Dancing with Nucleobases: Unveiling the Self-Assembly Properties of DNA and RNA Base-Containing Molecules for Gel Formation
Abstract
:1. Introduction
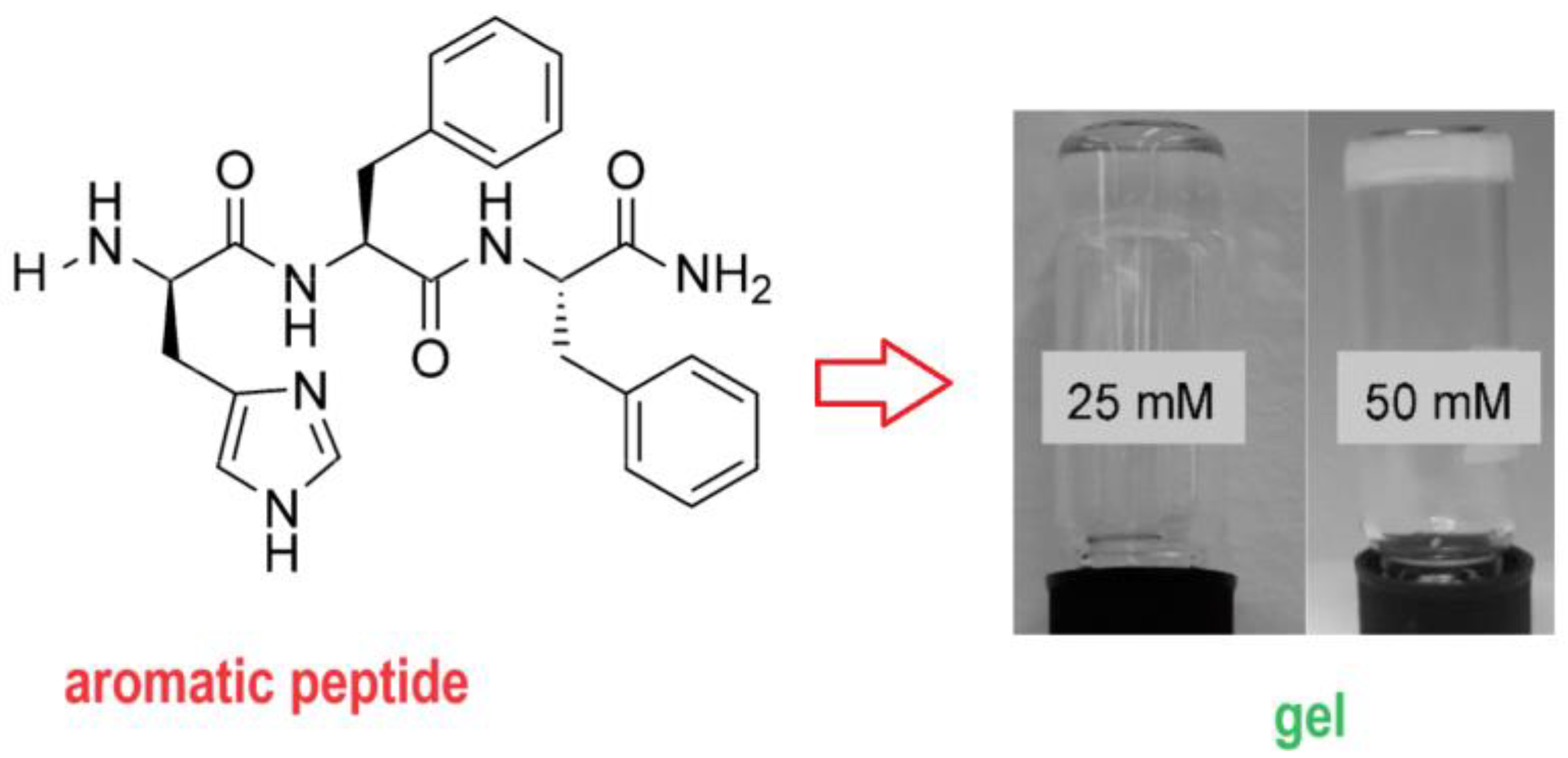
2. Nucleobase-Bearing Amino Acid Systems and Self-Assembly
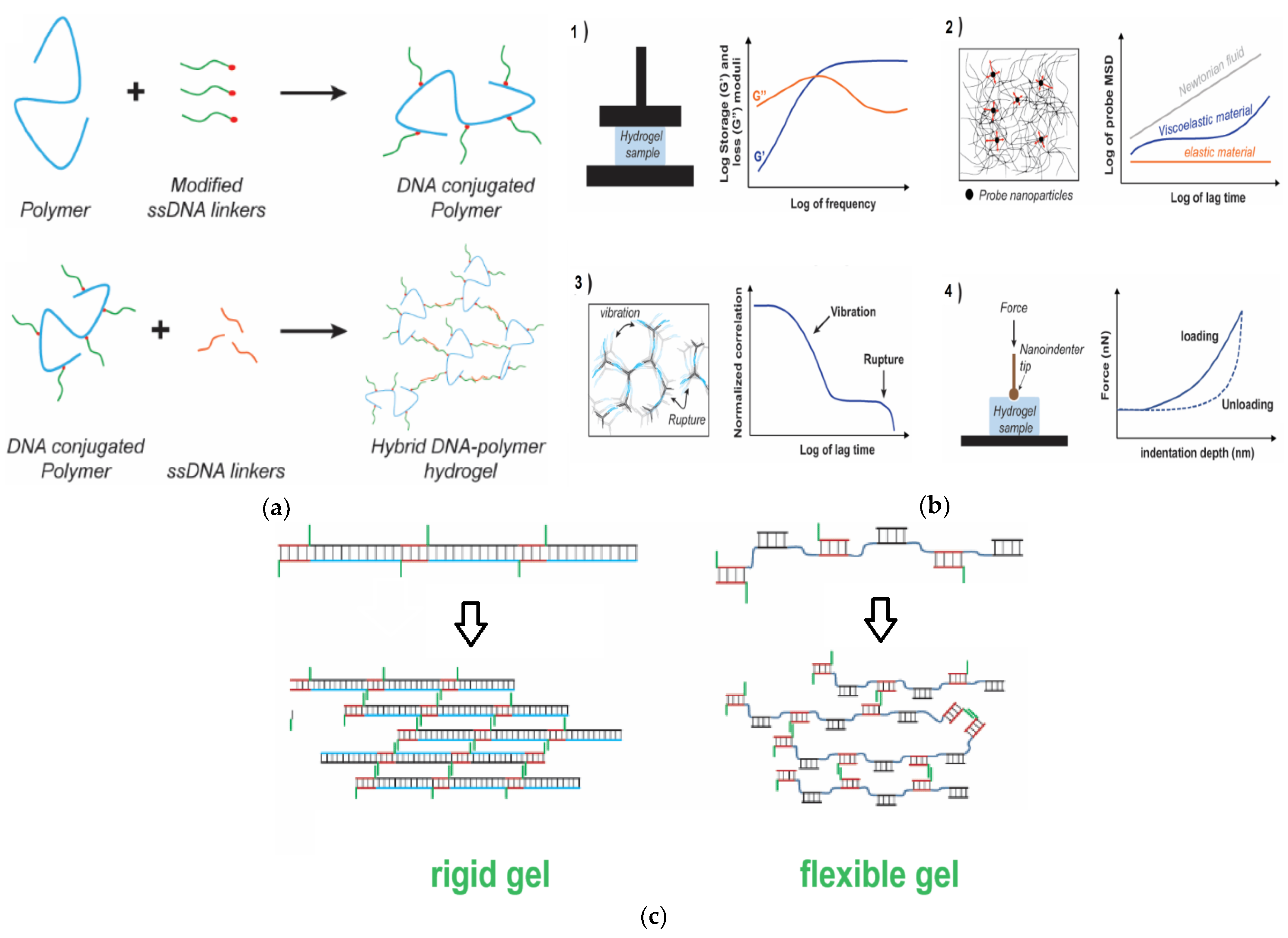
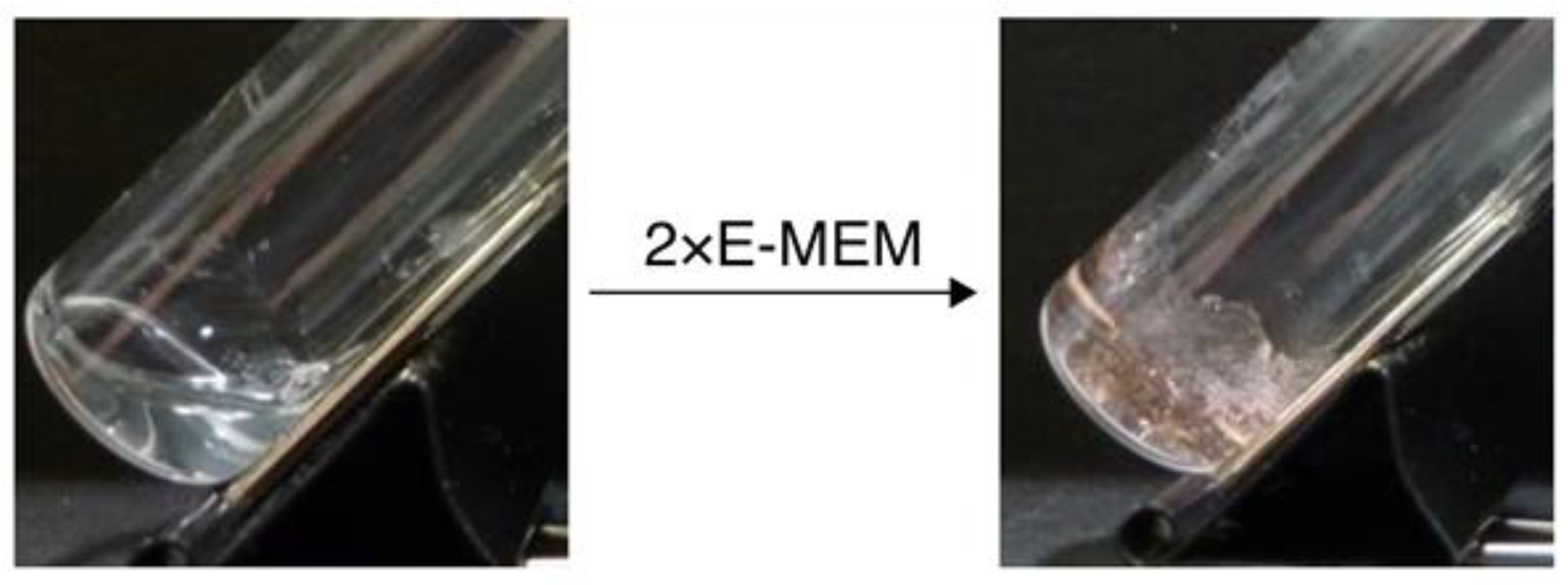
3. Nucleic Acid-Based Gels
3.1. G-Quartet-Based Gels
3.2. RNA-Based Gels
3.3. Comparing DNA and RNA-Based Systems
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Djabourov, M. Gelation—A review. Polym. Int. 1991, 25, 135–143. [Google Scholar] [CrossRef]
- Rogovina, L.Z.; Vasil’ev, V.G.; Braudo, E. Definition of the concept of polymer gel. Polym. Sci. Ser. C 2008, 50, 85–92. [Google Scholar] [CrossRef]
- Yamauchi, A. Gels: Introduction. In Gels Handbook; Elsevier: Amsterdam, The Netherlands, 2001; pp. 4–12. [Google Scholar]
- Draper, E.R.; Adams, D.J. Low-molecular-weight gels: The state of the art. Chem 2017, 3, 390–410. [Google Scholar] [CrossRef]
- Banerjee, S.; Bhattacharya, S. Food gels: Gelling process and new applications. Crit. Rev. Food Sci. Nutr. 2012, 52, 334–346. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, S.A.; Alvi, T.; Biswas, A.; Shityakov, S.; Gusinskaia, T.; Lavrentev, F.; Dutta, K.; Khan, M.K.I.; Stephen, J.; Radhakrishnan, M. Food gels: Principles, interaction mechanisms and its microstructure. Crit. Rev. Food Sci. Nutr. 2022, 2, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Nazir, A.; Asghar, A.; Maan, A.A. Food gels: Gelling process and new applications. In Advances in Food Rheology and Its Applications; Elsevier: Amsterdam, The Netherlands, 2017; pp. 335–353. [Google Scholar]
- Nayak, A.K.; Das, B. Introduction to polymeric gels. In Polymeric Gels; Elsevier: Amsterdam, The Netherlands, 2018; pp. 3–27. [Google Scholar]
- Vashist, A.; Vashist, A.; Gupta, Y.; Ahmad, S. Recent advances in hydrogel based drug delivery systems for the human body. J. Mater. Chem. B 2014, 2, 147–166. [Google Scholar] [CrossRef] [PubMed]
- Chamkouri, H.; Chamkouri, M. A review of hydrogels, their properties and applications in medicine. Am. J. Biomed. Sci. Res. 2021, 11, 485–493. [Google Scholar] [CrossRef]
- Hwang, H.S.; Lee, C.-S. Recent progress in hyaluronic-acid-based hydrogels for bone tissue engineering. Gels 2023, 9, 588. [Google Scholar] [CrossRef]
- Kopeček, J. Hydrogel biomaterials: A smart future? Biomaterials 2007, 28, 5185–5192. [Google Scholar] [CrossRef]
- Shakeel, S.; Karim, S.; Ali, A. Peptide nucleic acid (PNA)—A review. J. Chem. Technol. Biotechnol. Int. Res. Process Environ. Clean Technol. 2006, 81, 892–899. [Google Scholar] [CrossRef]
- Rodrigues, T.; Curti, F.; Leroux, Y.R.; Barras, A.; Pagneux, Q.; Happy, H.; Kleber, C.; Boukherroub, R.; Hasler, R.; Volpi, S. Discovery of a Peptide Nucleic Acid (PNA) aptamer for cardiac troponin I: Substituting DNA with neutral PNA maintains picomolar affinity and improves performances for electronic sensing with graphene field-effect transistors (gFET). Nano Today 2023, 50, 101840. [Google Scholar] [CrossRef]
- Pradeep, S.P.; Malik, S.; Slack, F.J.; Bahal, R. Unlocking the potential of chemically modified peptide nucleic acids for RNA-based therapeutics. RNA 2023, 29, 434–445. [Google Scholar] [CrossRef] [PubMed]
- Chu, T.-W.; Feng, J.; Yang, J.; Kopeček, J. Hybrid polymeric hydrogels via peptide nucleic acid (PNA)/DNA complexation. J. Control. Release 2015, 220, 608–616. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Park, S.M.; Kim, W.-k.; Lee, J. Hydrogel-based thermosensor using peptide nucleic acid and PEGylated graphene oxide. Anal. Chim. Acta 2023, 1239, 340708. [Google Scholar] [CrossRef] [PubMed]
- Langford, G.J.; Raeburn, J.; Ferrier, D.C.; Hands, P.J.; Shaver, M.P. Morpholino oligonucleotide cross-linked hydrogels as portable optical oligonucleotide biosensors. ACS Sens. 2018, 4, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, N.K.; Allen, P.; Song, Y.H.; Wachs, R.A.; Du, Y.; Ellington, A.D.; Schmidt, C.E. Oligonucleotide-functionalized hydrogels for sustained release of small molecule (aptamer) therapeutics. Acta Biomater. 2020, 102, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Liu, J. Oligonucleotide-functionalized hydrogels as stimuli responsive materials and biosensors. Soft Matter 2011, 7, 6757–6767. [Google Scholar] [CrossRef]
- Bhattacharyya, T.; Saha, P.; Dash, J. Guanosine-derived supramolecular hydrogels: Recent developments and future opportunities. ACS Omega 2018, 3, 2230–2241. [Google Scholar] [CrossRef]
- Ye, X.; Li, X.; Shen, Y.; Chang, G.; Yang, J.; Gu, Z. Self-healing pH-sensitive cytosine-and guanosine-modified hyaluronic acid hydrogels via hydrogen bonding. Polymer 2017, 108, 348–360. [Google Scholar] [CrossRef]
- Merino-Gómez, M.; Godoy-Gallardo, M.; Wendner, M.; Mateos-Timoneda, M.A.; Gil, F.J.; Perez, R.A. Optimization of guanosine-based hydrogels with boric acid derivatives for enhanced long-term stability and cell survival. Front. Bioeng. Biotechnol. 2023, 11, 1147943. [Google Scholar] [CrossRef]
- Godoy-Gallardo, M.; Merino-Gómez, M.; Mateos-Timoneda, M.A.; Eckhard, U.; Gil, F.J.; Perez, R.A. Advanced Binary Guanosine and Guanosine 5′-Monophosphate Cell-Laden Hydrogels for Soft Tissue Reconstruction by 3D Bioprinting. ACS Appl. Mater. Interfaces 2023. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, M.; Sharma, R.; Hussain, A.; Kumar, I.; Sharma, A.K.; Sarkar, A. Hydrogels and their combination with lipids and nucleotides. In Sustainable Hydrogels; Elsevier: Amsterdam, The Netherlands, 2023; pp. 471–487. [Google Scholar]
- Peters, G.M.; Davis, J.T. Supramolecular gels made from nucleobase, nucleoside and nucleotide analogs. Chem. Soc. Rev. 2016, 45, 3188–3206. [Google Scholar] [CrossRef] [PubMed]
- Godeau, G.; Brun, C.; Arnion, H.; Staedel, C.; Barthélémy, P. Glycosyl-nucleoside fluorinated amphiphiles as components of nanostructured hydrogels. Tetrahedron Lett. 2010, 51, 1012–1015. [Google Scholar] [CrossRef]
- Godoy-Gallardo, M.; Merino-Gómez, M.; Matiz, L.C.; Mateos-Timoneda, M.A.; Gil, F.J.; Perez, R.A. Nucleoside-based supramolecular hydrogels: From synthesis and structural properties to biomedical and tissue engineering applications. ACS Biomater. Sci. Eng. 2022, 9, 40–61. [Google Scholar] [CrossRef] [PubMed]
- Ignatowska, J.; Mironiuk-Puchalska, E.; Grześkowiak, P.; Wińska, P.; Wielechowska, M.; Bretner, M.; Karatsai, O.; Rędowicz, M.J.; Koszytkowska-Stawińska, M. New insight into nucleo α-amino acids–Synthesis and SAR studies on cytotoxic activity of β-pyrimidine alanines. Bioorg. Chem. 2020, 100, 103864. [Google Scholar] [CrossRef] [PubMed]
- More, J.C.; Troop, H.M.; Dolman, N.P.; Jane, D.E. Structural requirements for novel willardiine derivatives acting as AMPA and kainate receptor antagonists. Br. J. Pharmacol. 2003, 138, 1093–1100. [Google Scholar] [CrossRef] [PubMed]
- Mik, V.; Mičková, Z.; Doležal, K.; Frébort, I.; Pospisil, T. Activity of (+)-Discadenine as a plant cytokinin. J. Nat. Prod. 2017, 80, 2136–2140. [Google Scholar] [CrossRef]
- Xu, Q.; Song, B.; Liu, F.; Song, Y.; Chen, P.; Liu, S.; Krishnan, H.B. Identification and characterization of β-Lathyrin, an abundant glycoprotein of grass pea (Lathyrus sativus L.), as a potential allergen. J. Agric. Food Chem. 2018, 66, 8496–8503. [Google Scholar] [CrossRef]
- Roviello, G.N.; Gaetano, S.D.; Capasso, D.; Cesarani, A.; Bucci, E.M.; Pedone, C. Synthesis, spectroscopic studies and biological activity of a novel nucleopeptide with Moloney murine leukemia virus reverse transcriptase inhibitory activity. Amino Acids 2010, 38, 1489–1496. [Google Scholar] [CrossRef]
- Roviello, G.N.; Musumeci, D.; De Cristofaro, A.; Capasso, D.; Di Gaetano, S.; Bucci, E.M.; Pedone, C. Alternate dab-aeg PNAs: Synthesis, nucleic acid binding studies and biological activity. Mol. Biosyst. 2009, 6, 199–205. [Google Scholar] [CrossRef]
- Roviello, G.; Musumeci, D.; Castiglione, M.; Bucci, E.; Pedone, C.; Benedetti, E. Solid phase synthesis and RNA-binding studies of a serum-resistant nucleo-ε-peptide. J. Pept. Sci. Off. Publ. Eur. Pept. Soc. 2009, 15, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Roviello, G.N.; Moccia, M.; Sapio, R.; Valente, M.; Bucci, E.; Castiglione, M.; Pedone, C.; Perretta, G.; Benedetti, E.; Musumeci, D. Synthesis, characterization and hybridization studies of new nucleo-γ-peptides based on diaminobutyric acid. J. Pept. Sci. Off. Publ. Eur. Pept. Soc. 2006, 12, 829–835. [Google Scholar] [CrossRef] [PubMed]
- Roviello, V.; Musumeci, D.; Mokhir, A.; Roviello, G.N. Evidence of protein binding by a nucleopeptide based on a thyminedecorated L-diaminopropanoic acid through CD and in silico studies. Curr. Med. Chem. 2021, 28, 5004–5015. [Google Scholar] [CrossRef] [PubMed]
- Hoschtettler, P.; Pickaert, G.; Carvalho, A.; Averlant-Petit, M.-C.; Stefan, L. Highly Synergistic Properties of Multicomponent Hydrogels Thanks to Cooperative Nucleopeptide Assemblies. Chem. Mater. 2023, 35, 4259–4275. [Google Scholar] [CrossRef]
- Musumeci, D.; Ullah, S.; Ikram, A.; Roviello, G.N. Novel insights on nucleopeptide binding: A spectroscopic and In Silico investigation on the interaction of a thymine-bearing tetrapeptide with a homoadenine DNA. J. Mol. Liq. 2022, 347, 117975. [Google Scholar] [CrossRef]
- Boback, K.; Bacchi, K.; O’Neill, S.; Brown, S.; Dorsainvil, J.; Smith-Carpenter, J.E. Impact of C-terminal chemistry on self-assembled morphology of guanosine containing nucleopeptides. Molecules 2020, 25, 5493. [Google Scholar] [CrossRef]
- Datta, A. Synthetic Studies on Antifungal Peptidyl Nucleoside Antibiotics. Chem. Synth. Nucleoside Analog. 2013, 819–846. [Google Scholar] [CrossRef]
- Swinehart, W.; Deutsch, C.; Sarachan, K.L.; Luthra, A.; Bacusmo, J.M.; de Crécy-Lagard, V.; Swairjo, M.A.; Agris, P.F.; Iwata-Reuyl, D. Specificity in the biosynthesis of the universal tRNA nucleoside N6-threonylcarbamoyl adenosine (t6A)—TsaD is the gatekeeper. RNA 2020, 26, 1094–1103. [Google Scholar] [CrossRef]
- Roviello, G.N.; Benedetti, E.; Pedone, C.; Bucci, E.M. Nucleobase-containing peptides: An overview of their characteristic features and applications. Amino Acids 2010, 39, 45–57. [Google Scholar] [CrossRef]
- Kurbasic, M.; Garcia, A.M.; Viada, S.; Marchesan, S. Tripeptide self-assembly into bioactive hydrogels: Effects of terminus modification on biocatalysis. Molecules 2021, 26, 173. [Google Scholar] [CrossRef]
- Snip, E.; Koumoto, K.; Shinkai, S. Gel formation properties of a uracil-appended cholesterol gelator and cooperative effects of the complementary nucleobases. Tetrahedron 2002, 58, 8863–8873. [Google Scholar] [CrossRef]
- Marchesan, S.; Vargiu, A.V.; Styan, K.E. The Phe-Phe motif for peptide self-assembly in nanomedicine. Molecules 2015, 20, 19775–19788. [Google Scholar] [CrossRef] [PubMed]
- Dinesh, B.; Squillaci, M.A.; Ménard-Moyon, C.; Samorì, P.; Bianco, A. Self-assembly of diphenylalanine backbone homologues and their combination with functionalized carbon nanotubes. Nanoscale 2015, 7, 15873–15879. [Google Scholar] [CrossRef] [PubMed]
- Roviello, G.N. Novel insights into nucleoamino acids: Biomolecular recognition and aggregation studies of a thymine-conjugated l-phenyl alanine. Amino Acids 2018, 50, 933–941. [Google Scholar] [CrossRef] [PubMed]
- Scognamiglio, P.L.; Riccardi, C.; Palumbo, R.; Gale, T.F.; Musumeci, D.; Roviello, G.N. Self-assembly of thyminyl l-tryptophanamide (TrpT) building blocks for the potential development of drug delivery nanosystems. J. Nanostructure Chem. 2023, 1–19. [Google Scholar] [CrossRef]
- Guida, S.; Arginelli, F.; Farnetani, F.; Ciardo, S.; Bertoni, L.; Manfredini, M.; Zerbinati, N.; Longo, C.; Pellacani, G. Clinical applications of in vivo and ex vivo confocal microscopy. Appl. Sci. 2021, 11, 1979. [Google Scholar] [CrossRef]
- Li, X.; Kuang, Y.; Lin, H.-C.; Gao, Y.; Shi, J.; Xu, B. Supramolecular nanofibers and hydrogels of nucleopeptides. Angew. Chem. Int. Ed. Engl. 2011, 50, 9365. [Google Scholar] [CrossRef]
- Yuan, D.; Du, X.; Shi, J.; Zhou, N.; Zhou, J.; Xu, B. Mixing biomimetic heterodimers of nucleopeptides to generate biocompatible and biostable supramolecular hydrogels. Angew. Chem. 2015, 127, 5797–5800. [Google Scholar] [CrossRef]
- Ewert, E.; Pospieszna-Markiewicz, I.; Szymańska, M.; Kurkiewicz, A.; Belter, A.; Kubicki, M.; Patroniak, V.; Fik-Jaskółka, M.A.; Roviello, G.N. New N4-Donor Ligands as Supramolecular Guests for DNA and RNA: Synthesis, Structural Characterization, In Silico, Spectrophotometric and Antimicrobial Studies. Molecules 2023, 28, 400. [Google Scholar] [CrossRef]
- Baek, K.; Noblett, A.D.; Ren, P.; Suggs, L.J. Self-assembled nucleo-tripeptide hydrogels provide local and sustained doxorubicin release. Biomater. Sci. 2020, 8, 3130–3137. [Google Scholar] [CrossRef]
- Giraud, T.; Bouguet-Bonnet, S.; Marchal, P.; Pickaert, G.; Averlant-Petit, M.-C.; Stefan, L. Improving and fine-tuning the properties of peptide-based hydrogels via incorporation of peptide nucleic acids. Nanoscale 2020, 12, 19905–19917. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, R.; Simonyan, H.; Roviello, G.N. Advances in Amino Acid-Based Chemistry. Pharmaceuticals 2023, 16, 1490. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Zhou, J.; Li, X.; Xu, B. Self-assembly of nucleopeptides to interact with DNAs. Interface Focus 2017, 7, 20160116. [Google Scholar] [CrossRef] [PubMed]
- Roviello, G.N.; Musumeci, D.; Bucci, E.M.; Pedone, C. Evidences for supramolecular organization of nucleopeptides: Synthesis, spectroscopic and biological studies of a novel dithymine L-serine tetrapeptide. Mol. BioSyst. 2011, 7, 1073–1080. [Google Scholar] [CrossRef] [PubMed]
- Roviello, G.N.; Ricci, A.; Bucci, E.M.; Pedone, C. Synthesis, biological evaluation and supramolecular assembly of novel analogues of peptidyl nucleosides. Mol. BioSyst. 2011, 7, 1773–1778. [Google Scholar] [CrossRef] [PubMed]
- Giraud, T.; Hoschtettler, P.; Pickaert, G.; Averlant-Petit, M.-C.; Stefan, L. Emerging low-molecular weight nucleopeptide-based hydrogels: State of the art, applications, challenges and perspectives. Nanoscale 2022, 14, 4908–4921. [Google Scholar] [CrossRef] [PubMed]
- Scognamiglio, P.L.; Platella, C.; Napolitano, E.; Musumeci, D.; Roviello, G.N. From prebiotic chemistry to supramolecular biomedical materials: Exploring the properties of self-assembling nucleobase-containing peptides. Molecules 2021, 26, 3558. [Google Scholar] [CrossRef]
- Wang, H.; Feng, Z.; Xu, B. Supramolecular assemblies of peptides or nucleopeptides for gene delivery. Theranostics 2019, 9, 3213. [Google Scholar] [CrossRef]
- Wang, H.; Feng, Z.; Qin, Y.; Wang, J.; Xu, B. Nucleopeptide assemblies selectively sequester ATP in cancer cells to increase the efficacy of doxorubicin. Angew. Chem. 2018, 130, 5025–5029. [Google Scholar] [CrossRef]
- Ghosh, S.; Ghosh, T.; Bhowmik, S.; Patidar, M.K.; Das, A.K. Nucleopeptide-coupled injectable bioconjugated guanosine-quadruplex hydrogel with inherent antibacterial activity. ACS Appl. Bio Mater. 2023, 6, 640–651. [Google Scholar] [CrossRef]
- Noblett, A.D.; Baek, K.; Suggs, L.J. Controlling Nucleopeptide Hydrogel Self-Assembly and Formation for Cell-Culture Scaffold Applications. ACS Biomater. Sci. Eng. 2021, 7, 2605–2614. [Google Scholar] [CrossRef] [PubMed]
- Baek, K.; Noblett, A.D.; Ren, P.; Suggs, L.J. Design and characterization of nucleopeptides for hydrogel self-assembly. ACS Appl. Bio Mater. 2019, 2, 2812–2821. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Han, J.; Pei, Y.; Fan, R.; Du, J. Chaperone copolymer-assisted aptamer-patterned DNA hydrogels for triggering spatiotemporal release of protein. ACS Appl. Bio Mater. 2018, 1, 1206–1214. [Google Scholar] [CrossRef] [PubMed]
- Morán, M.C.; Infante, M.R.; Miguel, M.G.; Lindman, B.; Pons, R. Novel biocompatible DNA gel particles. Langmuir 2010, 26, 10606–10613. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Shi, Z.; Dong, Y.; Wu, F.; Liu, D. Responsive DNA-based supramolecular hydrogels. ACS Appl. Bio Mater. 2020, 3, 2827–2837. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Wang, K.; Luo, S.; Li, F.; Zuo, X.; Fan, C.; Li, Q. Programmable DNA hydrogels as Artificial extracellular matrix. Small 2022, 18, 2107640. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Zhang, K.; Wang, H.; Liang, Z.; Zhou, L.; Yan, B. Versatile synthesis of a highly porous DNA/CNT hydrogel for the adsorption of the carcinogen PAH. Chem. Commun. 2021, 57, 2289–2292. [Google Scholar] [CrossRef]
- Bush, J.; Hu, C.-H.; Veneziano, R. Mechanical properties of DNA hydrogels: Towards highly programmable biomaterials. Appl. Sci. 2021, 11, 1885. [Google Scholar] [CrossRef]
- Gao, C.; Zhang, Z.; Zhang, X.; Chen, J.; Chen, Y.; Zhao, C.; Zhao, L.; Feng, L. A molecular crowding thermo-switchable chiral G-quartet hydrogel with circularly polarized luminescence property. Soft Matter 2022, 18, 3125–3129. [Google Scholar] [CrossRef]
- Yu, Y.; Nakamura, D.; DeBoyace, K.; Neisius, A.W.; McGown, L.B. Tunable thermoassociation of binary guanosine gels. J. Phys. Chem. B 2008, 112, 1130–1134. [Google Scholar] [CrossRef]
- Davis, J.T.; Spada, G.P. Supramolecular architectures generated by self-assembly of guanosine derivatives. Chem. Soc. Rev. 2007, 36, 296–313. [Google Scholar] [CrossRef] [PubMed]
- Longhi, G.; Castiglioni, E.; Koshoubu, J.; Mazzeo, G.; Abbate, S. Circularly polarized luminescence: A review of experimental and theoretical aspects. Chirality 2016, 28, 696–707. [Google Scholar] [CrossRef] [PubMed]
- Imai, Y. Generation of Circularly Polarized Luminescence by Symmetry Breaking. Symmetry 2020, 12, 1786. [Google Scholar] [CrossRef]
- Yang, G.; Zhang, S.; Hu, J.; Fujiki, M.; Zou, G. The chirality induction and modulation of polymers by circularly polarized light. Symmetry 2019, 11, 474. [Google Scholar] [CrossRef]
- Zou, C.; Qu, D.; Jiang, H.; Lu, D.; Ma, X.; Zhao, Z.; Xu, Y. Bacterial cellulose: A versatile chiral host for circularly polarized luminescence. Molecules 2019, 24, 1008. [Google Scholar] [CrossRef] [PubMed]
- Le Bideau, J.; Viau, L.; Vioux, A. Ionogels, ionic liquid based hybrid materials. Chem. Soc. Rev. 2011, 40, 907–925. [Google Scholar] [CrossRef] [PubMed]
- Qi, P.; Li, X.; Huang, Z.; Liu, Y.; Song, A.; Hao, J. G-quadruplex-based ionogels with controllable chirality for circularly polarized luminescence. Colloids Surf. A Physicochem. Eng. Asp. 2021, 629, 127411. [Google Scholar] [CrossRef]
- Li, Y.; Chi, J.; Xu, P.; Dong, X.; Le, A.-T.; Shi, K.; Liu, Y.; Xiao, J. Supramolecular G-quadruplex hydrogels: Bridging fabrication to biomedical application. J. Mater. Sci. Technol. 2023, 155, 238–252. [Google Scholar] [CrossRef]
- Fang, J.; Zheng, L.; Liu, Y.; Peng, Y.; Yang, Q.; Huang, Y.; Zhang, J.; Luo, L.; Shen, D.; Tan, Y. Smart G-quadruplex hydrogels: From preparations to comprehensive applications. Int. J. Biol. Macromol. 2023, 247, 125614. [Google Scholar] [CrossRef]
- Roxo, C.; Kotkowiak, W.; Pasternak, A. G-quadruplex-forming aptamers—Characteristics, applications, and perspectives. Molecules 2019, 24, 3781. [Google Scholar] [CrossRef]
- Bidzinska, J.; Cimino-Reale, G.; Zaffaroni, N.; Folini, M. G-quadruplex structures in the human genome as novel therapeutic targets. Molecules 2013, 18, 12368–12395. [Google Scholar] [CrossRef] [PubMed]
- Asamitsu, S.; Obata, S.; Yu, Z.; Bando, T.; Sugiyama, H. Recent progress of targeted G-quadruplex-preferred ligands toward cancer therapy. Molecules 2019, 24, 429. [Google Scholar] [CrossRef] [PubMed]
- Alessandrini, I.; Recagni, M.; Zaffaroni, N.; Folini, M. On the road to fight cancer: The potential of G-quadruplex ligands as novel therapeutic agents. Int. J. Mol. Sci. 2021, 22, 5947. [Google Scholar] [CrossRef] [PubMed]
- Santos, T.; Salgado, G.F.; Cabrita, E.J.; Cruz, C. G-quadruplexes and their ligands: Biophysical methods to unravel G-quadruplex/ligand interactions. Pharmaceuticals 2021, 14, 769. [Google Scholar] [CrossRef] [PubMed]
- Ruggiero, E.; Zanin, I.; Terreri, M.; Richter, S.N. G-quadruplex targeting in the fight against viruses: An update. Int. J. Mol. Sci. 2021, 22, 10984. [Google Scholar] [CrossRef] [PubMed]
- Marzano, M.; Falanga, A.P.; Marasco, D.; Borbone, N.; D’Errico, S.; Piccialli, G.; Roviello, G.N.; Oliviero, G. Evaluation of an analogue of the marine ε-PLL peptide as a ligand of G-quadruplex DNA structures. Mar. Drugs 2020, 18, 49. [Google Scholar] [CrossRef]
- Tanaka, S.; Yukami, S.; Hachiro, Y.; Ohya, Y.; Kuzuya, A. Application of DNA quadruplex hydrogels prepared from polyethylene glycol-oligodeoxynucleotide conjugates to cell culture media. Polymers 2019, 11, 1607. [Google Scholar] [CrossRef]
- Huang, Z.; Kangovi, G.N.; Wen, W.; Lee, S.; Niu, L. An RNA aptamer capable of forming a hydrogel by self-assembly. Biomacromolecules 2017, 18, 2056–2063. [Google Scholar] [CrossRef]
- Ahn, S.Y.; Kim, J.; Vellampatti, S.; Oh, S.; Lim, Y.T.; Park, S.H.; Luo, D.; Chung, J.; Um, S.H. Protein-Encoding Free-Standing RNA Hydrogel for Sub-Compartmentalized Translation. Adv. Mater. 2022, 34, 2110424. [Google Scholar] [CrossRef]
- Han, S.; Park, Y.; Kim, H.; Nam, H.; Ko, O.; Lee, J.B. Double controlled release of therapeutic RNA modules through injectable DNA–RNA hybrid hydrogel. ACS Appl. Mater. Interfaces 2020, 12, 55554–55563. [Google Scholar] [CrossRef]
- Brown, J.A. Unraveling the structure and biological functions of RNA triple helices. Wiley Interdiscip. Rev. RNA 2020, 11, e1598. [Google Scholar] [CrossRef] [PubMed]
- Conrad, N.K. The emerging role of triple helices in RNA biology. Wiley Interdiscip. Rev. RNA 2014, 5, 15–29. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, R.; Längst, G. The chromatin–triple helix connection. Biol. Chem. 2023, 404, 1037–1049. [Google Scholar] [CrossRef] [PubMed]
- Conde, J.; Oliva, N.; Atilano, M.; Song, H.S.; Artzi, N. Self-assembled RNA-triple-helix hydrogel scaffold for microRNA modulation in the tumour microenvironment. Nat. Mater. 2016, 15, 353–363. [Google Scholar] [CrossRef]
- Li, J.; Yuan, D.; Zheng, X.; Zhang, X.; Li, X.; Zhang, S. A triple-combination nanotechnology platform based on multifunctional RNA hydrogel for lung cancer therapy. Sci. China Chem. 2020, 63, 546–553. [Google Scholar] [CrossRef]
- Wang, W.; Liu, X.; Ding, L.; Jin, H.J.; Li, X. Rna hydrogel combined with MnO2 nanoparticles as a nano-vaccine to treat triple negative breast cancer. Front. Chem. 2021, 9, 797094. [Google Scholar] [CrossRef]
- Ma, Y.; Duan, X.; Huang, J. DNA Hydrogels as Functional Materials and Their Biomedical Applications. Adv. Funct. Mater. 2023, 2309070. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scognamiglio, P.L.; Vicidomini, C.; Roviello, G.N. Dancing with Nucleobases: Unveiling the Self-Assembly Properties of DNA and RNA Base-Containing Molecules for Gel Formation. Gels 2024, 10, 16. https://doi.org/10.3390/gels10010016
Scognamiglio PL, Vicidomini C, Roviello GN. Dancing with Nucleobases: Unveiling the Self-Assembly Properties of DNA and RNA Base-Containing Molecules for Gel Formation. Gels. 2024; 10(1):16. https://doi.org/10.3390/gels10010016
Chicago/Turabian StyleScognamiglio, Pasqualina Liana, Caterina Vicidomini, and Giovanni N. Roviello. 2024. "Dancing with Nucleobases: Unveiling the Self-Assembly Properties of DNA and RNA Base-Containing Molecules for Gel Formation" Gels 10, no. 1: 16. https://doi.org/10.3390/gels10010016
APA StyleScognamiglio, P. L., Vicidomini, C., & Roviello, G. N. (2024). Dancing with Nucleobases: Unveiling the Self-Assembly Properties of DNA and RNA Base-Containing Molecules for Gel Formation. Gels, 10(1), 16. https://doi.org/10.3390/gels10010016







