Transglutaminase-Induced Polymerization of Pea and Chickpea Protein to Enhance Functionality
Abstract
:1. Introduction
2. Results and Discussion
2.1. Impact of the TG Treatment on the Protein Structural Characteristics
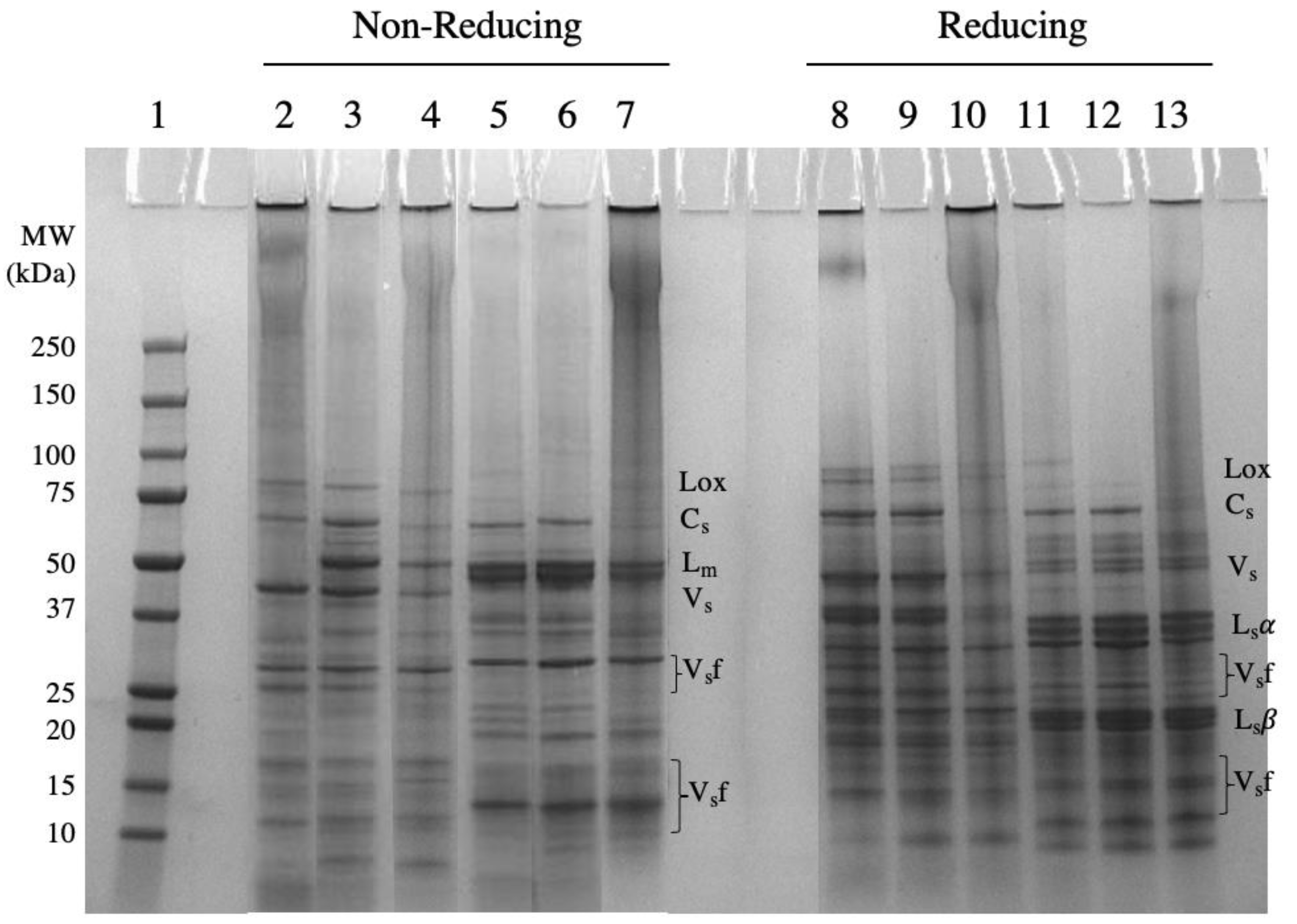
2.1.1. Protein Profile
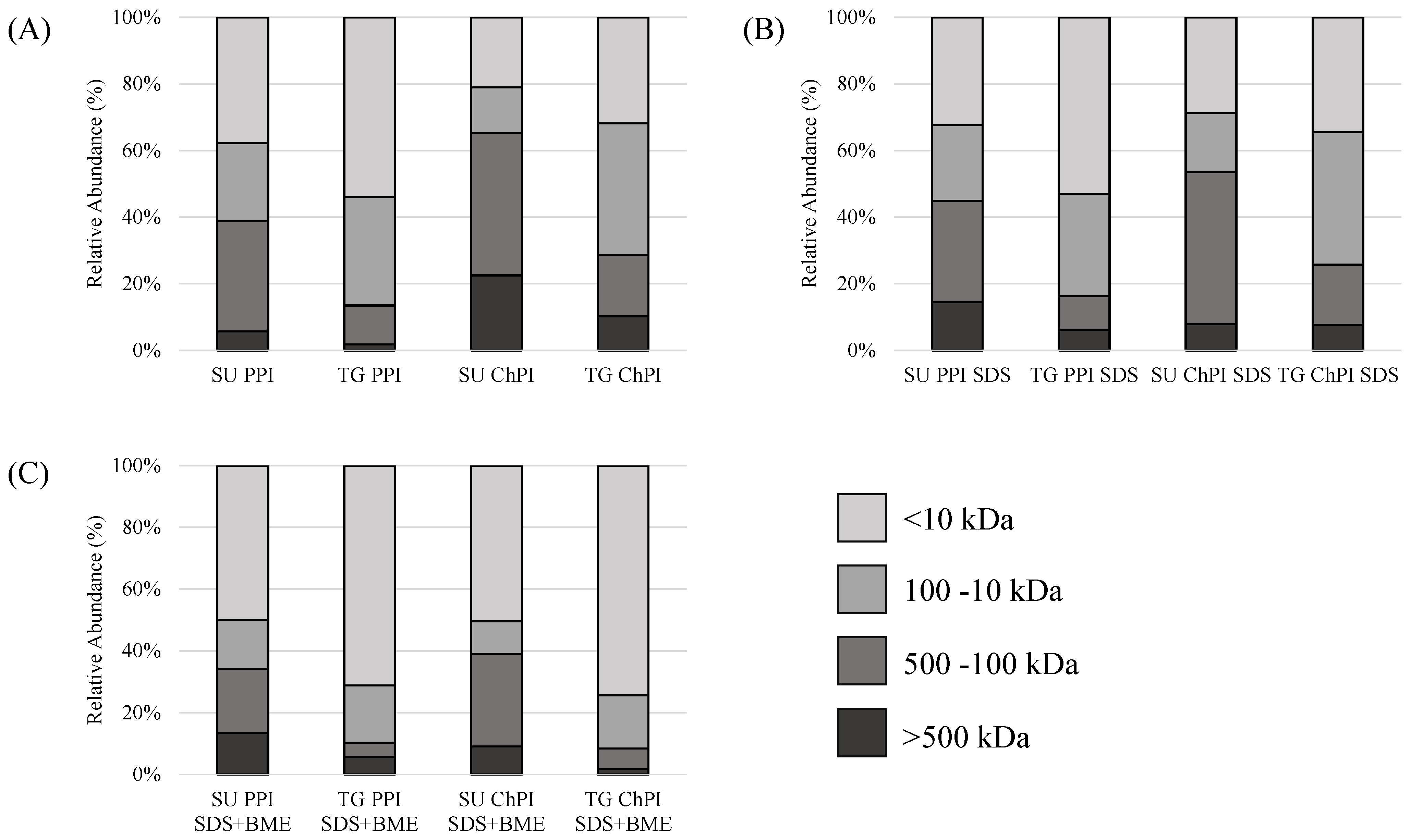
2.1.2. Protein Molecular Weight Distribution and Polymerization
2.1.3. Protein Denaturation
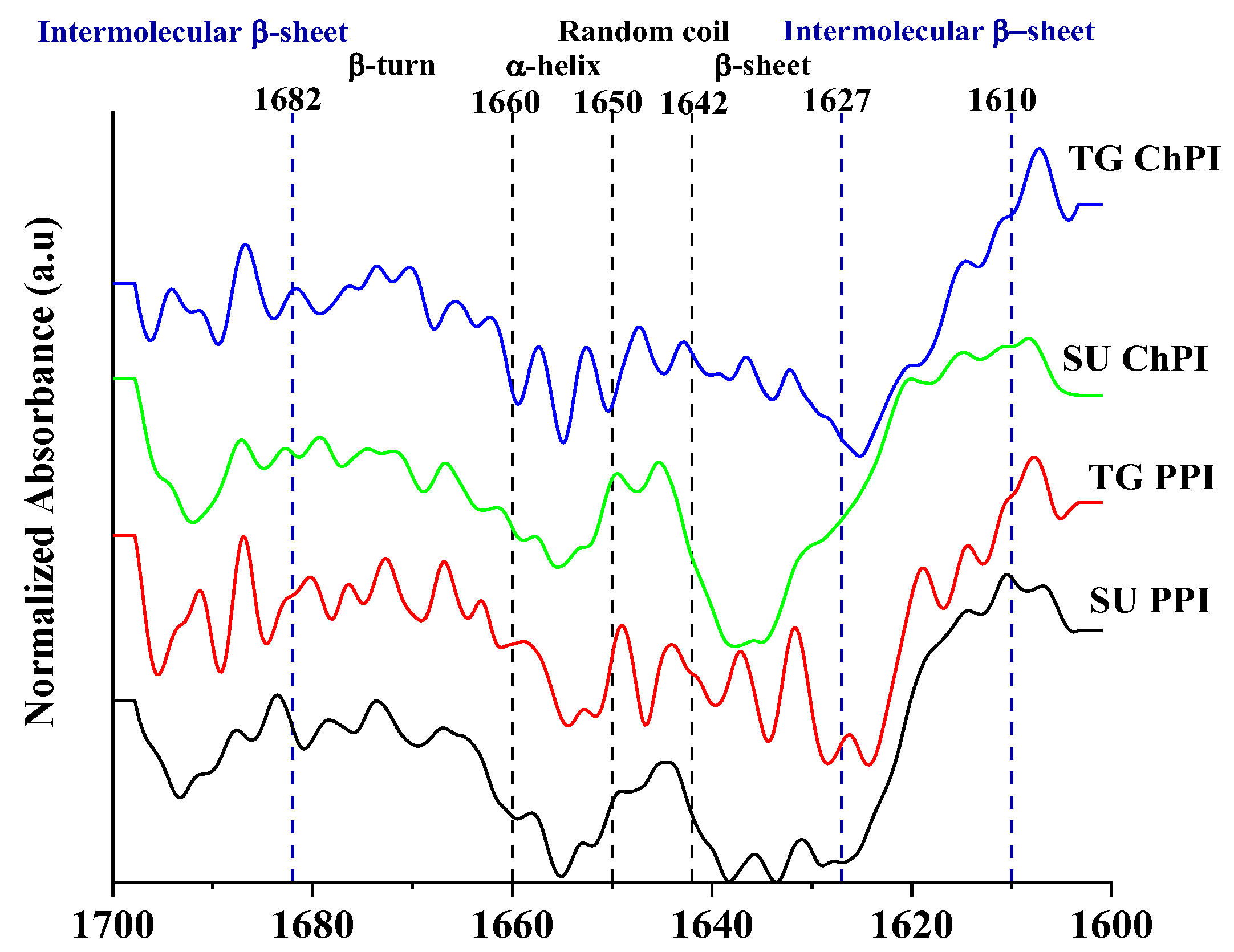
2.1.4. Protein Secondary Structure
2.1.5. Protein Surface Properties
2.2. Impact of the TG-Induced Polymerization on the Protein Functional Properties
2.2.1. Solubility
2.2.2. Gel Strength
2.2.3. Emulsion Capacity
3. Conclusions
4. Materials and Methods
4.1. Materials
4.2. Production of the PPI and ChPI
4.3. Selection of the TGase Treatment Conditions and Production of the TG PPI and TG ChPI
4.4. Protein Structural Characterization
4.4.1. Protein Profiling by Gel Electrophoresis
4.4.2. Protein Molecular Weight Distribution via Size-Exclusion High-Performance Liquid Chromatography (SE-HPLC)
4.4.3. Protein Denaturation
4.4.4. Protein Secondary Structure by Attenuated Total Reflectance Fourier Transform Infrared Spectroscopy (ATR-FTIR)
4.4.5. Surface Properties of the Protein Ingredients
4.5. Protein Functional Characterization
4.5.1. Protein Solubility
4.5.2. Gel Strength
4.5.3. Emulsion Capacity
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Global Newswire. Plant-based Food Market Size More Than Doubles to Touch USD 22.3 Billion with the CAGR of 11.82% by 2029|BlueWeave Consulting. 2023. Available online: https://www.globenewswire.com/en/news-release/2023/04/25/2654175/0/en/Plant-based-Food-Market-Size-More-Than-Doubles-to-Touch-USD-22-3-Billion-with-the-CAGR-of-11-82-by-2029-BlueWeave-Consulting.html (accessed on 30 August 2023).
- Plant Based Foods. 2022 U.S. Retail Sales Data for The Plant-Based Foods Industry. 2023. Available online: https://www.plantbasedfoods.org/2022-u-s-retail-sales-data-for-the-plant-based-foods-industry/ (accessed on 28 August 2023).
- Ignaszewski, E. 2021 U.S. Retail Market Insights Plant-based Foods. Good Food Institute. 2022. Available online: https://gfi.org/wp-content/uploads/2022/03/2021-U.S.-retail-market-insights_Plant-based-foods-GFI.pdf (accessed on 30 August 2022).
- Samard, S.; Gu, B.; Ryu, G.-H. Effects of extrusion types, screw speed and addition of wheat gluten on physicochemical characteristics and cooking stability of meat analogues. J. Sci. Food Agric. 2019, 99, 4922–4931. [Google Scholar] [CrossRef] [PubMed]
- Yuliarti, O.; Kovis, T.J.K.; Yi, N.J. Structuring the meat analogue by using plant-based derived composites. J. Food Eng. 2021, 288, 110138. [Google Scholar] [CrossRef]
- Chiang, J.H.; Loveday, S.M.; Hardacre, A.K.; Parker, M.E. Effects of soy protein to wheat gluten ratio on the physicochemical properties of extruded meat analogues. Food Struct. 2019, 19, 100102. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, Y.; Zhang, T.; Zhang, Y.; Jiang, L.; Sui, X. High moisture extrusion of soy protein and wheat gluten blend: An underlying mechanism for the formation of fibrous structures. LWT 2022, 163, 113561. [Google Scholar] [CrossRef]
- Doxzen, K.; Henderson, H. Is this safe? Addressing societal concerns about crispr-edited foods without reinforcing GMO framing. Environ. Commun. 2020, 14, 865–871. [Google Scholar] [CrossRef]
- Zheng, Q.; Wang, H.H. Do consumers view the genetically modified food labeling systems differently? “contains GMO” versus “Non-GMO” labels. Chin. Econ. 2021, 54, 376–388. [Google Scholar] [CrossRef]
- Igbinedion, S.O.; Ansari, J.; Vasikaran, A.; Gavins, F.N.; Jordan, P.; Boktor, M.; Alexander, J.S. Non-celiac gluten sensitivity: All wheat attack is not celiac. World J. Gastroenterol. 2017, 23, 7201–7210. [Google Scholar] [CrossRef]
- Rubio-Tapia, A.; Ludvigsson, J.F.; Brantner, T.L.; Murray, J.A.; Everhart, J.E. The prevalence of celiac disease in the United States. Off. J. Am. Coll. Gastroenterol. |ACG 2012, 107, 1538–1544. [Google Scholar] [CrossRef]
- Hansen, L.; Bu, F.; Ismail, B.P. Structure-function guided extraction and scale-up of pea protein isolate production. Foods 2022, 11, 3773. [Google Scholar] [CrossRef]
- Ladjal-Ettoumi, Y.; Boudries, H.; Chibane, M.; Romero, A. Pea, Chickpea and lentil protein isolates: Physicochemical characterization and emulsifying properties. Food Biophys. 2016, 11, 43–51. [Google Scholar] [CrossRef]
- Yaputri, B.P.; Bu, F.; Ismail, B.P. Salt Solubilization Coupled with Membrane Filtration-Impact on the Structure/Function of Chickpea Compared to Pea Protein. Foods 2023, 12, 1694. [Google Scholar] [CrossRef] [PubMed]
- Sajib, M.; Forghani, B.; Vate, N.K.; Abdollahi, M. Combined effects of isolation temperature and pH on functionality and beany flavor of pea protein isolates for meat analogue applications. Food Chem. 2023, 412, 135585. [Google Scholar] [CrossRef] [PubMed]
- Schreuders, F.K.; Dekkers, B.L.; Bodnár, I.; Erni, P.; Boom, R.M.; van der Goot, A.J. Comparing structuring potential of pea and soy protein with gluten for meat analogue preparation. J. Food Eng. 2019, 261, 32–39. [Google Scholar] [CrossRef]
- Webb, D.; Plattner, B.J.; Donald, E.; Funk, D.; Plattner, B.S.; Alavi, S. Role of chickpea flour in texturization of extruded pea protein. J. Food Sci. 2020, 85, 4180–4187. [Google Scholar] [CrossRef]
- Ma, K.K.; Greis, M.; Lu, J.; Nolden, A.A.; McClements, D.J.; Kinchla, A.J. Functional performance of plant proteins. Foods 2022, 11, 594. [Google Scholar] [CrossRef] [PubMed]
- Withana-Gamage, T.S.; Wanasundara, J.P.; Pietrasik, Z.; Shand, P.J. Physicochemical, thermal and functional characterisation of protein isolates from Kabuli and Desi chickpea (Cicer arietinum L.): A comparative study with soy (Glycine max) and pea (Pisum sativum L.). J. Sci. Food Agric. 2011, 91, 1022–1031. [Google Scholar] [CrossRef] [PubMed]
- Stone, A.K.; Karalash, A.; Tyler, R.T.; Warkentin, T.D.; Nickerson, M.T. Functional attributes of pea protein isolates prepared using different extraction methods and cultivars. Food Res. Int. 2015, 76, 31–38. [Google Scholar] [CrossRef]
- Yang, J.; Zamani, S.; Liang, L.; Chen, L. Extraction methods significantly impact pea protein composition, structure and gelling properties. Food Hydrocoll. 2021, 117, 106678. [Google Scholar] [CrossRef]
- Husband, H.; Ferreira, S.; Bu, F.; Feyzi, S.; Ismail, B.P. Pea protein globulins: Does their relative ratio matter? Food Hydrocoll. 2023, 148, 109429. [Google Scholar] [CrossRef]
- Bu, F.; Nayak, G.; Bruggeman, P.; Annor, G.; Ismail, B.P. Impact of plasma reactive species on the structure and functionality of pea protein isolate. Food Chem. 2022, 371, 131135. [Google Scholar] [CrossRef]
- Bu, F.; Feyzi, S.; Nayak, G.; Mao, Q.; Kondeti, V.S.K.; Bruggeman, P.; Chen, C.; Ismail, B.P. Investigation of novel cold atmospheric plasma sources and their impact on the structural and functional characteristics of pea protein. Innov. Food Sci. Emerg. Technol. 2023, 83, 103248. [Google Scholar] [CrossRef]
- Hall, A.E.; Moraru, C.I. Structure and function of pea, lentil and faba bean proteins treated by high pressure processing and heat treatment. LWT 2021, 152, 112349. [Google Scholar] [CrossRef]
- Zhang, S.; Han, J.; Chen, L. Fabrication of pea protein gels with modulated rheological properties using high pressure processing. Food Hydrocoll. 2023, 144, 109002. [Google Scholar] [CrossRef]
- Chao, D.; Aluko, R.E. Modification of the structural, emulsifying, and foaming properties of an isolated pea protein by thermal pretreatment. CyTA-J. Food 2018, 16, 357–366. [Google Scholar] [CrossRef]
- Habiba, R. Changes in anti-nutrients, protein solubility, digestibility, and HCl-extractability of ash and phosphorus in vegetable peas as affected by cooking methods. Food Chem. 2002, 77, 187–192. [Google Scholar] [CrossRef]
- Mirmoghtadaie, L.; Aliabadi, S.S.; Hosseini, S.M. Recent approaches in physical modification of protein functionality. Food Chem. 2016, 199, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Han, M.Y.; Zu, H.Z.; Xu, X.L.; Zhou, G.H. Microbial transglutaminase catalyzed the cross-linking of myofibrillar/soy protein isolate mixtures. J. Food Process. Preserv. 2015, 39, 309–317. [Google Scholar] [CrossRef]
- Shand, P.; Ya, H.; Pietrasik, Z.; Wanasundara, P. Transglutaminase treatment of pea proteins: Effect on physicochemical and rheological properties of heat-induced protein gels. Food Chem. 2008, 107, 692–699. [Google Scholar] [CrossRef]
- Sun, X.D.; Arntfield, S.D. Gelation properties of salt-extracted pea protein isolate catalyzed by microbial transglutaminase cross-linking. Food Hydrocoll. 2011, 25, 25–31. [Google Scholar] [CrossRef]
- Kieliszek, M.; Misiewicz, A. Microbial transglutaminase and its application in the food industry. A review. Folia Microbiol. 2014, 59, 241–250. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, P.; Zou, M.; Yang, R.; Tian, M.; Gu, Z. Microbial transglutaminase-modified protein network and its importance in enhancing the quality of high-fiber tofu with okara. Food Chem. 2019, 289, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.; Xiong, S.; Hu, Y.; Yin, T.; You, J. In vitro pepsin digestion of silver carp (Hypophthalmichthys molitrix) surimi gels after cross-linking by Microbial Transglutaminase (MTGase). Food Hydrocoll. 2019, 95, 152–160. [Google Scholar] [CrossRef]
- Forghani, Z.; Eskandari, M.H.; Aminlari, M.; Shekarforoush, S.S. Effects of microbial transglutaminase on physicochemical properties, electrophoretic patterns and sensory attributes of veggie burger. J. Food Sci. Technol. 2017, 54, 2203–2213. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Damodaran, S.; Heinonen, M. Effects of microbial transglutaminase treatment on physiochemical properties and emulsifying functionality of faba bean protein isolate. LWT 2019, 99, 396–403. [Google Scholar] [CrossRef]
- Mostafa, H.S. Microbial transglutaminase: An overview of recent applications in food and packaging. Biocatal. Biotransformation 2020, 38, 161–177. [Google Scholar] [CrossRef]
- Yasir, S.B.M.; Sutton, K.; Newberry, M.; Andrews, N.; Gerrard, J. The impact of transglutaminase on soy proteins and tofu texture. Food Chem. 2007, 104, 1491–1501. [Google Scholar] [CrossRef]
- Djoullah, A.; Djemaoune, Y.; Husson, F.; Saurel, R. Native-state pea albumin and globulin behavior upon transglutaminase treatment. Process. Biochem. 2015, 50, 1284–1292. [Google Scholar] [CrossRef]
- Espinosa-Ramírez, J.; Serna-Saldívar, S.O. Wet-milled chickpea coproduct as an alternative to obtain protein isolates. LWT 2019, 115, 108468. [Google Scholar] [CrossRef]
- Glusac, J.; Isaschar-Ovdat, S.; Fishman, A. Transglutaminase modifies the physical stability and digestibility of chickpea protein-stabilized oil-in-water emulsions. Food Chem. 2020, 315, 126301. [Google Scholar] [CrossRef]
- Chang, Y.-W.; Alli, I.; Molina, A.T.; Konishi, Y.; Boye, J.I. Isolation and characterization of chickpea (Cicer arietinum L.) seed protein fractions. Food Bioprocess Technol. 2012, 5, 618–625. [Google Scholar] [CrossRef]
- Gao, Z.; Shen, P.; Lan, Y.; Cui, L.; Ohm, J.-B.; Chen, B.; Rao, J. Effect of alkaline extraction pH on structure properties, solubility, and beany flavor of yellow pea protein isolate. Food Res. Int. 2020, 131, 109045. [Google Scholar] [CrossRef] [PubMed]
- Damodaran, S.; Parkin, K.L. Amino acids, peptides, and proteins. In Fennema’s Food Chemistry; CRC Press: Boca Raton, FL, USA, 2017; pp. 235–356. [Google Scholar]
- Phillips, G.O.; Williams, P.A. (Eds.) Handbook of Food Proteins; Elsevier: Amsterdam, The Netherlands, 2011. [Google Scholar]
- Gouvêa, L.d.P.; Caldeira, R.; Azevedo, T.d.L.; Galdeano, M.C.; Felberg, I.; Lima, J.R.; Mellinger, C.G. Physical and techno-functional properties of a common bean protein concentrate compared to commercial legume ingredients for the plant-based market. Food Hydrocoll. 2023, 137, 108351. [Google Scholar] [CrossRef]
- Romano, A.; Giosafatto, C.V.L.; Di Pierro, P.; Romano, R.; Masi, P.; Mariniello, L. Impact of transglutaminase treatment on properties and in vitro digestibility of white bean (Phaseolus vulgaris L.) flour. Food Res. Int. 2016, 88, 239–246. [Google Scholar] [CrossRef]
- Zhao, L.; Chen, M.H.; Bi, X.; Du, J. Physicochemical properties, structural characteristics and in vitro digestion of brown rice–pea protein isolate blend treated by microbial transglutaminase. Food Hydrocoll. 2023, 141, 108673. [Google Scholar] [CrossRef]
- O’Kane, F.E.; Happe, R.P.; Vereijken, J.M.; Gruppen, H.; van Boekel, M.A.J.S. Heat-induced gelation of pea legumin: Comparison with soybean glycinin. J. Agric. Food Chem. 2004, 52, 5071–5078. [Google Scholar] [CrossRef] [PubMed]
- Carbonaro, M.; Nucara, A. Secondary structure of food proteins by Fourier transform spectroscopy in the mid-infrared region. Amino Acids 2010, 38, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Long, G.; Ji, Y.; Pan, H.; Sun, Z.; Li, Y.; Qin, G. Characterization of thermal denaturation structure and morphology of soy glycinin by FTIR and SEM. Int. J. Food Prop. 2015, 18, 763–774. [Google Scholar] [CrossRef]
- Feyzi, S.; Varidi, M.; Zare, F.; Varidi, M.J. Effect of drying methods on the structure, thermo and functional properties of fenugreek (Trigonella foenum graecum) protein isolate. J. Sci. Food Agric. 2018, 98, 1880–1888. [Google Scholar] [CrossRef]
- Sadat, A.; Joye, I.J. Peak fitting applied to fourier transform infrared and raman spectroscopic analysis of proteins. Appl. Sci. 2020, 10, 5918. [Google Scholar] [CrossRef]
- Carbonaro, M.; Maselli, P.; Nucara, A. Relationship between digestibility and secondary structure of raw and thermally treated legume proteins: A Fourier transform infrared (FT-IR) spectroscopic study. Amino Acids 2012, 43, 911–921. [Google Scholar] [CrossRef]
- Moreno, H.M.; Domínguez-Timón, F.; Díaz, M.T.; Pedrosa, M.M.; Borderías, A.J.; Tovar, C.A. Evaluation of gels made with different commercial pea protein isolate: Rheological, structural and functional properties. Food Hydrocoll. 2020, 99, 105375. [Google Scholar] [CrossRef]
- Cao, Y.; Mezzenga, R. Food protein amyloid fibrils: Origin, structure, formation, characterization, applications and health implications. Adv. Colloid Interface Sci. 2019, 269, 334–356. [Google Scholar] [CrossRef] [PubMed]
- Shevkani, K.; Singh, N.; Kaur, A.; Rana, J.C. Structural and functional characterization of kidney bean and field pea protein isolates: A comparative study. Food Hydrocoll. 2015, 43, 679–689. [Google Scholar] [CrossRef]
- Zhu, X.; Zhang, J.; Liu, S.; Gu, Y.; Yu, X.; Gao, F.; Wang, R. Relationship between Molecular Structure and Heat-Induced Gel Properties of Duck Myofibrillar Proteins Affected by the Addition of Pea Protein Isolate. Foods 2022, 11, 1040. [Google Scholar] [CrossRef]
- Nivala, O.; Nordlund, E.; Kruus, K.; Ercili-Cura, D. The effect of heat and transglutaminase treatment on emulsifying and gelling properties of faba bean protein isolate. LWT 2021, 139, 110517. [Google Scholar] [CrossRef]
- Nivala, O.; Mäkinen, O.E.; Kruus, K.; Nordlund, E.; Ercili-Cura, D. Structuring colloidal oat and faba bean protein particles via enzymatic modification. Food Chem. 2017, 231, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, A.L.C.; de Góes-Favoni, S.P. Action of microbial transglutaminase (MTGase) in the modification of food proteins: A review. Food Chem. 2015, 171, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Tanger, C.; Engel, J.; Kulozik, U. Influence of extraction conditions on the conformational alteration of pea protein extracted from pea flour. Food Hydrocoll. 2020, 107, 105949. [Google Scholar] [CrossRef]
- Shen, Y.; Hong, S.; Singh, G.; Koppel, K.; Li, Y. Improving functional properties of pea protein through “green” modifications using enzymes and polysaccharides. Food Chem. 2022, 385, 132687. [Google Scholar] [CrossRef]
- Babiker, E.E. Effect of transglutaminase treatment on the functional properties of native and chymotrypsin-digested soy protein. Food Chem. 2000, 70, 139–145. [Google Scholar] [CrossRef]
- Popović, L.M.; Peričin, D.M.; Vaštag, Ž.G.; Popović, S.Z. Optimization of transglutaminase cross-linking of pumpkin oil cake globulin; improvement of the solubility and gelation properties. Food Bioprocess Technol. 2013, 6, 1105–1111. [Google Scholar] [CrossRef]
- Motoki, M.; Seguro, K.; Nio, N.; Takinami, K. Glutamine-specific deamidation of α S1-casein by transglutaminase. Agric. Biol. Chem. 1986, 50, 3025–3030. [Google Scholar]
- Papalamprou, E.; Doxastakis, G.; Biliaderis, C.; Kiosseoglou, V. Influence of preparation methods on physicochemical and gelation properties of chickpea protein isolates. Food Hydrocoll. 2009, 23, 337–343. [Google Scholar] [CrossRef]
- Sun, X.D.; Arntfield, S.D. Gelation properties of salt-extracted pea protein induced by heat treatment. Food Res. Int. 2010, 43, 509–515. [Google Scholar] [CrossRef]
- Ding, X.; Zeng, N.; Zhang, G.; Pan, J.; Hu, X.; Gong, D. Influence of transglutaminase-assisted ultrasound treatment on the structure and functional properties of soy protein isolate. J. Food Process. Preserv. 2019, 43, e14203. [Google Scholar] [CrossRef]
- Moreno, H.M.; Tovar, C.A.; Dominguez-Timon, F.; Cano-Baez, J.; Díaz, M.T.; Pedrosa, M.M.; Boderías, A.J. Gelation of commercial pea protein isolate: Effect of microbial transglutaminase and thermal processing. Food Science and Technology 2020, 40, 800–809. [Google Scholar] [CrossRef]
- O’Kane, F.E.; Vereijken, J.M.; Gruppen, H.; Van Boekel, M.A. Gelation behavior of protein isolates extracted from 5 cultivars of Pisum sativum L. J. Food Sci. 2005, 70, C132–C137. [Google Scholar] [CrossRef]
- Qin, X.; Luo, S.; Cai, J.; Zhong, X.; Jiang, S.; Zheng, Z.; Zhao, Y. Effects of microwave pretreatment and transglutaminase crosslinking on the gelation properties of soybean protein isolate and wheat gluten mixtures. J. Sci. Food Agric. 2016, 96, 3559–3566. [Google Scholar] [CrossRef]
- Lam, A.C.Y.; Can Karaca, A.; Tyler, R.T.; Nickerson, M.T. Pea protein isolates: Structure, extraction, and functionality. Food Rev. Int. 2018, 34, 126–147. [Google Scholar] [CrossRef]
- Mession, J.-L.; Chihi, M.L.; Sok, N.; Saurel, R. Effect of globular pea proteins fractionation on their heat-induced aggregation and acid cold-set gelation. Food Hydrocoll. 2015, 46, 233–243. [Google Scholar] [CrossRef]
- Karaca, A.C.; Low, N.; Nickerson, M. Emulsifying properties of chickpea, faba bean, lentil and pea proteins produced by isoelectric precipitation and salt extraction. Food Res. Int. 2011, 44, 2742–2750. [Google Scholar] [CrossRef]
- Tang, C.-H.; Sun, X.; Yin, S.-W.; Ma, C.-Y. Transglutaminase-induced cross-linking of vicilin-rich kidney protein isolate: Influence on the functional properties and in vitro digestibility. Food Res. Int. 2008, 41, 941–947. [Google Scholar] [CrossRef]
- Liang, H.-N.; Tang, C.-H. Emulsifying and Interfacial properties of vicilins: Role of conformational flexibility at quaternary and/or tertiary levels. J. Agric. Food Chem. 2013, 61, 11140–11150. [Google Scholar] [CrossRef] [PubMed]
- Boyle, C.; Hansen, L.; Hinnenkamp, C.; Ismail, B.P. Emerging camelina protein: Extraction, modification, and structural/functional characterization. J. Am. Oil Chem. Soc. 2018, 95, 1049–1062. [Google Scholar] [CrossRef]
| Relative Abundance 1 (%) of Protein Fractions | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Samples | Phosphate Buffer 2 | Phosphate Buffer (0.1% SDS) 3 | Phosphate Buffer (0.1% SDS + 2.5% βME) 4 | |||||||||
| Soluble Aggregates (600–1200 kDa) | Legumin (~450 kDa) | Convicilin (~250 kDa) | Vicilin (~160 kDa) | Soluble Aggregates | Legumin | Convicilin | Vicilin | Soluble Aggregates | Legumin | Convicilin | Vicilin | |
| PPI | 5.73 ± 0.90 a,5 | 21.2 ± 0.15 a | 6.16 ± 0.23 a | 5.01 ± 0.17 a | 14.4 ± 1.53 a | 19.5 ± 0.47 a | 5.54 ± 0.09 a | 4.64 ± 0.08 a | 13.4 ± 1.29 a | 13.0 ± 0.15 a | 3.92 ± 0.04 a | 3.17 ± 0.07 a |
| TG PPI | 1.77 ± 0.07 b | 5.83 ± 0.02 b | 3.84 ± 0.01 b | 2.03 ± 0.01 b | 6.21 ± 0.02 b | 4.05 ± 0.05 b | 3.81 ± 0.04 b | 2.19 ± 0.02 b | 4.71 ± 0.01 b | 2.00 ± 0.01 b | 2.19 ± 0.01 a | 1.34 ± 0.00 b |
| ChPI | 22.5 ± 1.67 A,6 | 32.9 ± 0.78 A | 2.47 ± 0.08 B | 7.42 ± 0.17 A | 7.82 ± 0.16 A | 32.6 ± 0.56 A | 2.98 ± 0.01 B | 10.2 ± 0.12 A | 9.08 ± 0.24 A | 22.6 ± 0.03 A | 1.57 ± 0.01 A | 5.80 ± 0.01 A |
| TG ChPI | 10.2 ± 0.15 B | 6.70 ± 0.19 B | 5.25 ± 0.13 A | 6.55 ± 0.16 A | 7.62 ± 0.03 A | 5.91 ± 0.01 B | 4.85 ± 0.02 A | 7.33 ± 0.04 B | 1.80 ± 0.01 B | 2.65 ± 0.04 B | 1.65 ± 0.02 A | 2.30 ± 0.00 B |
| Samples | Denaturation Temperature and Enthalpy | Surface Properties | |||
|---|---|---|---|---|---|
| Denaturation Temperature | H) | Surface Hydrophobicity | Surface Charge | ||
| C | J g−1 | RFI 1 | mV | ||
| -conglycinin | Glycinin | ||||
| cSPI | * 2 | * | * | 10,800 ± 530.3 c | −41.3 ± 0.20 a |
| Vicilin (7S) | Legumin (11S) | ||||
| cPPI | * | * | * | 13,800 ± 434.4 a | −30.2 ± 0.13 cd |
| PPI | 82.6 ± 0.13 a,3 | 89.9 ± 0.16 c | 5.45 ± 0.07 b | 14,200 ± 105.9 a | −27.2 ± 0.07 ef |
| TG PPI | * | * | * | 12,900 ± 203.6 ab | −28.8 ± 0.28 de |
| cChPC | 81.5 ± 0.09 b | 99.6 ± 0.02 a | 3.77 ± 0.09 c | 13,300 ± 450.4 a | −25.7 ± 0.33 f |
| ChPI | 80.5 ± 0.07 c | 90.8 ± 0.17 b | 8.61 ± 0.14 a | 8970 ± 186.5 d | −30.9 ± 0.21 bc |
| TG ChPI | * | * | * | 11,600 ± 255.3 bc | −32.2 ± 0.17 b |
| Sample | Intermolecular β-Sheet | β-Sheet | α-Helix | β-Turn | Random Coil |
|---|---|---|---|---|---|
| PPI | 39.8 ± 0.39 * | 32.1 ± 0.53 * | 18.7 ± 0.28 * | 8.10 ± 0.72 * | 2.13 ± 0.50 * |
| TG PPI | 58.1 ± 2.26 | 11.7 ± 2.08 | 13.3 ± 0.34 | 12.1 ± 0.50 | 4.71 ± 0.02 |
| ChPI | 17.5 ± 1.04 * | 55.7 ± 1.01 * | 17.9 ± 1.29 | 7.81 ± 0.40 * | 1.08 ± 0.11 * |
| TG ChPI | 47.8 ± 0.54 | 11.6 ± 0.09 | 16.1 ± 0.29 | 17.9 ± 0.56 | 6.59 ± 0.68 |
| Samples | % Protein Solubility | Gel Strength 1 (N) | Emulsion Capacity (mL oil/g Protein) | |
|---|---|---|---|---|
| Nonheated | Heated 2 | |||
| cSPI | 66.8 ± 0.40 c,3 | 78.5 ± 0.39 b,*,4 | 19.2 ± 0.09 a | 1120 ± 10.7 a |
| cPPI | 29.5 ± 0.85 d | 57.1 ± 0.64 e,* | 11.5 ± 0.16 d | 769 ± 6.20 d |
| PPI | 68.9 ± 0.42 bc | 69.9 ± 0.90 c | 10.7 ± 0.11 d | 856 ± 10.7 b |
| TG PPI | 20.8 ± 1.07 e | 25.6 ± 0.44 g,* | 16.2 ± 0.26 b | 843 ± 6.20 bc |
| cChPC | 70.7 ± 0.31 b | 66.7 ± 0.47 d,* | 13.3 ± 0.33 c | 750 ± 16.4 d |
| ChPI | 94.3 ± 0.69 a | 92.5 ± 0.76 a | 15.3 ± 0.17 b | 794 ± 12.4 cd |
| TG ChPI | 23.4 ± 1.5 e | 32.6 ± 0.39 f,* | 18.4 ± 0.42 a | 688 ± 11 e |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yaputri, B.P.; Feyzi, S.; Ismail, B.P. Transglutaminase-Induced Polymerization of Pea and Chickpea Protein to Enhance Functionality. Gels 2024, 10, 11. https://doi.org/10.3390/gels10010011
Yaputri BP, Feyzi S, Ismail BP. Transglutaminase-Induced Polymerization of Pea and Chickpea Protein to Enhance Functionality. Gels. 2024; 10(1):11. https://doi.org/10.3390/gels10010011
Chicago/Turabian StyleYaputri, Brigitta P., Samira Feyzi, and Baraem P. Ismail. 2024. "Transglutaminase-Induced Polymerization of Pea and Chickpea Protein to Enhance Functionality" Gels 10, no. 1: 11. https://doi.org/10.3390/gels10010011
APA StyleYaputri, B. P., Feyzi, S., & Ismail, B. P. (2024). Transglutaminase-Induced Polymerization of Pea and Chickpea Protein to Enhance Functionality. Gels, 10(1), 11. https://doi.org/10.3390/gels10010011







