Self-assembly and Hydrogelation Properties of Peptides Derived from Peptic Cleavage of Aggregation-prone Regions of Ovalbumin
Abstract
1. Introduction
2. Results and Discussion
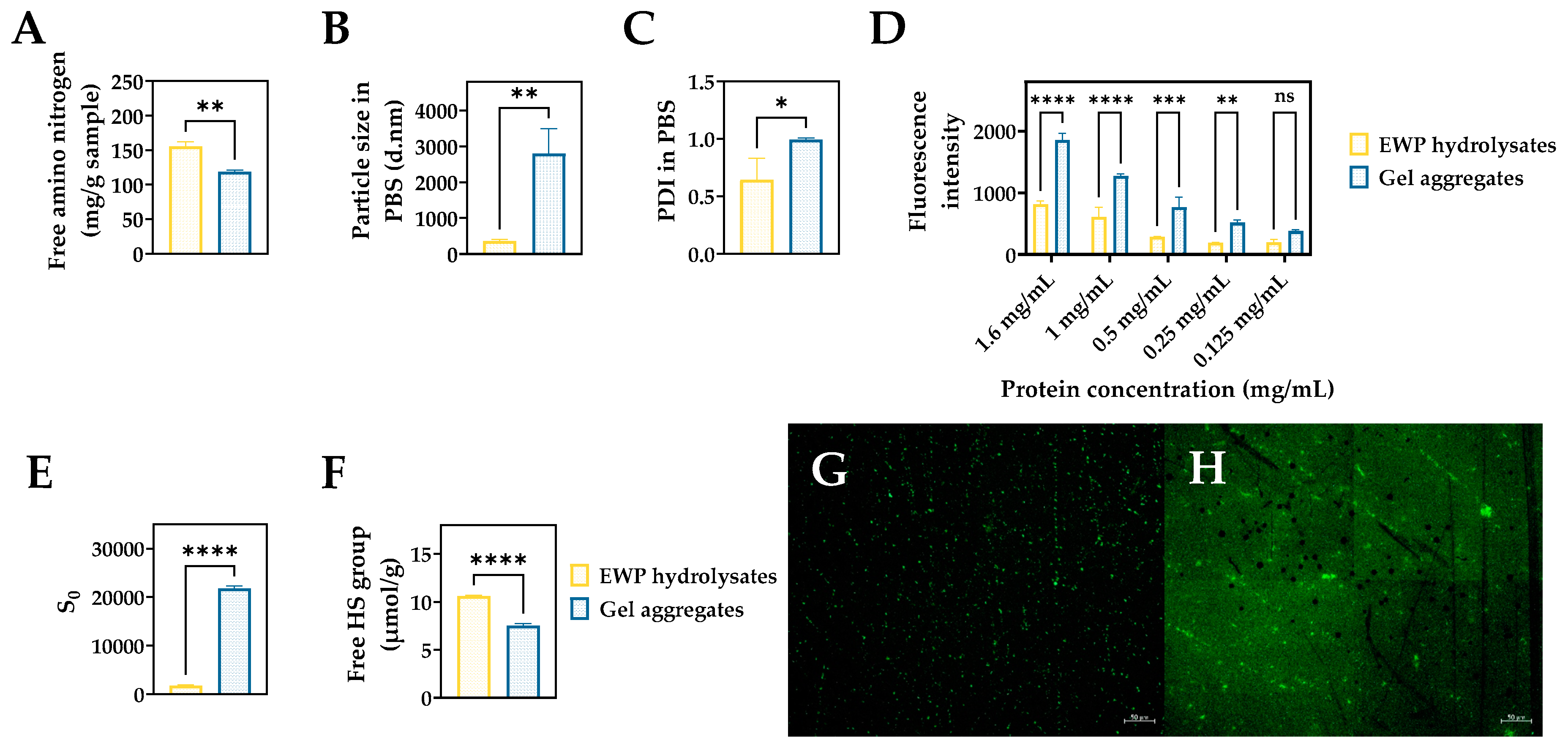
2.1. Physicochemical and Surface Properties of EWP Hydrolysate and Aggregate Samples
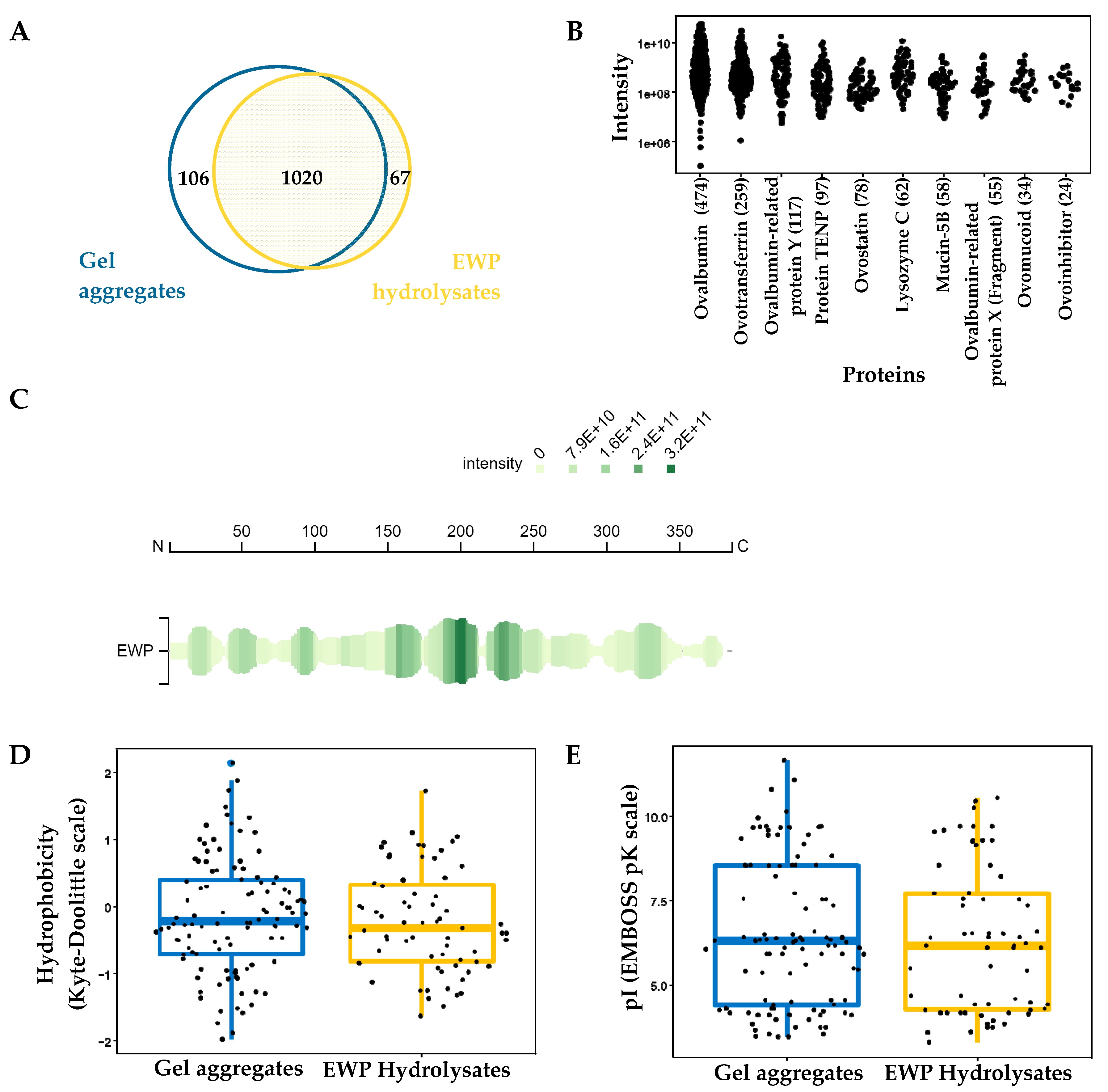
2.2. Peptidomic Profile of EWP Hydrolysate and Aggregate Samples
2.3. Predicted Aggregation Propensity of Identified Peptides
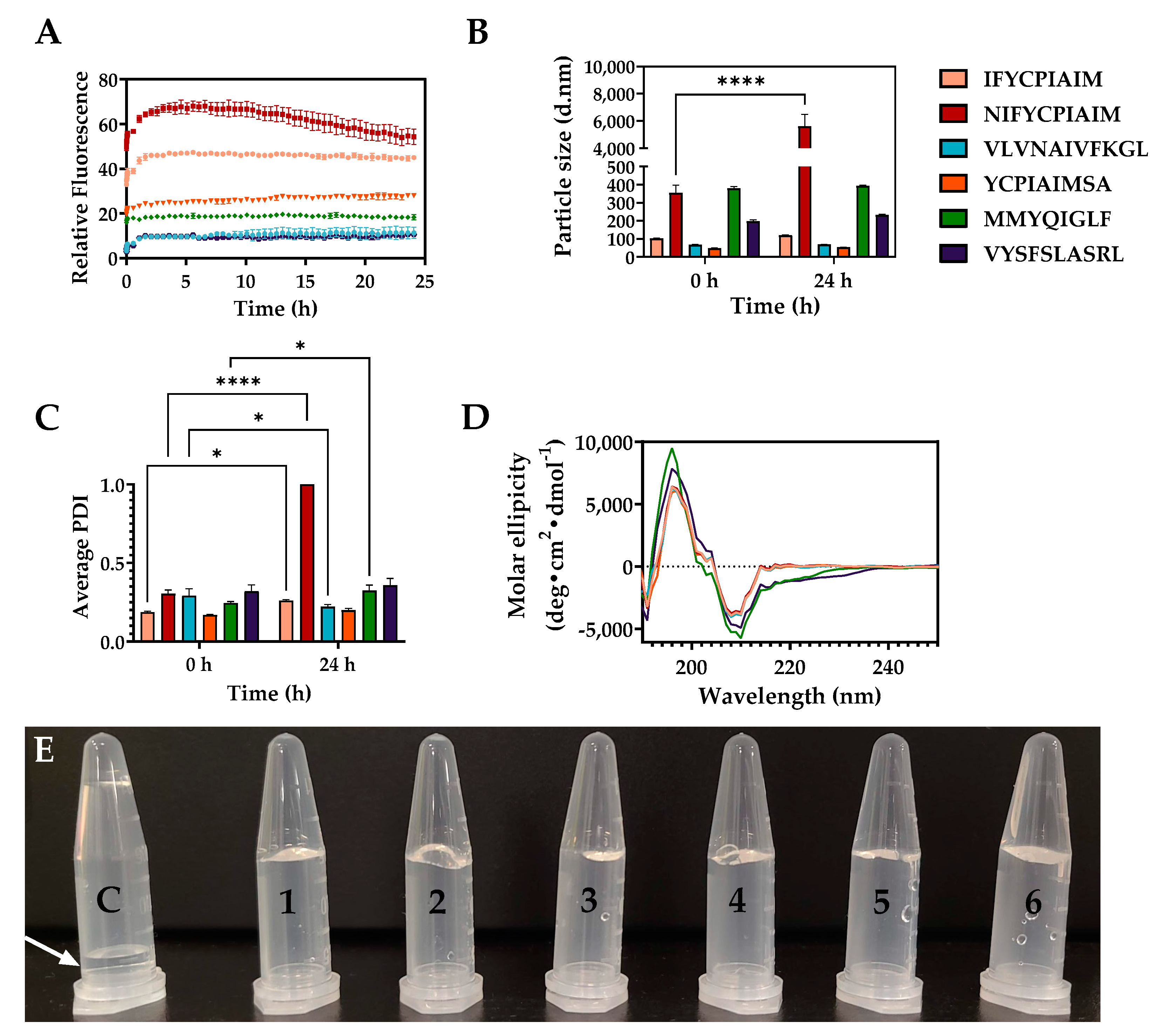
2.4. Self-Assembly Properties of the Aggregation-Prone Peptides
2.5. Microstructures of the Self-Assembling Ovalbumin Peptides
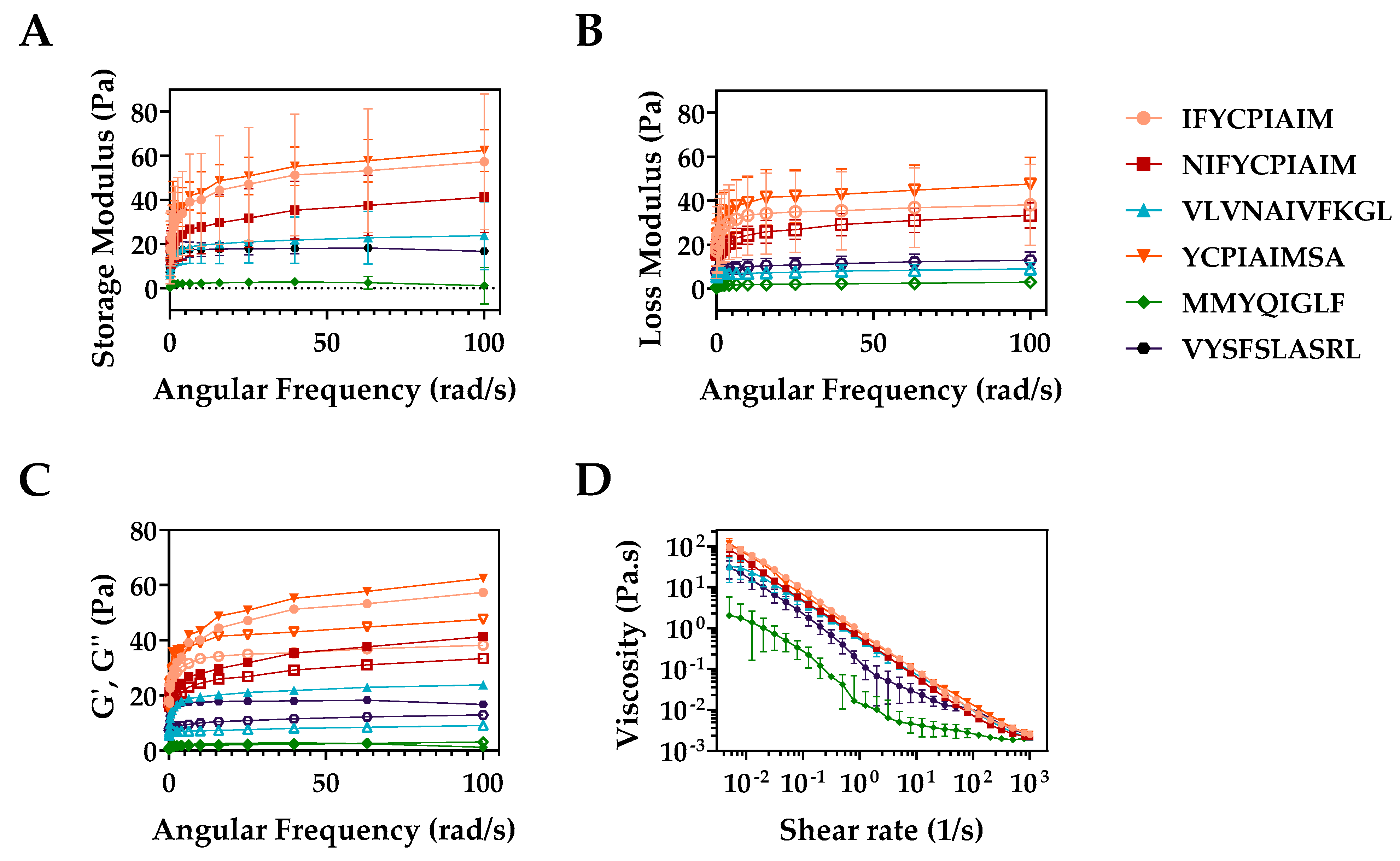
2.6. Mechanical Properties of the Peptide Hydrogels
3. Conclusions
4. Materials and Methods
4.1. Materials and Chemicals
4.2. Preparation of Egg White Protein (EWP) Hydrolysate and Aggregate Samples
4.3. Characterization of EWP Hydrolysate and Aggregate Samples
4.4. LC-MS/MS Analysis
4.5. Prediction of Aggregation Propensity of Ovalbumin-derived Peptides Using AGGRESCAN
4.6. Peptide Synthesis
4.7. Peptide Hydrogel Formation
4.8. Thioflavin T Assays
4.9. Dynamic Light Scattering
4.10. Circular Dichroism Spectroscopy
4.11. Transmission Electron Microscopy
4.12. Rheological Measurements
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chang, C.; Lahti, T.; Tanaka, T.; Nickerson, M.T. Egg proteins: Fractionation, bioactive peptides and allergenicity. J. Sci. Food Agric. 2018, 98, 5547–5558. [Google Scholar] [CrossRef] [PubMed]
- Bhat, Z.F.; Morton, J.D.; Bekhit, A.E.D.A.; Kumar, S.; Bhat, H.F. Effect of processing technologies on the digestibility of egg proteins. Compr. Rev. Food Sci. Food Saf. 2021, 20, 4703–4738. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Liang, R.; Xing, Q.; Lozano-Ojalvo, D. Can food processing produce hypoallergenic egg? J. Food Sci. 2020, 85, 2635–2644. [Google Scholar] [CrossRef]
- Mahdavi, S.; Amirsadeghi, A.; Jafari, A.; Niknezhad, S.V.; Bencherif, S.A. Avian egg: A multifaceted biomaterial for tissue engineering. Ind. Eng. Chem. Res. 2021, 60, 17348–17364. [Google Scholar] [CrossRef] [PubMed]
- Farjami, T.; Babaei, J.; Nau, F.; Dupont, D.; Madadlou, A. Effects of thermal, non-thermal and emulsification processes on the gastrointestinal digestibility of egg white proteins. Trends Food Sci. Technol. 2021, 107, 45–56. [Google Scholar] [CrossRef]
- Lv, X.; Huang, X.; Ma, B.; Chen, Y.; Batool, Z.; Fu, X.; Jin, Y. Modification methods and applications of egg protein gel properties: A review. Compr. Rev. Food Sci. Food Saf. 2022, 21, 2233–2252. [Google Scholar] [CrossRef]
- Pugliese, R.; Gelain, F.; Arnoldi, A.; Lammi, C. Chapter 6: Chemistry and functional roles of food protein hydrogels. In Food Proteins and Peptides: Emerging Biofunctions, Food and Biomaterial Applications; Udenigwe, C.C., Ed.; Royal Society of Chemistry: Cambridge, UK, 2021; Volume 27, pp. 157–172. [Google Scholar]
- Rathnapala, E.C.N.; Ahn, D.U.; Abeyrathne, S. Functional properties of ovotransferrin from chicken egg white and its derived peptides: A review. Food Sci. Biotechnol. 2021, 30, 619–630. [Google Scholar] [CrossRef]
- Benedé, S.; Molina, E. Chicken egg proteins and derived peptides with antioxidant properties. Foods 2020, 9, 735. [Google Scholar] [CrossRef]
- Mildenberger, J.; Remm, M.; Atanassova, M. Self-assembly potential of bioactive peptides from norwegian sea cucumber parastichopus tremulus for development of functional hydrogels. LWT 2021, 148, 111678. [Google Scholar] [CrossRef]
- de Leon Rodriguez, L.M.; Hemar, Y. Prospecting the applications and discovery of peptide hydrogels in food. Trends Food Sci. Technol. 2020, 104, 37–48. [Google Scholar] [CrossRef]
- Mondal, S.; Das, S.; Nandi, A.K. A review on recent advances in polymer and peptide hydrogels. Soft Matter 2020, 16, 1404. [Google Scholar] [CrossRef] [PubMed]
- Raza, F.; Zhu, Y.; Chen, L.; You, X.; Zhang, J.; Khan, A.; Khan, M.W.; Hasnat, M.; Zafar, H.; Wu, J.; et al. Paclitaxel-loaded pH responsive hydrogel based on self-assembled peptides for tumor targeting. Biomater. Sci. 2019, 7, 2023–2036. [Google Scholar] [CrossRef]
- Lammi, C.; Bollati, C.; Gelain, F.; Arnoldi, A.; Pugliese, R. Enhancement of the stability and anti-DPPIV activity of hempseed hydrolysates through self-assembling peptide-based hydrogels. Front. Chem. 2019, 7, 670. [Google Scholar] [CrossRef]
- Pramanik, B.; Ahmed, S. Peptide-based low molecular weight photosensitive supramolecular gelators. Gels 2022, 8, 533. [Google Scholar] [CrossRef] [PubMed]
- Nandi, N.; Gayen, K.; Ghosh, S.; Bhunia, D.; Kirkham, S.; Sen, S.K.; Ghosh, S.; Hamley, I.W.; Banerjee, A. Amphiphilic peptide-based supramolecular, noncytotoxic, stimuli-responsive hydrogels with antibacterial activity. Biomacromolecules 2017, 18, 3621–3629. [Google Scholar] [CrossRef] [PubMed]
- Pugliese, R.; Fontana, F.; Marchini, A.; Gelain, F. Branched peptides integrate into self-assembled nanostructures and enhance biomechanics of peptidic hydrogels. Acta Biomater. 2018, 66, 258–271. [Google Scholar] [CrossRef]
- Omana, D.A.; Liang, Y.; Kav, N.N.V.; Wu, J. Proteomic analysis of egg white proteins during storage. Proteomics 2011, 11, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Wan, M.; Huang, Z.; Yang, X.; Chen, Q.; Chen, L.; Liang, S.; Zeng, Q.; Zhang, R.; Dong, L.; Su, D. Fabrication and interaction mechanism of ovalbumin-based nanocarriers for metallic ion encapsulation. Int. J. Food Sci. Technol. 2022, 57, 643–652. [Google Scholar] [CrossRef]
- Hassan, N.; Messina, P.V.; Dodero, V.I.; Ruso, J.M. Rheological properties of ovalbumin hydrogels as affected by surfactants addition. Int. J. Biol. Macromol. 2011, 48, 495–500. [Google Scholar] [CrossRef]
- Yu, S.; Hu, J.; Pan, X.; Yao, P.; Jiang, M. Stable and pH-sensitive nanogels prepared by self-assembly of chitosan and ovalbumin. Langmuir 2006, 22, 2754–2759. [Google Scholar] [CrossRef]
- Kamalov, M.; Kählig, H.; Rentenberger, C.; Müllner, A.R.M.; Peterlik, H.; Becker, C.F.W. Ovalbumin epitope SIINFEKL self-assembles into a supramolecular hydrogel. Sci. Rep. 2019, 9, 2696. [Google Scholar] [CrossRef]
- Malmos, K.G.; Blancas-Mejia, L.M.; Weber, B.; Buchner, J.; Ramirez-Alvarado, M.; Naiki, H.; Otzen, D. ThT 101: A primer on the use of thioflavin t to investigate amyloid formation. Amyloid 2017, 24, 1–16. [Google Scholar] [CrossRef]
- Wang, Q.; Jiang, J.; Xiong, Y.L. Genipin-aided protein cross-linking to modify structural and rheological properties of emulsion-filled hempseed protein hydrogels. J. Agric. Food Chem. 2019, 67, 12895–12903. [Google Scholar] [CrossRef]
- Pugliese, R.; Bartolomei, M.; Bollati, C.; Boschin, G.; Arnoldi, A.; Lammi, C. Gel-forming of self-assembling peptides functionalized with food bioactive motifs modulate DPP-IV and ACE inhibitory activity in human intestinal caco-2 cells. Biomedicines 2022, 10, 330. [Google Scholar] [CrossRef]
- Sun, X.; Acquah, C.; Gazme, B.; Boachie, R.T.; Nwachukwu, I.D.; Udenigwe, C.C. Mechanisms of plastein formation influence the IGE-binding activity of egg white protein hydrolysates after simulated static digestion. Food Chem. 2021, 345, 128783. [Google Scholar] [CrossRef]
- Shen, W.; Lammertink, R.G.H.; Sakata, J.K.; Kornfield, J.A.; Tirrell, D.A. Assembly of an artificial protein hydrogel through leucine zipper aggregation and bisulfide bond formation. Macromolecules 2005, 38, 3909–3916. [Google Scholar] [CrossRef]
- Mirzaei, M.; Mirdamadi, S.; Safavi, M.; Soleymanzadeh, N. The stability of antioxidant and ACE-inhibitory peptides as influenced by peptide sequences. LWT 2020, 130, 109710. [Google Scholar] [CrossRef]
- Conchillo-Solé, O.; de Groot, N.S.; Avilés, F.X.; Vendrell, J.; Daura, X.; Ventura, S. AGGRESCAN: A server for the prediction and evaluation of “hot spots” of aggregation in polypeptides. BMC Bioinform. 2007, 8, 65. [Google Scholar] [CrossRef] [PubMed]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Osorio, D.; Rondon-Villarreal, P.; Torres, R. Peptides: A package for data mining of antimicrobial peptides. The R Journal 2015, 7, 4–14. [Google Scholar] [CrossRef]
- Micsonai, A.; Wien, F.; Bulyáki, É.; Kun, J.; Moussong, É.; Lee, Y.H.; Goto, Y.; Réfrégiers, M.; Kardos, J. BeStSel: A web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucleic Acids Res. 2018, 46, W315. [Google Scholar] [CrossRef] [PubMed]
- LeVine, H. Quantification of β-sheet amyloid fibril structures with thioflavin t. Methods Enzymol. 1999, 309, 274–284. [Google Scholar]
- Braun, G.A.; Ary, B.E.; Dear, A.J.; Rohn, M.C.H.; Payson, A.M.; Lee, D.S.M.; Parry, R.C.; Friedman, C.; Knowles, T.P.J.; Linse, S.; et al. On the mechanism of self-assembly by a hydrogel-forming peptide. Biomacromolecules 2020, 21, 4781–4794. [Google Scholar] [CrossRef]
- Yuan, C.; Levin, A.; Chen, W.; Xing, R.; Zou, Q.; Herling, T.W.; Challa, P.K.; Knowles, T.P.J.; Yan, X. Nucleation and growth of amino acid and peptide supramolecular polymers through liquid–liquid phase separation. Angew. Chem. 2019, 131, 18284–18291. [Google Scholar] [CrossRef]
- Gallo, E.; Diaferia, C.; Rosa, E.; Smaldone, G.; Morelli, G.; Accardo, A. Peptide-based hydrogels and nanogels for delivery of doxorubicin. Int. J. Nanomed. 2021, 16, 1617. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, L.; Yang, Z.; Zhao, X. Controlled release of paclitaxel from a self-assembling peptide hydrogel formed in situ and antitumor study in vitro. Int. J. Nanomed. 2011, 6, 2143. [Google Scholar] [CrossRef]
- Baek, K.; Noblett, A.D.; Ren, P.; Suggs, L.J. Design and characterization of nucleopeptides for hydrogel self-assembly. ACS Appl Bio. Mater. 2019, 2, 2812–2821. [Google Scholar] [CrossRef]
- Hule, R.A.; Nagarkar, R.P.; Hammouda, B.; Schneider, J.P.; Pochan, D.J. Dependence of self-assembled peptide hydrogel network structure on local fibril nanostructure. Macromolecules 2009, 42, 7137–7145. [Google Scholar] [CrossRef]
- Yan, C.; Pochan, D.J. Rheological properties of peptide-based hydrogels for biomedical and other applications. Chem. Soc. Rev. 2010, 39, 3528–3540. [Google Scholar] [CrossRef]
- Stojkov, G.; Niyazov, Z.; Picchioni, F.; Bose, R.K. Relationship between structure and rheology of hydrogels for various applications. Gels 2021, 7, 255. [Google Scholar] [CrossRef]
- Adams, D.J. Personal perspective on understanding low molecular weight gels. J. Am. Chem. Soc. 2022, 144, 11047–11053. [Google Scholar] [CrossRef] [PubMed]
- Sathaye, S.; Mbi, A.; Sonmez, C.; Chen, Y.; Blair, D.L.; Schneider, J.P.; Pochan, D.J. Rheology of peptide- and protein-based physical hydrogels: Are everyday measurements just scratching the surface? Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2015, 7, 34–68. [Google Scholar] [CrossRef] [PubMed]
- Draper, E.R.; Su, H.; Brasnett, C.; Poole, R.J.; Rogers, S.; Cui, H.; Seddon, A.; Adams, D.J. Opening a can of worm(-like micelle)s: The effect of temperature of solutions of functionalized dipeptides. Angew. Chem. Int. Ed. 2017, 56, 10467–10470. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, G.; Liang, Q.; Cai, W.; Zhang, Q. Rheological and microstructural properties of gelatin b/tara gum hydrogels: Effect of protein/polysaccharide ratio, pH and salt addition. LWT 2019, 103, 108–115. [Google Scholar] [CrossRef]
- Nielsen, P.M.; Petersen, D.; Dambmann, C. Improved method for determining food protein degree of hydrolysis. J. Food Sci. 2001, 66, 642–646. [Google Scholar] [CrossRef]
- Hong, G.P.; Avramenko, N.; Stone, A.; Abbott, D.; Classen, H.; Nickerson, M. Extractability and molecular modifications of gliadin and glutenin proteins withdrawn from different stages of a commercial ethanol fuel/distillers dried grains with solubles process using a wheat feedstock. Cereal Chem. 2012, 89, 276–283. [Google Scholar] [CrossRef]
- Hayakawa, S.; Nakai, S. Relationships of hydrophobicity and net charge to the solubility of milk and soy proteins. J. Food Sci. 1985, 50, 486–491. [Google Scholar] [CrossRef]
- Boachie, R.T.; Okagu, O.D.; Abioye, R.; Hüttmann, N.; Oliviero, T.; Capuano, E.; Fogliano, V.; Udenigwe, C.C. Lentil protein and tannic acid interaction limits in vitro peptic hydrolysis and alters peptidomic profiles of the proteins. J. Agric. Food Chem. 2022, 70, 6519–6529. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef] [PubMed]
- de Groot, N.S.; Castillo, V.; Graña-Montes, R.; Ventura, S. AGGRESCAN: Method, application, and perspectives for drug design. Methods Mol. Biol. 2012, 819, 199–220. [Google Scholar] [PubMed]
- Tjernberg, A.; Markova, N.; Griffiths, W.J.; Hallén, D. DMSO-related effects in protein characterization. SLAS Discov. 2006, 11, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Miles, A.J.; Wallace, B.A. CDtoolX, a downloadable software package for processing and analyses of circular dichroism spectroscopic data. Protein Sci. 2018, 27, 1717–1722. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]

| Sequence | Location in ovalbumin | Experimental MW (Da) | Na4vSS | Hydrophobicity score | β-sheet % |
| IFYCPIAIM | 28–36 | 1069.53 | 79.1 | 2.18 | 54.3 |
| NIFYCPIAIM | 27–36 | 1183.58 | 71.6 | 1.61 | 53.6 |
| VLVNAIVFKGL | 174–184 | 1171.73 | 54.2 | 1.95 | 52.7 |
| YCPIAIMSA | 30–38 | 967.451 | 52.8 | 1.48 | 51.0 |
| MMYQIGLF | 211–218 | 1001.47 | 46.0 | 1.21 | 31.0 |
| VYSFSLASRL | 97–106 | 1141.61 | 32.0 | 0.82 | 52.0 |
| Sequence | Flow index (n) × 10−3 | Consistency (K) | G′/G′′ (tan δ) |
|---|---|---|---|
| IFYCPIAIM | 1.23 ± 0.06 c | 0.824 ± 0.045 a | 0.928 ± 0.165 a |
| NIFYCPIAIM | 1.53 ± 0.06 c | 0.542 ± 0.036 c | 0.978 ± 0.050 a |
| VLVNAIVFKGL | 1.77 ± 0.38 c | 0.506 ± 0.121 c | 0.521 ± 0.162 a |
| YCPIAIMSA | 1.40 ± 0.10 c | 0.795 ± 0.050 b | 1.106 ± 0.431 a |
| MMYQIGLF | 5.35 ± 0.07 a | 0.032 ± 0.002 e | 1.182 ± 1.152 a |
| VYSFSLASRL | 3.10 ± 0.53 b | 0.245 ± 0.095 d | 0.657 ± 0.138 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abioye, R.O.; Acquah, C.; Hsu, P.C.Q.; Hüttmann, N.; Sun, X.; Udenigwe, C.C. Self-assembly and Hydrogelation Properties of Peptides Derived from Peptic Cleavage of Aggregation-prone Regions of Ovalbumin. Gels 2022, 8, 641. https://doi.org/10.3390/gels8100641
Abioye RO, Acquah C, Hsu PCQ, Hüttmann N, Sun X, Udenigwe CC. Self-assembly and Hydrogelation Properties of Peptides Derived from Peptic Cleavage of Aggregation-prone Regions of Ovalbumin. Gels. 2022; 8(10):641. https://doi.org/10.3390/gels8100641
Chicago/Turabian StyleAbioye, Raliat O., Caleb Acquah, Pei Chun Queenie Hsu, Nico Hüttmann, Xiaohong Sun, and Chibuike C. Udenigwe. 2022. "Self-assembly and Hydrogelation Properties of Peptides Derived from Peptic Cleavage of Aggregation-prone Regions of Ovalbumin" Gels 8, no. 10: 641. https://doi.org/10.3390/gels8100641
APA StyleAbioye, R. O., Acquah, C., Hsu, P. C. Q., Hüttmann, N., Sun, X., & Udenigwe, C. C. (2022). Self-assembly and Hydrogelation Properties of Peptides Derived from Peptic Cleavage of Aggregation-prone Regions of Ovalbumin. Gels, 8(10), 641. https://doi.org/10.3390/gels8100641










