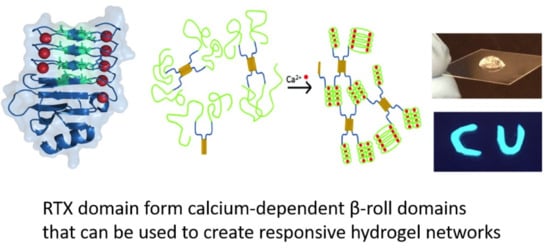
Calcium-Dependent RTX Domains in the Development of Protein Hydrogels
Abstract
1. Introduction
2. Repeats-in-Toxin (RTX) Domains and the β-Roll Fold
2.1. Adenylate Cyclase from Bordetella Pertussis
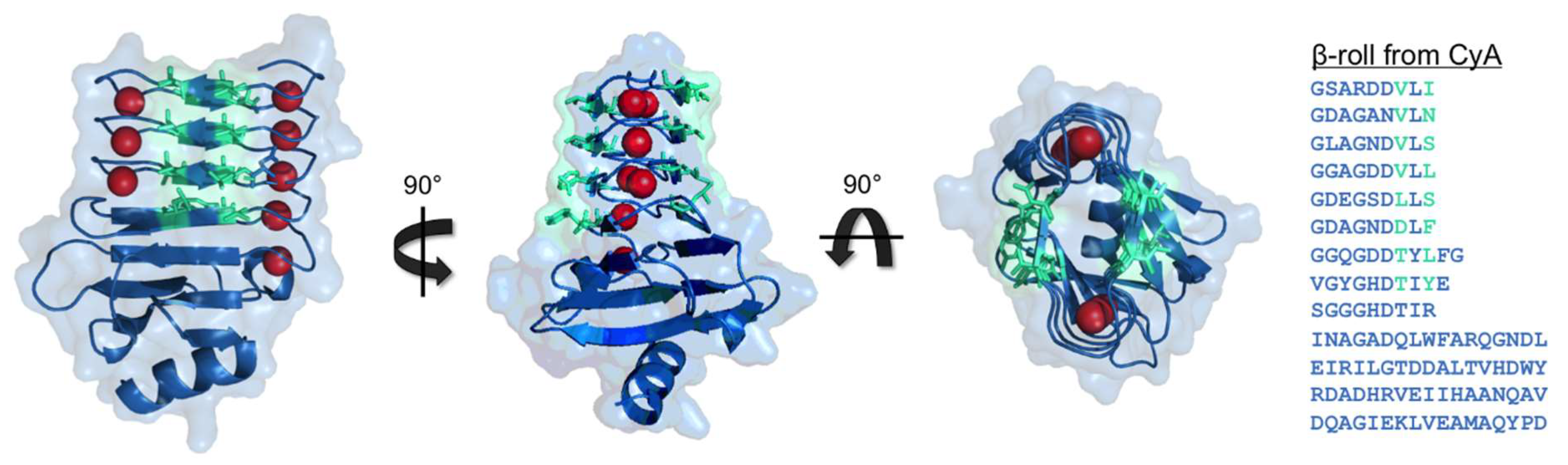
2.2. Serralysin from Serratia Marcescens and Other RTX Proteins
3. RTX Domain-Based Calcium Responsive Hydrogels
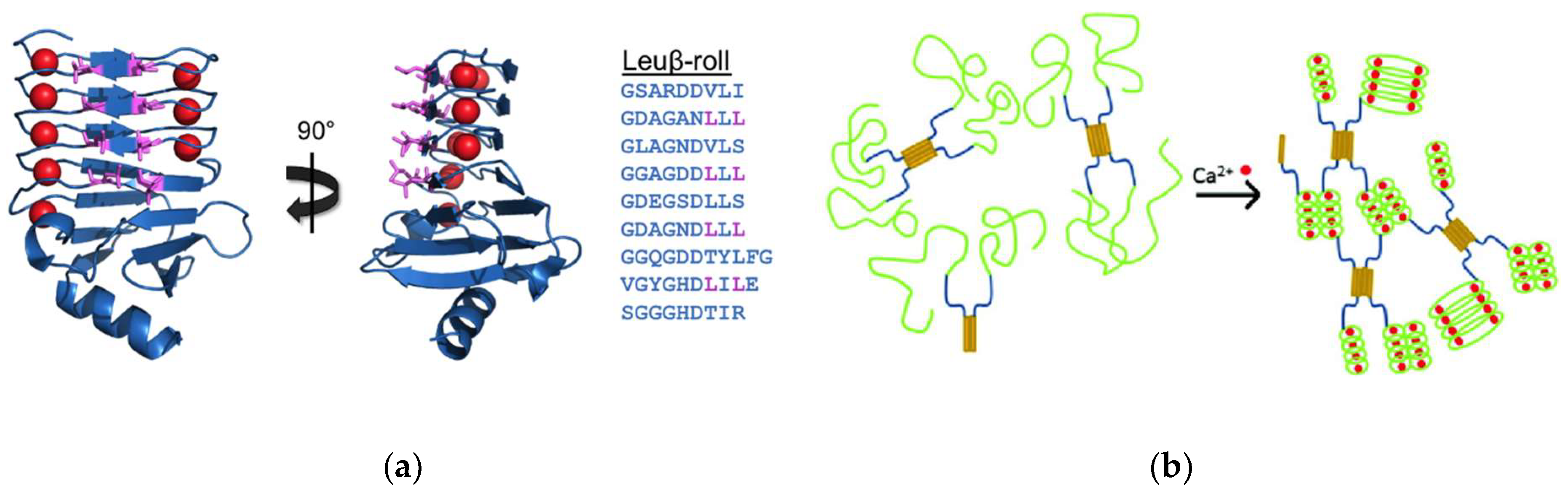
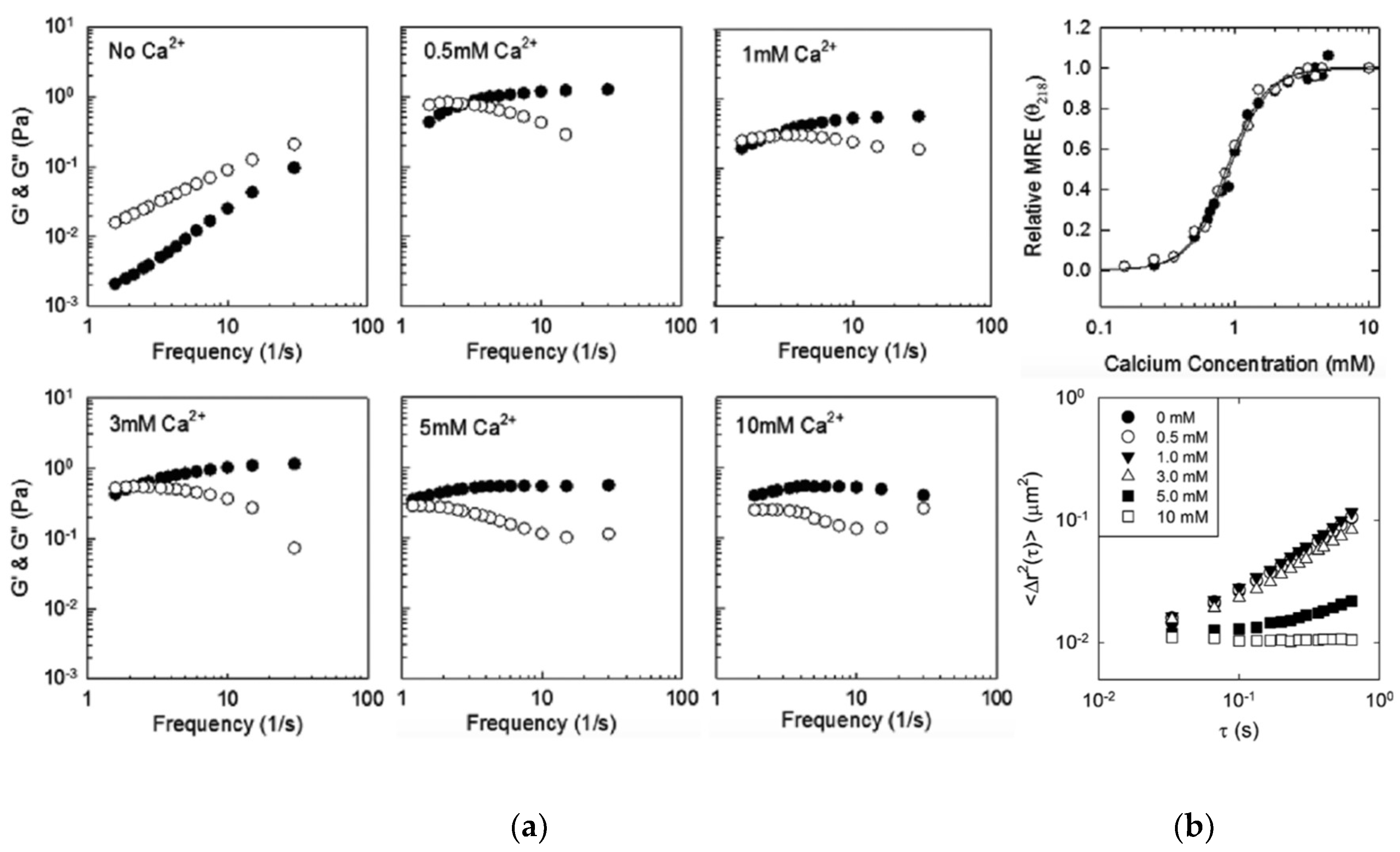
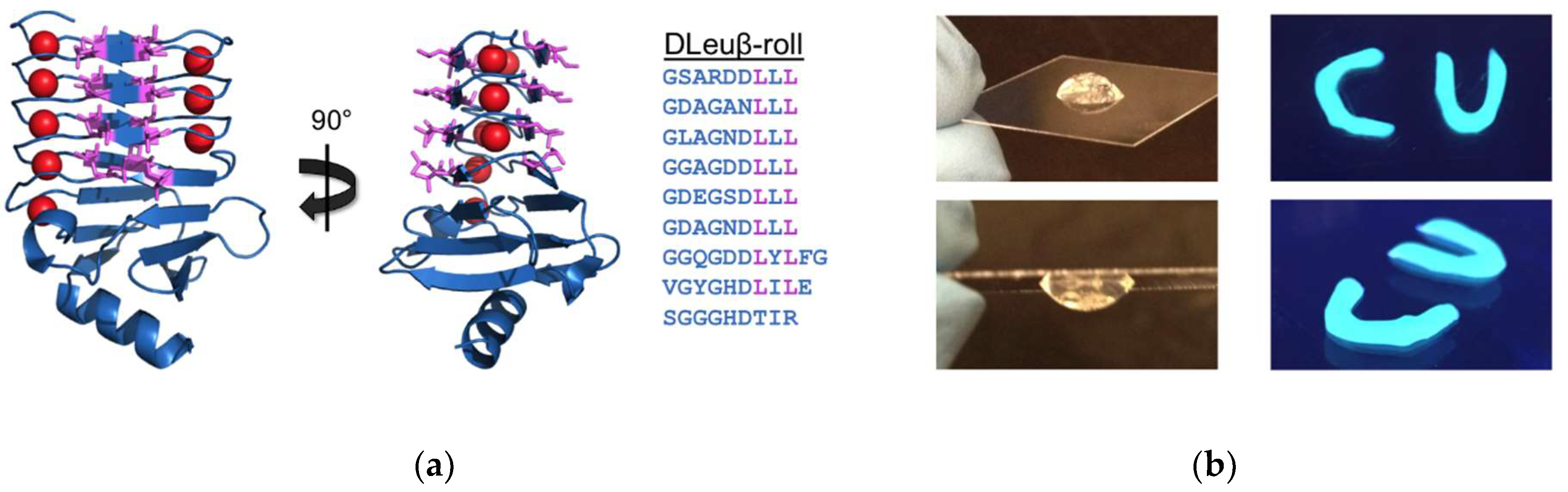
3.1. β-Roll Mutants Fused to α-Helical Leucine Zipper Domain
3.2. DLeuβ-Roll as a Stand-Alone Crosslinking Domain
3.3. RTX Domain Fused to Elastomeric Proteins
3.4. RTX Domain Fused to 6-Phospho-β-Galactosidase
3.5. RTX Domain Conjugated to Palmitic Acid
4. Summary and Conclusions
Funding
Conflicts of Interest
References
- Banta, S.; Wheeldon, I.; Blenner, M. Protein engineering in the development of functional hydrogels. Annu. Rev. Biomed. Eng. 2010, 12, 167–186. [Google Scholar] [CrossRef] [PubMed]
- Hendrickson, G.R.; Lyon, L.A. Bioresponsive hydrogels for sensing applications. Soft Matter 2009, 5, 29–35. [Google Scholar] [CrossRef]
- Qiu, Y.; Park, K. Environment-sensitive hydrogels for drug delivery. Adv. Drug Deliv. Rev. 2001, 53, 321–339. [Google Scholar] [CrossRef]
- Klouda, L.; Mikos, A.G. Thermoresponsive hydrogels in biomedical applications. Eur. J. Pharm. Biopharm. 2008, 68, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Mano, J.F. Stimuli-responsive polymeric systems for biomedical applications. Adv. Eng. Mater. 2008, 10, 515–527. [Google Scholar] [CrossRef]
- Koetting, M.C.; Peters, J.T.; Steichen, S.D.; Peppas, N.A. Stimulus-responsive hydrogels: Theory, modern advances, and applications. Mater. Sci. Eng. R. Rep. 2015, 93, 1–49. [Google Scholar] [CrossRef] [PubMed]
- Chilkoti, A.; Christensen, T.; MacKay, J.A. Stimulus responsive elastin biopolymers: Applications in medicine and biotechnology. Curr. Opin. Chem. Biol. 2006, 10, 652–657. [Google Scholar] [CrossRef] [PubMed]
- Chilkoti, A.; Dreher, M.R.; Meyer, D.E. Design of thermally responsive, recombinant polypeptide carriers for targeted drug delivery. Adv. Drug Deliv. Rev. 2002, 54, 1093–1111. [Google Scholar] [CrossRef]
- Petka, W.A.; Harden, J.L.; McGrath, K.P.; Wirtz, D.; Tirrell, D.A. Reversible hydrogels from self-assembling artificial proteins. Science 1998, 281, 389–392. [Google Scholar] [CrossRef] [PubMed]
- Wheeldon, I.R.; Gallaway, J.W.; Barton, S.C.; Banta, S. Bioelectrocatalytic hydrogels from electron-conducting metallopolypeptides coassembled with bifunctional enzymatic building blocks. Proc. Natl. Acad. Sci. USA 2008, 105, 15275–15280. [Google Scholar] [CrossRef] [PubMed]
- Ehrick, J.D.; Deo, S.K.; Browning, T.W.; Bachas, L.G.; Madou, M.J.; Daunert, S. Genetically engineered protein in hydrogels tailors stimuli-responsive characteristics. Nat. Mater. 2005, 4, 298–302. [Google Scholar] [CrossRef] [PubMed]
- Murphy, W.L.; Dillmore, W.S.; Modica, J.; Mrksich, M. Dynamic hydrogels: Translating a protein conformational change into macroscopic motion. Angew. Chem. Int. Ed. Engl. 2007, 46, 3066–3069. [Google Scholar] [CrossRef] [PubMed]
- Schneider, J.P.; Pochan, D.J.; Ozbas, B.; Rajagopal, K.; Pakstis, L.; Kretsinger, J. Responsive hydrogels from the intramolecular folding and self-assembly of a designed peptide. JACS 2002, 124, 15030–15037. [Google Scholar] [CrossRef]
- Branco, M.C.; Pochan, D.J.; Wagner, N.J.; Schneider, J.P. Macromolecular diffusion and release from self-assembled beta-hairpin peptide hydrogels. Biomaterials 2009, 30, 1339–1347. [Google Scholar] [CrossRef] [PubMed]
- Collier, J.H.; Hu, B.-H.; Ruberti, J.W.; Zhang, J.; Shum, P.; Thompson, D.H.; Messersmith, P.B. Thermally and photochemically triggered self-assembly of peptide hydrogels. J. Am. Chem. Soc. JACS 2001, 123, 9463–9464. [Google Scholar] [CrossRef]
- Welch, R.A. RTX toxin structure and function: A story of numerous anomalies and few analogies in toxin biology. In Pore-Forming Toxins; Springer: Berlin/Heidelberg, Germany, 2001; Volume 257, pp. 85–111. [Google Scholar]
- Chenal, A.; Karst, J.C.; Sotomayor Pérez, A.C.; Wozniak, A.K.; Baron, B.; England, P.; Ladant, D. Calcium-induced folding and stabilization of the intrinsically disordered RTX domain of the CyaA toxin. Biophys. J. 2010, 99, 3744–3753. [Google Scholar] [CrossRef] [PubMed]
- Bauche, C.; Chenal, A.; Knapp, O.; Bodenreider, C.; Benz, R.; Chaffotte, A.; Ladant, D. Structural and functional characterization of an essential RTX subdomain of Bordetella pertussis adenylate cyclase toxin. J. Biol. Chem. 2006, 281, 16914–16926. [Google Scholar] [CrossRef] [PubMed]
- Rose, T.; Sebo, P.; Bellalou, J.; Ladant, D. Interaction of calcium with Bordetella pertussis adenylate cyclase toxin. J. Biol. Chem. 1995, 270, 26370–26376. [Google Scholar] [CrossRef] [PubMed]
- Sotomayor-Pérez, A.C.; Ladant, D.; Chenal, A. Disorder-to-order transition in the CyaA toxin RTX domain: Implications for toxin secretion. Toxins 2015, 7, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Bulutoglu, B.; Dooley, K.; Szilvay, G.R.; Blenner, M.; Banta, S. Catch and release: Engineered allosterically-regulated β-roll peptides enable on/off biomolecular recognition. ACS Synth. Biol. 2017, 6, 1732–1741. [Google Scholar] [CrossRef] [PubMed]
- Bulutoglu, B.; Yang, S.J.; Banta, S. Conditional network assembly and targeted protein retention via environmentally responsive, engineered β-roll peptides. Biomacromolecules 2017, 18, 2139–2145. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, W.; Solanki, K.; Banta, S. Insertion of a calcium-responsive β-roll domain into a thermostable alcohol dehydrogenase enables tunable control over cofactor selectivity. ACS Catal. 2018, 8, 1602–1613. [Google Scholar] [CrossRef]
- Meier, R.; Drepper, T.; Svensson, V.; Jaeger, K.-E.; Baumann, U. A calcium-gated lid and a large beta roll sandwich are revealed by the crystal structure of extracellular lipase from Serratia marcescens. J. Biol. Chem. 2007, 282, 31477–31483. [Google Scholar] [CrossRef] [PubMed]
- Pojanapotha, P.; Thamwiriyasati, N.; Powthongchin, B.; Katzenmeier, G.; Angsuthanasombat, C. Bordetella pertussis CyaA-RTX subdomain requires calcium ions for structural stability against proteolytic degradation. Protein Expr. Purif. 2011, 75, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Ladant, D.; Ullmann, A. Bordetella pertussis adenylate cyclase: A toxin with multiple talents. Trends Microbiol. 1999, 7, 172–176. [Google Scholar] [CrossRef]
- Zhang, L.; Conway, J.F.; Thibodeau, P.H. Calcium-induced folding and stabilization of the Pseudomonas aeruginosa alkaline protease. J. Biol. Chem. 2012, 287, 4311–4322. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Functional roles of transiently and intrinsically disordered regions within proteins. FEBS J. 2015, 282, 1182–1189. [Google Scholar] [CrossRef] [PubMed]
- Chenal, A.; Guijarro, J.I.; Raynal, B.; Delepierre, M.; Ladant, D. RTX calcium binding motifs are intrinsically disordered in the absence of calcium. J. Biol. Chem. 2009, 284, 1781–1789. [Google Scholar] [CrossRef] [PubMed]
- Cheung, G.Y.; Kelly, S.M.; Jess, T.J.; Prior, S.; Price, N.C.; Parton, R.; Coote, J.G. Functional and structural studies on different forms of the adenylate cyclase toxin of Bordetella pertussis. Microb. Pathog. 2009, 46, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Coote, J.G. Structural and functional relationships among the RTX toxin determinants of gram-negative bacteria. FEMS Microbiol. Rev. 1992, 88, 137–162. [Google Scholar] [CrossRef] [PubMed]
- Short, E.C.; Kurtz, H.J. Properties of the hemolytic activities of Escherichia coli. Infect. Immun. 1971, 3, 678–687. [Google Scholar] [CrossRef] [PubMed]
- Hanski, E.; Farfel, Z. Bordetella pertussis invasive adenylate cyclase: Partial resolution and properties of its cellular penetration. J. Biol. Chem. 1985, 260, 5526–5532. [Google Scholar] [CrossRef] [PubMed]
- Bumba, L.; Masin, J.; Macek, P.; Wald, T.; Motlova, L.; Bibova, I.; Klimova, N.; Bednarova, L.; Veverka, V.; Kachala, M.; et al. Calcium-driven folding of RTX domain β-rolls ratchets translocation of RTX proteins through type I secretion ducts. Mol. Cell 2016, 62, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Blenner, M.A.; Shur, O.; Szilvay, G.R.; Cropek, D.M.; Banta, S. Calcium-induced folding of a beta roll motif requires C-terminal entropic stabilization. J. Mol. Biol. 2010, 400, 244–256. [Google Scholar] [CrossRef] [PubMed]
- Sotomayor Pérez, A.C.; Karst, J.C.; Davi, M.; Guijarro, J.I.; Ladant, D.; Chenal, A. Characterization of the regions involved in the calcium-induced folding of the intrinsically disordered RTX motifs from the Bordetella pertussis adenylate cyclase toxin. J. Mol. Biol. 2010, 397, 534–549. [Google Scholar] [CrossRef] [PubMed]
- Shur, O.; Banta, S. Rearranging and concatenating a native RTX domain to understand sequence modularity. Protein Eng. Des. Sel. 2013, 26, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Baumann, U. Crystal structure of the 50 kDa metallo protease from Serratia marcescens. J. Mol. Biol. 1994, 242, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Baumann, U.; Bauer, M.; Letoffe, S.; Delepelaire, P.; Wandersman, C. Crystal structure of a complex between Serratia marcescens metallo-protease and an inhibitor from Erwinia chrysanthemi. J. Mol. Biol. 1995, 248, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Stocker, W.; Grams, F.; Baumann, U.; Reinemer, P.; Gomis-Ruth, F.X.; McKay, D.B.; Bode, W. The metzincins-Topological and sequential relations between the astacins, adamalysins, serralysins, and matrix-ins (collagenases) define a superfamily of zinc-peptidase. Protein Sci. 1995, 4, 823–840. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Morrison, A.J.; Thibodeau, P.H. Interdomain contacts and the stability of serralysin protease from Serratia marcescens. PLoS ONE 2015, 10, e0138419. [Google Scholar] [CrossRef] [PubMed]
- Baumann, U.; Wu, S.; Flaherty, K.M.; McKay, D.B. Three-dimensional structure of the alkaline prote-ase of Pseudomonas aeruginosa: A two-domain protein with a calcium binding parallel beta roll motif. EMBO J. 1993, 12, 3357–3364. [Google Scholar] [CrossRef] [PubMed]
- Linhartova, I.; Bumba, L.; Masin, J.; Basler, M.; Osicka, R.; Kamanova, J.; Prochazkova, K.; Adkins, I.; Hejnova-Holubova, J.; Sadilkova, L.; et al. RTX proteins: A highly diverse family secreted by a common mechanism. FEMS Microbiol. Rev. 2010, 34, 1076–1112. [Google Scholar] [CrossRef] [PubMed]
- Hyland, C.; Vuillard, L.; Hughes, C.; Koronakis, V. Membrane interaction of Escherichia coli hemolysin: Flotation and insertion-dependent labeling by phospholipid vesicles. J. Bacteriol. 2001, 183, 5364–5370. [Google Scholar] [CrossRef] [PubMed]
- Dooley, K.; Kim, Y.H.; Lu, H.D.; Tu, R.; Banta, S. Engineering of an environmentally responsive beta roll peptide for use as a calcium-dependent cross-linking domain for peptide hydrogel formation. Biomacromolecules 2012, 13, 1758–1764. [Google Scholar] [CrossRef] [PubMed]
- Dooley, K.; Bulutoglu, B.; Banta, S. Doubling the cross-linking interface of a rationally designed beta roll peptide for calcium-dependent proteinaceous hydrogel formation. Biomacromolecules 2014, 15, 3617–3624. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, H.; Han, Y.; Lv, S.; Chen, J. Using single molecule force spectroscopy to facilitate a rational design of Ca2+-responsive β-roll peptide-based hydrogels. J. Mater. Chem. B 2018, 6, 5303–5312. [Google Scholar] [CrossRef]
- Ringler, P.; Schulz, G.E. Self-assembly of proteins into designed networks. Science 2003, 302, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.R.; Ge, R.; Luo, S.Z. Self-assembly of pH and calcium dual-responsive peptide-amphiphilic hydrogel. J. Pept. Sci. 2013, 19, 737–744. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bulutoglu, B.; Banta, S. Calcium-Dependent RTX Domains in the Development of Protein Hydrogels. Gels 2019, 5, 10. https://doi.org/10.3390/gels5010010
Bulutoglu B, Banta S. Calcium-Dependent RTX Domains in the Development of Protein Hydrogels. Gels. 2019; 5(1):10. https://doi.org/10.3390/gels5010010
Chicago/Turabian StyleBulutoglu, Beyza, and Scott Banta. 2019. "Calcium-Dependent RTX Domains in the Development of Protein Hydrogels" Gels 5, no. 1: 10. https://doi.org/10.3390/gels5010010
APA StyleBulutoglu, B., & Banta, S. (2019). Calcium-Dependent RTX Domains in the Development of Protein Hydrogels. Gels, 5(1), 10. https://doi.org/10.3390/gels5010010