Ao, W.; Han, X.; Ye, H. Meta-Analysis of Peripheral Blood Transcriptome Datasets Reveals a Biomarker Panel for Tuberculosis in Patients Infected with HIV. Front. Cell Infect. Microbiol. 2021, 11, 585919. [Google Scholar] [CrossRef]
- Ma, Y.; Xu, Y.; Cao, X.; Chen, X.; Zhong, Y. Diagnostic value of interferon-gamma release assay in HIV-infected individuals complicated with active tuberculosis: A systematic review and meta-analysis. Epidemiol. Infect. 2021, 149, e204. [Google Scholar] [CrossRef]
- Anderson, S.T.; Kaforou, M.; Brent, A.J.; Wright, V.J.; Banwell, C.M.; Chagaluka, G.; Crampin, A.C.; Dockrell, H.M.; French, N.; Hamilton, M.S.; et al. Diagnosis of childhood tuberculosis and host RNA expression in Africa. N. Engl. J. Med. 2014, 370, 1712–1723. [Google Scholar] [CrossRef] [PubMed]
- Dawany, N.; Showe, L.C.; Kossenkov, A.V.; Chang, C.; Ive, P.; Conradie, F.; Stevens, W.; Sanne, I.; Azzoni, L.; Montaner, L.J. Identification of a 251 gene expression signature that can accurately detect M. tuberculosis in patients with and without HIV co-infection. PLoS ONE 2014, 9, e89925. [Google Scholar] [CrossRef] [PubMed]
- Darboe, F.; Mbandi, S.K.; Thompson, E.G.; Fisher, M.; Rodo, M.; van Rooyen, M.; Filander, E.; Bilek, N.; Mabwe, S.; Hatherill, M.; et al. Diagnostic performance of an optimized transcriptomic signature of risk of tuberculosis in cryopreserved peripheral blood mononuclear cells. Tuberculosis (Edinb) 2018, 108, 124–126. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Shi, W.; Wang, F.; Shen, X.; Qi, J.; Cong, H.; Yuan, J.; Shi, L.; Zhu, B.; Luo, X.; et al. Long non-coding RNA HULC as a novel serum biomarker for diagnosis and prognosis prediction of gastric cancer. Oncotarget 2016, 7, 51763–51772. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Chen, Q.; Chen, Y.; Ge, X.; Leng, W.; Tang, Q.; Ren, M.; Chen, L.; Yuan, D.; Zhang, Y.; et al. The lncRNA MALAT1 is a novel biomarker for gastric cancer metastasis. Oncotarget 2016, 7, 56209–56218. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Huang, Y.; Chen, X.; Chen, T.; Hu, W.; Hou, W.; Zhang, Q.; Xiong, Y. Transcriptomic study reveals changes of lncRNAs in PBMCs from HIV-1 patients before and after ART. Sci. Rep. 2023, 13, 22493. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yang, J.; Wang, J.; Wen, Q.; Wang, H.; He, J.; Hu, S.; He, W.; Du, X.; Liu, S.; et al. Microarray analysis of long noncoding RNA and mRNA expression profiles in human macrophages infected with Mycobacterium tuberculosis. Sci. Rep. 2016, 6, 38963. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, C.; Xu, Y. Long Non-coding RNAs in Tuberculosis: From Immunity to Biomarkers. Front. Microbiol. 2022, 13, 883513. [Google Scholar] [CrossRef]
- Yu, Z.; Wang, Y.; Deng, J.; Liu, D.; Zhang, L.; Shao, H.; Wang, Z.; Zhu, W.; Zhao, C.; Ke, Q. Long non-coding RNA COL4A2-AS1 facilitates cell proliferation and glycolysis of colorectal cancer cells via miR-20b-5p/hypoxia inducible factor 1 alpha subunit axis. Bioengineered 2021, 12, 6251–6263. [Google Scholar] [CrossRef]
- Zhan, Y.; Guan, X.Y.; Li, Y. MAFA-AS1, a long non-coding RNA, predicts for poor survival of hepatocellular carcinoma. Transl. Cancer Res. 2020, 9, 2449–2459. [Google Scholar] [CrossRef]
- Chung, I.H.; Lu, P.H.; Lin, Y.H.; Tsai, M.M.; Lin, Y.W.; Yeh, C.T.; Lin, K.H. The long non-coding RNA LINC01013 enhances invasion of human anaplastic large-cell lymphoma. Sci. Rep. 2017, 7, 295. [Google Scholar] [CrossRef]
- Sasindran, S.J.; Torrelles, J.B. Mycobacterium Tuberculosis Infection and Inflammation: What is Beneficial for the Host and for the Bacterium? Front. Microbiol. 2011, 2, 2. [Google Scholar] [CrossRef]
- Kaufmann, S.H.; Dorhoi, A. Inflammation in tuberculosis: Interactions, imbalances and interventions. Curr. Opin. Immunol. 2013, 25, 441–449. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef]
- McLean, C.Y.; Bristor, D.; Hiller, M.; Clarke, S.L.; Schaar, B.T.; Lowe, C.B.; Wenger, A.M.; Bejerano, G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010, 28, 495–501. [Google Scholar] [CrossRef]
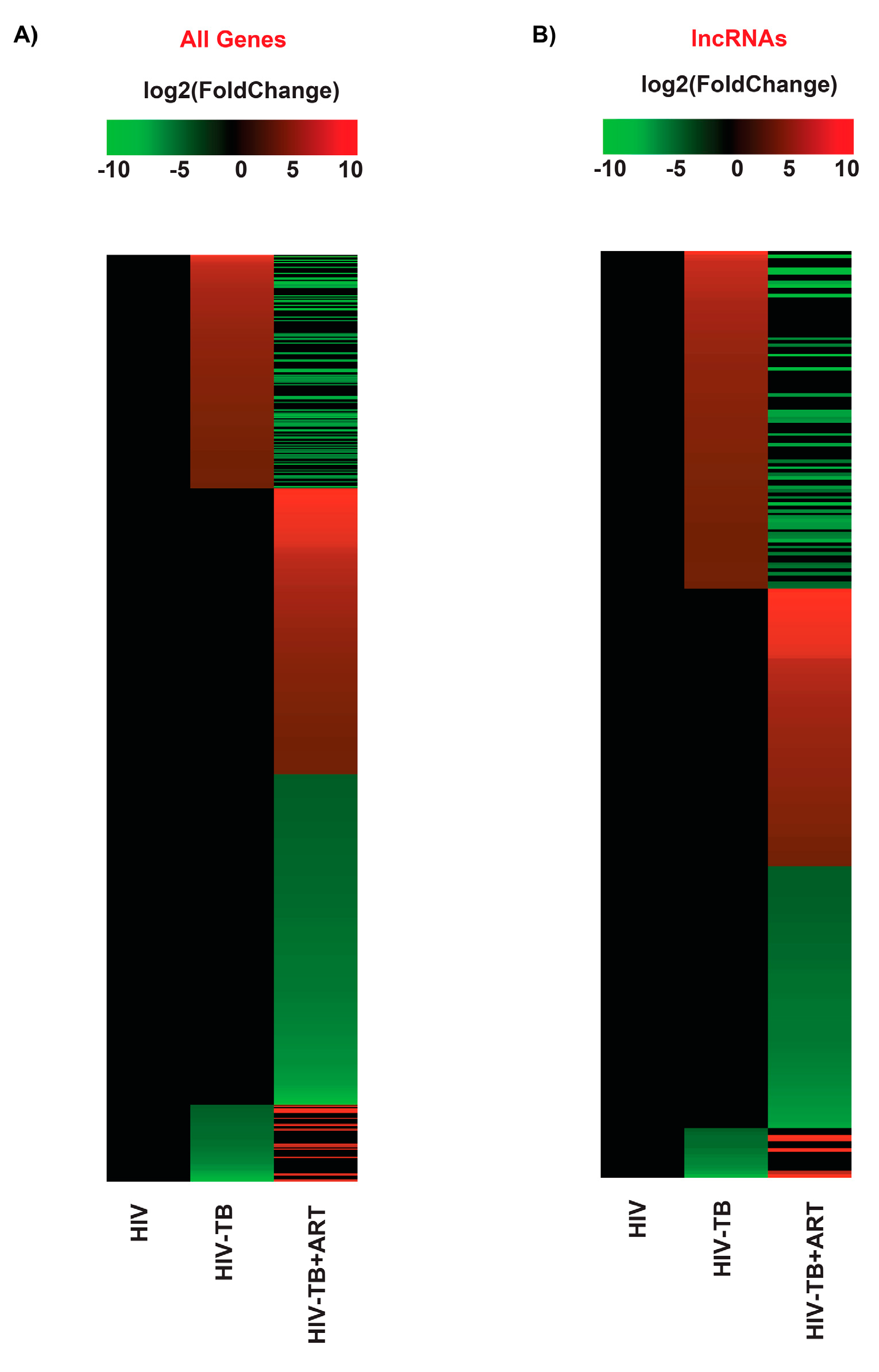
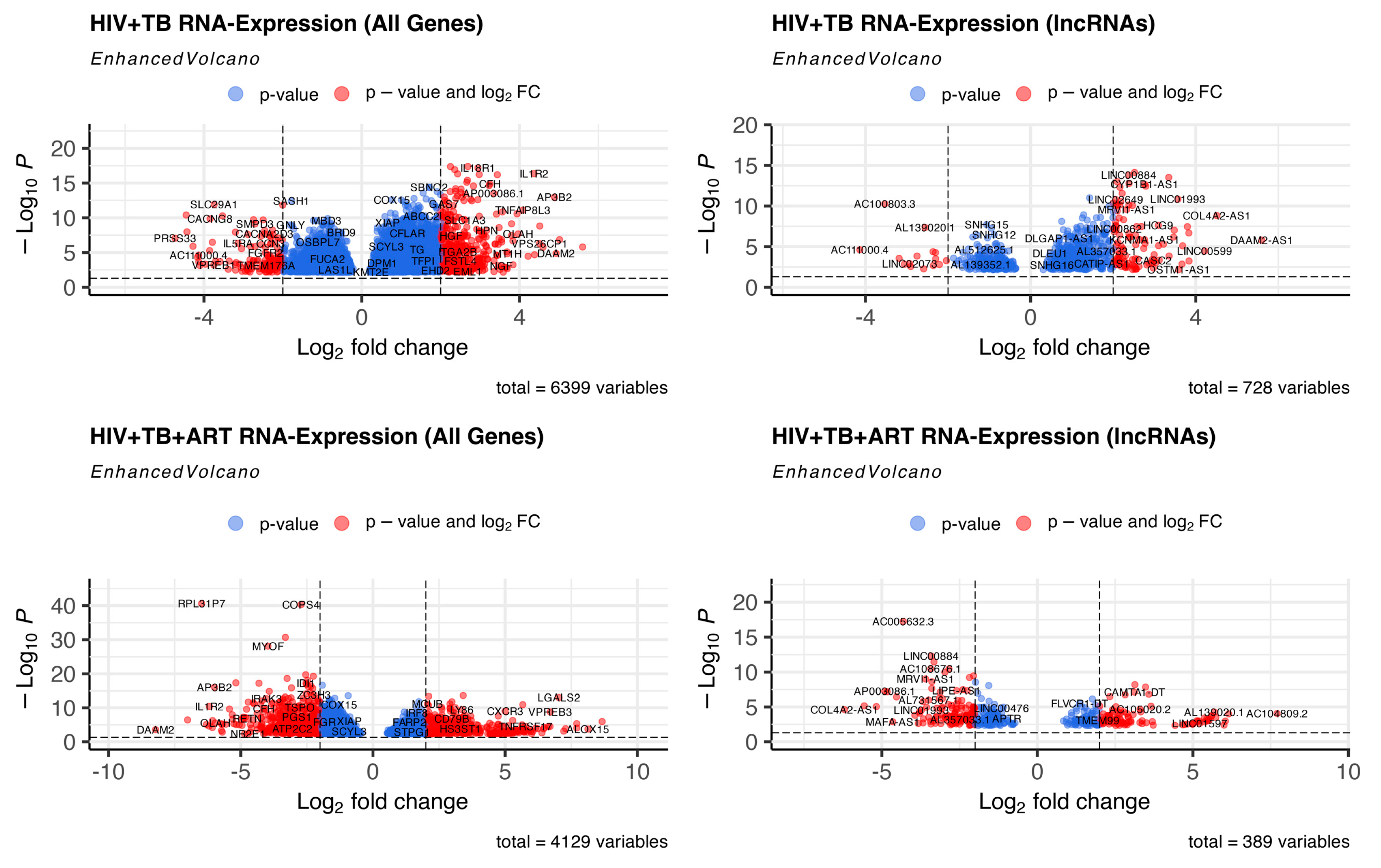
| Up-Regulated Genes | Down-Regulated Genes | |||
|---|---|---|---|---|
| S. No. | Gene ID | log2(Fold Change) | Gene ID | log2(Fold Change) |
| 1 | DAAM2-AS1 | 5.599065321 | AC111000.4 | −4.13637139 |
| 2 | COL4A2-AS1 | 4.51193714 | AC100803.3 | −3.530569923 |
| 3 | LINC00599 | 4.216277843 | AC016168.2 | −3.184558686 |
| 4 | AC008592.1 | 3.835650792 | AC245100.7 | −3.054170863 |
| 5 | CLRN1-AS1 | 3.831798567 | LINC02073 | −2.927654567 |
| 6 | AC016831.5 | 3.797296573 | AC005277.2 | −2.903194655 |
| 7 | MAFA-AS1 | 3.681848495 | AL645608.7 | −2.750602911 |
| 8 | FAM83A-AS1 | 3.666535159 | LINC01996 | −2.589747063 |
| 9 | OSTM1-AS1 | 3.58676319 | AL139020.1 | −2.555607503 |
| 10 | LINC01993 | 3.566680578 | AC025279.1 | −2.362957614 |
| 11 | AC079210.1 | 3.479032992 | AL109659.2 | −2.356363243 |
| 12 | AC021594.2 | 3.461800659 | LINC01013 | −2.303911575 |
| 13 | AC010980.2 | 3.353776771 | AC007922.2 | −2.299321335 |
| 14 | AP003086.1 | 3.346010527 | AC008966.2 | −2.209235807 |
| 15 | AC009229.3 | 3.21655715 | AC069503.1 | −2.040937496 |
| Up-Regulated Genes | Down-Regulated Genes | |||
|---|---|---|---|---|
| S. No | Gene ID | log2(Fold Change) | Gene ID | log2(Fold Change) |
| 1 | AC104809.2 | 7.706535903 | COL4A2-AS1 | −6.199159707 |
| 2 | AC092068.2 | 6.151251928 | AC079210.1 | −5.566015767 |
| 3 | AC007922.2 | 6.043128911 | AC016831.5 | −5.175141591 |
| 4 | AC111000.4 | 6.002785695 | AP003086.1 | −4.904392854 |
| 5 | AC012511.1 | 5.735772764 | LINC01482 | −4.827203602 |
| 6 | AL139020.1 | 5.689201588 | MAFA-AS1 | −4.647752828 |
| 7 | MIR600HG | 5.604156733 | LINC01093 | −4.528892547 |
| 8 | AL022329.2 | 5.59066406 | AC005632.3 | −4.312134927 |
| 9 | AC010998.2 | 5.514527786 | AL133353.1 | −3.936394158 |
| 10 | LINC01730 | 5.513093748 | LINC01579 | −3.8268448 |
| 11 | CLYBL-AS1 | 5.451063622 | AC103740.1 | −3.78711058 |
| 12 | AL121929.3 | 5.446269531 | SCN1A-AS1 | −3.780568776 |
| 13 | LINC01013 | 5.37460914 | LINC01993 | −3.726868434 |
| 14 | AC110285.2 | 5.371066891 | AC093278.2 | −3.677140638 |
| 15 | LINC01597 | 5.225006302 | AL078604.2 | −3.675382142 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reid, V.A.; Ramos, E.I.; Veerapandian, R.; Carmona, A.; Gadad, S.S.; Dhandayuthapani, S. Differential Expression of lncRNAs in HIV Patients with TB and HIV-TB with Anti-Retroviral Treatment. Non-Coding RNA 2024, 10, 40. https://doi.org/10.3390/ncrna10040040
Reid VA, Ramos EI, Veerapandian R, Carmona A, Gadad SS, Dhandayuthapani S. Differential Expression of lncRNAs in HIV Patients with TB and HIV-TB with Anti-Retroviral Treatment. Non-Coding RNA. 2024; 10(4):40. https://doi.org/10.3390/ncrna10040040
Chicago/Turabian StyleReid, Victoria A., Enrique I. Ramos, Raja Veerapandian, Areanna Carmona, Shrikanth S. Gadad, and Subramanian Dhandayuthapani. 2024. "Differential Expression of lncRNAs in HIV Patients with TB and HIV-TB with Anti-Retroviral Treatment" Non-Coding RNA 10, no. 4: 40. https://doi.org/10.3390/ncrna10040040
APA StyleReid, V. A., Ramos, E. I., Veerapandian, R., Carmona, A., Gadad, S. S., & Dhandayuthapani, S. (2024). Differential Expression of lncRNAs in HIV Patients with TB and HIV-TB with Anti-Retroviral Treatment. Non-Coding RNA, 10(4), 40. https://doi.org/10.3390/ncrna10040040








