Complete Genome Sequence of Pithoascus kurdistanensis CBS 149789, an Endophytic Fungus Isolated from Papaver bracteatum
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungus Growth Conditions
2.2. DNA and RNA Preparations
2.3. Whole Genome Sequencing Using Illumina, Nanopore
2.4. Hybrid Genome Assembly
2.5. Genome Completeness and Ploidy
2.6. Gene Prediction
2.7. Gene Product Annotation
2.8. Isoquinoline Alkaloid Biosynthesis Pathway Coverage
2.9. Mitochondrial Genome Annotation
2.10. Genomic Phylogenetic Tree
3. Results
3.1. Genome Characteristics
3.2. Repetitive DNA Contents
3.3. Genome Completeness
3.4. Taxonomic Assignment and Phylogenetic Analysis
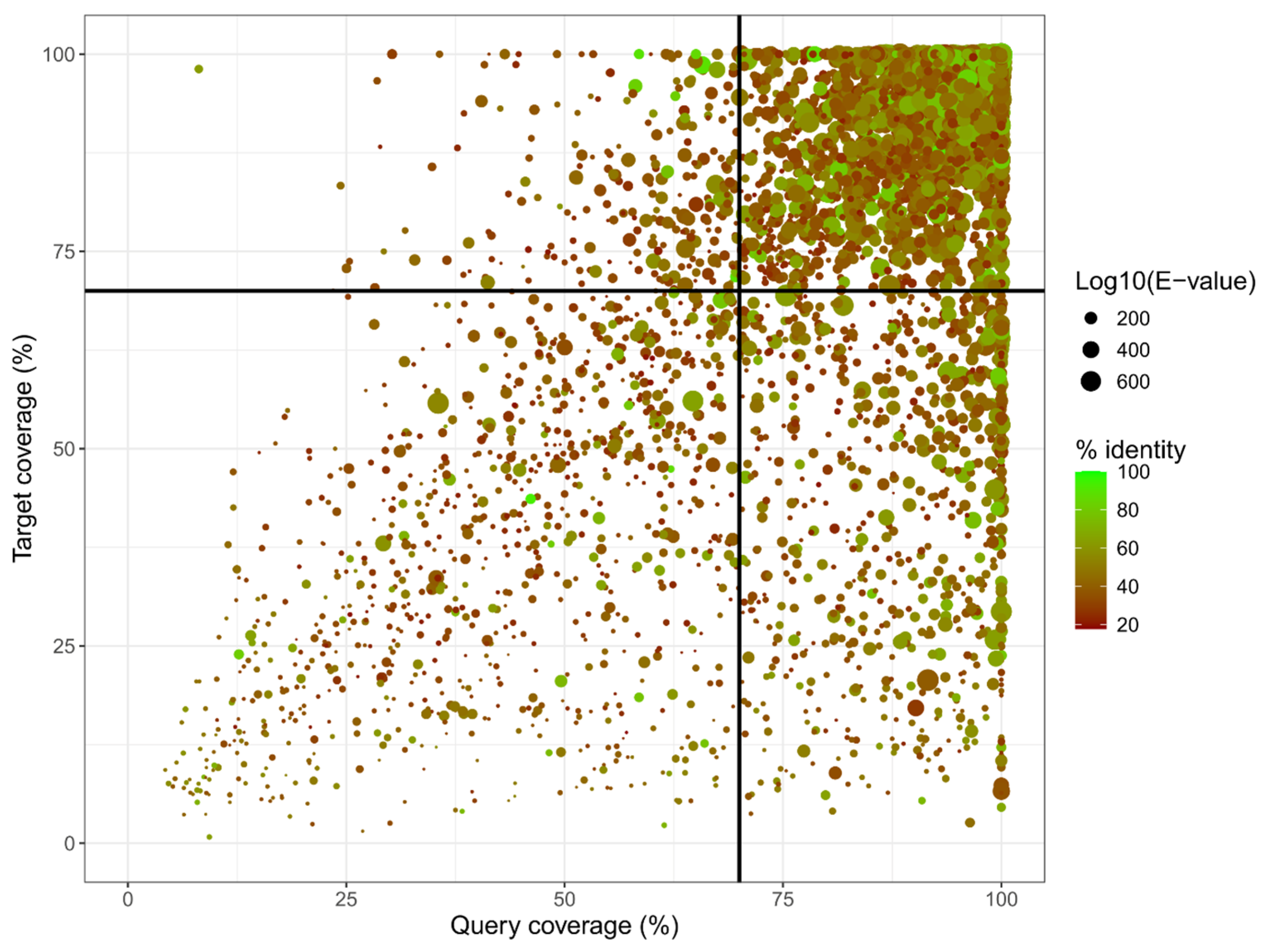
3.5. Gene Prediction and Annotation
3.6. Genes Potentially Involved in Alkaloid Biosynthesis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roberts, M.F.; Kutchan, T.M.; Brown, R.T.; Coscia, C.J. Implication of tyramine in the biosynthesis of morphinan alkaloids in Papaver. Planta 1987, 172, 230–237. [Google Scholar] [CrossRef]
- Tian, Y.; Kong, L.; Li, Q.; Wang, Y.; Wang, Y.; An, Z.; Ma, Y.; Tian, L.; Duan, B.; Sun, W.; et al. Structural diversity, evolutionary origin, and metabolic engineering of plant specialized benzylisoquinoline alkaloids. Nat. Prod. Rep. 2024, 41, 1787–1810. [Google Scholar] [CrossRef]
- Bhambhani, S.; Kondhare, K.R.; Giri, A.P. Diversity in chemical structures and biological properties of plant alkaloids. Molecules 2021, 26, 3374. [Google Scholar] [CrossRef]
- Fossati, E.; Ekins, A.; Narcross, L.; Zhu, Y.; Falgueyret, J.-P.; Beaudoin, G.A.; Facchini, P.J.; Martin, V.J. Reconstitution of a 10-gene pathway for synthesis of the plant alkaloid dihydrosanguinarine in Saccharomyces cerevisiae. Nat. Commun. 2014, 5, 3283. [Google Scholar] [CrossRef]
- Chaudhary, P.; Agri, U.; Chaudhary, A.; Kumar, A.; Kumar, G. Endophytes and their potential in biotic stress management and crop production. Front. Microbiol. 2022, 13, 933017. [Google Scholar] [CrossRef] [PubMed]
- Schardl, C.; Craven, K. Interspecific hybridization in plant-associated fungi and oomycetes: A review. Mol. Ecol. 2003, 12, 2861–2873. [Google Scholar] [CrossRef] [PubMed]
- Pecundo, M.H.; dela Cruz, T.E.E.; Chen, T.; Notarte, K.I.; Ren, H.; Li, N. Diversity, phylogeny and antagonistic activity of fungal endophytes associated with endemic species of Cycas (Cycadales) in China. J. Fungi 2021, 7, 572. [Google Scholar] [CrossRef]
- Toppo, P.; Jangir, P.; Mehra, N.; Kapoor, R.; Mathur, P. Bioprospecting of endophytic fungi from medicinal plant Anisomeles indica L. for their diverse role in agricultural and industrial sectors. Sci. Rep. 2024, 14, 588. [Google Scholar] [CrossRef]
- Khare, E.; Mishra, J.; Arora, N.K. Multifaceted interactions between endophytes and plant: Developments and prospects. Front. Microbiol. 2018, 9, 2732. [Google Scholar] [CrossRef]
- Stierle, A.; Strobel, G.; Stierle, D. Taxol and taxane production by Taxomyces andreanae, an endophytic fungus of Pacific yew. Science 1993, 260, 214–216. [Google Scholar] [CrossRef] [PubMed]
- Kusari, S.; Hertweck, C.; Spiteller, M. Chemical ecology of endophytic fungi: Origins of secondary metabolites. Chem. Biol. 2012, 19, 792–798. [Google Scholar] [CrossRef]
- Toghueo, R.M.K. Bioprospecting endophytic fungi from Fusarium genus as sources of bioactive metabolites. Mycology 2020, 11, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Ray, T.; Pandey, S.S.; Pandey, A.; Srivastava, M.; Shanker, K.; Kalra, A. Endophytic consortium with diverse gene-regulating capabilities of benzylisoquinoline alkaloids biosynthetic pathway can enhance endogenous morphine biosynthesis in Papaver somniferum. Front. Microbiol. 2019, 10, 925. [Google Scholar] [CrossRef]
- Marra, R.; Gutiérrez, S.; Woo, S.L.; Bonanomi, G.; Vinale, F. Editorial: Designing Bio-Formulations Based on Organic Amendments, Beneficial Microbes and Their Metabolites. Front. Microbiol. 2022, 12, 832149. [Google Scholar] [CrossRef]
- Mohammadi, S.; Bahramnejad, B.; Abdollahzadeh, J.; Bashiri, S.; Vincent, A.T.; Majdi, M.; Soltani, J.; Levesque, R.C. Novel endophytic fungal species Pithoascus kurdistanensis producing morphine compounds. Sci. Rep. 2024, 14, 22747. [Google Scholar] [CrossRef]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016, 17, 333–351. [Google Scholar] [CrossRef]
- Clarke, J.; Wu, H.-C.; Jayasinghe, L.; Patel, A.; Reid, S.; Bayley, H. Continuous base identification for single-molecule nanopore DNA sequencing. Nat. Nanotechnol. 2009, 4, 265–270. [Google Scholar] [CrossRef]
- Bielecka, M.; Pencakowski, B.; Nicoletti, R. Using next-generation sequencing technology to explore genetic pathways in endophytic fungi in the syntheses of plant bioactive metabolites. Agriculture 2022, 12, 187. [Google Scholar] [CrossRef]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Simão, F.A.; Zdobnov, E.M. BUSCO update: Novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef]
- Smit, A.; Hubley, R.; Green, P. RepeatMasker Open-4.0. 2013–2015. 2015. Available online: http://www.repeatmasker.org (accessed on 1 September 2023).
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Roach, M.J.; Schmidt, S.A.; Borneman, A.R. Purge Haplotigs: Allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinform. 2018, 19, 460. [Google Scholar] [CrossRef]
- Bourras, S.; Vélëz, H.; Ihrmark, K.; Corrales Gutiérrez, M.Á.; Elfstrand, M.; Garkava-Gustavsson, L.; Falk, K.D. Genome sequence resources from three isolates of the apple canker pathogen Neonectria ditissima infecting forest trees. PhytoFrontiers 2025, 5, 117–119. [Google Scholar] [CrossRef]
- Jain, M.; Abu-Shumays, R.; Olsen, H.E.; Akeson, M. Advances in nanopore direct RNA sequencing. Nat. Methods 2022, 19, 1160–1164. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Boutet, E.; Lieberherr, D.; Tognolli, M.; Schneider, M.; Bairoch, A. UniProtKB/Swiss-Prot: The manually annotated section of the UniProt KnowledgeBase. In Plant Bioinformatics: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2007; pp. 89–112. [Google Scholar]
- Rost, B. Twilight zone of protein sequence alignments. Protein Eng. 1999, 12, 85–94. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016; Available online: http://www.R-project.org (accessed on 1 November 2025).
- Bruccoleri, R.; Russo, M.; Smith, A.; Chasalow, S. Docker Image Builder: A Robust Method for Constructing Docker Images for Reproducible Research. OSF 2025. [Google Scholar] [CrossRef]
- Aramaki, T.; Blanc-Mathieu, R.; Endo, H.; Ohkubo, K.; Kanehisa, M.; Goto, S.; Ogata, H. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 2020, 36, 2251–2252. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020, 29, 28–35. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.-y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef]
- Li, Y.; Steenwyk, J.L.; Chang, Y.; Wang, Y.; James, T.Y.; Stajich, J.E.; Spatafora, J.W.; Groenewald, M.; Dunn, C.W.; Hittinger, C.T.; et al. A genome-scale phylogeny of the kingdom Fungi. Curr. Biol. 2021, 31, 1653–1665.e5. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Kück, P.; Longo, G.C. FASconCAT-G: Extensive functions for multiple sequence alignment preparations concerning phylogenetic studies. Front. Zool. 2014, 11, 81. [Google Scholar] [CrossRef] [PubMed]
- Criscuolo, A.; Gribaldo, S. BMGE (Block Mapping and Gathering with Entropy): A new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 2010, 10, 210. [Google Scholar] [CrossRef]
- Pearson, W.R. Finding protein and nucleotide similarities with FASTA. Curr. Protoc. Bioinform. 2016, 53, 3.9.1–3.9.25. [Google Scholar] [CrossRef]
- Rice, P.M.; Rice, P.M.; Bleasby, A.J.; Ison, J.C. EMBOSS User’s Guide: Practical Bioinformatics with EMBOSS; Cambridge University Press: Cambridge, UK, 2011. [Google Scholar]
- Muszewska, A.; Steczkiewicz, K.; Stepniewska-Dziubinska, M.; Ginalski, K. Transposable elements contribute to fungal genes and impact fungal lifestyle. Sci. Rep. 2019, 9, 4307. [Google Scholar] [CrossRef]
- Wang, H.; Dong, Y.; Liao, W.; Zhang, X.; Wang, Q.; Li, G.; Xu, J.-R.; Liu, H. High-quality genome resource of Clonostachys rosea strain CanS41 by oxford nanopore long-read sequencing. Plant Dis. 2021, 105, 2231–2234. [Google Scholar] [CrossRef]
- Cheng, J.; Zeng, D.; Zhang, T.; Zhang, L.; Han, X.; Zhou, P.; Wang, L.; He, J.; Han, Q. Microascus cirrosus SZ 2021: A potentially new genotype of Microascus cirrosus, which can cause fatal pulmonary infection in patients with acute leukemia following haplo-HSCT. Exp. Ther. Med. 2023, 26, 404. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Henrissat, B.; Arvas, M.; Syed, M.F.; Thieme, N.; Benz, J.P.; Sørensen, J.L.; Record, E.; Poeggeler, S.; Kempken, F. De novo assembly and genome analyses of the marine-derived Scopulariopsis brevicaulis strain LF580 unravels life-style traits and anticancerous scopularide biosynthetic gene cluster. PLoS ONE 2015, 10, e0140398. [Google Scholar] [CrossRef]
- Tavares, S.; Ramos, A.P.; Pires, A.S.; Azinheira, H.G.; Caldeirinha, P.; Link, T.; Abranches, R.; Silva, M.d.C.; Voegele, R.T.; Loureiro, J.; et al. Genome size analyses of Pucciniales reveal the largest fungal genomes. Front. Plant Sci. 2014, 5, 422. [Google Scholar] [CrossRef] [PubMed]
- Gregory, T.R.; Nicol, J.A.; Tamm, H.; Kullman, B.; Kullman, K.; Leitch, I.J.; Murray, B.G.; Kapraun, D.F.; Greilhuber, J.; Bennett, M.D. Eukaryotic genome size databases. Nucleic Acids Res. 2007, 35, D332–D338. [Google Scholar] [CrossRef]
- Paun, L.; Kempken, F. Fungal transposable elements. In Genetic Transformation Systems in Fungi; Springer: Berlin/Heidelberg, Germany, 2015; Volume 2, pp. 79–96. [Google Scholar]
- Gupta, S.; Bhatt, P.; Chaturvedi, P. Determination and quantification of asiaticoside in endophytic fungus from Centella asiatica (L.) Urban. World J. Microbiol. Biotechnol. 2018, 34, 111. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.-J.; Fang, X.; Li, C.-Y.; Yang, L.; Chen, X.-Y. General and specialized tyrosine metabolism pathways in plants. Abiotech 2020, 1, 97–105. [Google Scholar] [CrossRef]
- Vincent, A.T. Bacterial hypothetical proteins may be of functional interest. Front. Bacteriol. 2024, 3, 1334712. [Google Scholar] [CrossRef]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef]
- Venter, J.C.; Adams, M.D.; Myers, E.W.; Li, P.W.; Mural, R.J.; Sutton, G.G.; Smith, H.O.; Yandell, M.; Evans, C.A.; Holt, R.A.; et al. The sequence of the human genome. Science 2001, 291, 1304–1351. [Google Scholar] [CrossRef]
- Liao, W.-W.; Asri, M.; Ebler, J.; Doerr, D.; Haukness, M.; Hickey, G.; Lu, S.; Lucas, J.K.; Monlong, J.; Abel, H.J.; et al. A draft human pangenome reference. Nature 2023, 617, 312–324. [Google Scholar] [CrossRef] [PubMed]
- Ijaq, J.; Chandrasekharan, M.; Poddar, R.; Bethi, N.; Sundararajan, V.S. Annotation and curation of uncharacterized proteins-challenges. Front. Genet. 2015, 6, 119. [Google Scholar] [CrossRef]
- Bairoch, A.; Apweiler, R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000, 28, 45–48. [Google Scholar] [CrossRef]
- Jablonowski, K. Hidden Markov Models for protein domain homology identification and analysis. In SH2 Domains: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2017; pp. 47–58. [Google Scholar] [CrossRef]
- Weissenow, K.; Rost, B. Are protein language models the new universal key? Curr. Opin. Struct. Biol. 2025, 91, 102997. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]



| Draft (Canu) | Pilon Round #1 | Pilon Round #2 | Final (Masked) | |
|---|---|---|---|---|
| Genome size (bp) | 33,982,329 | 34,011,236 | 34,009,754 | 34,009,754 |
| Number of contigs | 13 | 13 | 13 | 13 |
| Longest contig (bp) | 6,631,532 | 6,637,200 | 6,636,903 | 6,636,903 |
| GC % | 58.61 | 58.64 | 58.64 | 58.64 |
| N count * | 0 | 0 | 0 | 2,165,484 |
| N50 size (bp) | 3,993,926 | 3,996,197 | 3,996,062 | 3,996,062 |
| Contigs above N50 size | 4 | 4 | 4 | 4 |
| N75 size (bp) | 2,860,699 | 2,863,255 | 2,863,137 | 2,863,137 |
| Contigs above N75 size | 6 | 6 | 6 | 6 |
| Fragment | Size (bp) | CDS | rRNA | tRNA |
|---|---|---|---|---|
| Chromosome 1 | 6,636,903 | 1647 | 3 | 35 |
| Chromosome 2 | 4,400,315 | 1061 | 1 | 30 |
| Chromosome 3 | 4,395,346 | 1050 | 1 | 21 |
| Chromosome 4 | 3,996,062 | 1041 | - | 18 |
| Chromosome 5 | 3,330,179 | 798 | - | 17 |
| Chromosome 6 | 2,863,137 | 643 | - | 10 |
| Chromosome 7 | 2,828,770 | 682 | - | 9 |
| Chromosome 8 | 2,813,902 | 656 | - | 14 |
| Chromosome 9 | 2,608,864 | 714 | - | 9 |
| TOTAL | 33,873,478 | 8292 | 5 | 163 |
| Number of Elements | Length Occupied | % of Genome Size | |
|---|---|---|---|
| Retroelements | 635 | 891,335 bp | 2.62% |
| LINEs: | 176 | 160,299 bp | 0.47% |
| CRE/SLACS | 57 | 53,664 bp | 0.16% |
| R1/LOA/Jockey | 12 | 1550 bp | 0.00% |
| LTR elements: | 459 | 731,036 bp | 2.15% |
| Ty1/Copia | 95 | 103,470 bp | 0.30% |
| Gypsy/DIRS1 | 364 | 627,566 bp | 1.85% |
| DNA transposons | 523 | 496,324 bp | 1.46% |
| Tc1-IS630-Pogo | 315 | 298,983 bp | 0.88% |
| MULE-MuDR | 208 | 197,341 bp | 0.58% |
| Unclassified | 1594 | 370,488 bp | 1.09% |
| Total interspersed repeats: | 4438 | 1,758,147 bp | 5.17% |
| Small RNA | 22 | 56,043 bp | 0.16% |
| Simple repeats | 6374 | 282,351 bp | 0.83% |
| Low complexity | 1331 | 68,943 bp | 0.20% |
| Total bases masked: | 12,165 | 2,165,484 bp | 6.37% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohammadi, S.; Gauthier, J.; Nguyen, G.Q.H.; Vincent, A.T.; Bahramnejad, B.; Levesque, R.C. Complete Genome Sequence of Pithoascus kurdistanensis CBS 149789, an Endophytic Fungus Isolated from Papaver bracteatum. J. Fungi 2025, 11, 861. https://doi.org/10.3390/jof11120861
Mohammadi S, Gauthier J, Nguyen GQH, Vincent AT, Bahramnejad B, Levesque RC. Complete Genome Sequence of Pithoascus kurdistanensis CBS 149789, an Endophytic Fungus Isolated from Papaver bracteatum. Journal of Fungi. 2025; 11(12):861. https://doi.org/10.3390/jof11120861
Chicago/Turabian StyleMohammadi, Sima, Jeff Gauthier, Guillaume Quang Henri Nguyen, Antony T. Vincent, Bahman Bahramnejad, and Roger C. Levesque. 2025. "Complete Genome Sequence of Pithoascus kurdistanensis CBS 149789, an Endophytic Fungus Isolated from Papaver bracteatum" Journal of Fungi 11, no. 12: 861. https://doi.org/10.3390/jof11120861
APA StyleMohammadi, S., Gauthier, J., Nguyen, G. Q. H., Vincent, A. T., Bahramnejad, B., & Levesque, R. C. (2025). Complete Genome Sequence of Pithoascus kurdistanensis CBS 149789, an Endophytic Fungus Isolated from Papaver bracteatum. Journal of Fungi, 11(12), 861. https://doi.org/10.3390/jof11120861







