Comparison of Fungal Genera Isolated from Cucumber Plants and Rhizosphere Soil by Using Various Cultural Media
Abstract
:1. Introduction
2. Materials and Methods
2.1. Trial Design for Plants and Soil Sampling
2.2. Cultural Media
2.3. Isolation of Endophytic and Rhizosphere Soil Fungi
2.4. Fungal Strain Identification
2.5. Data Analysis
3. Results
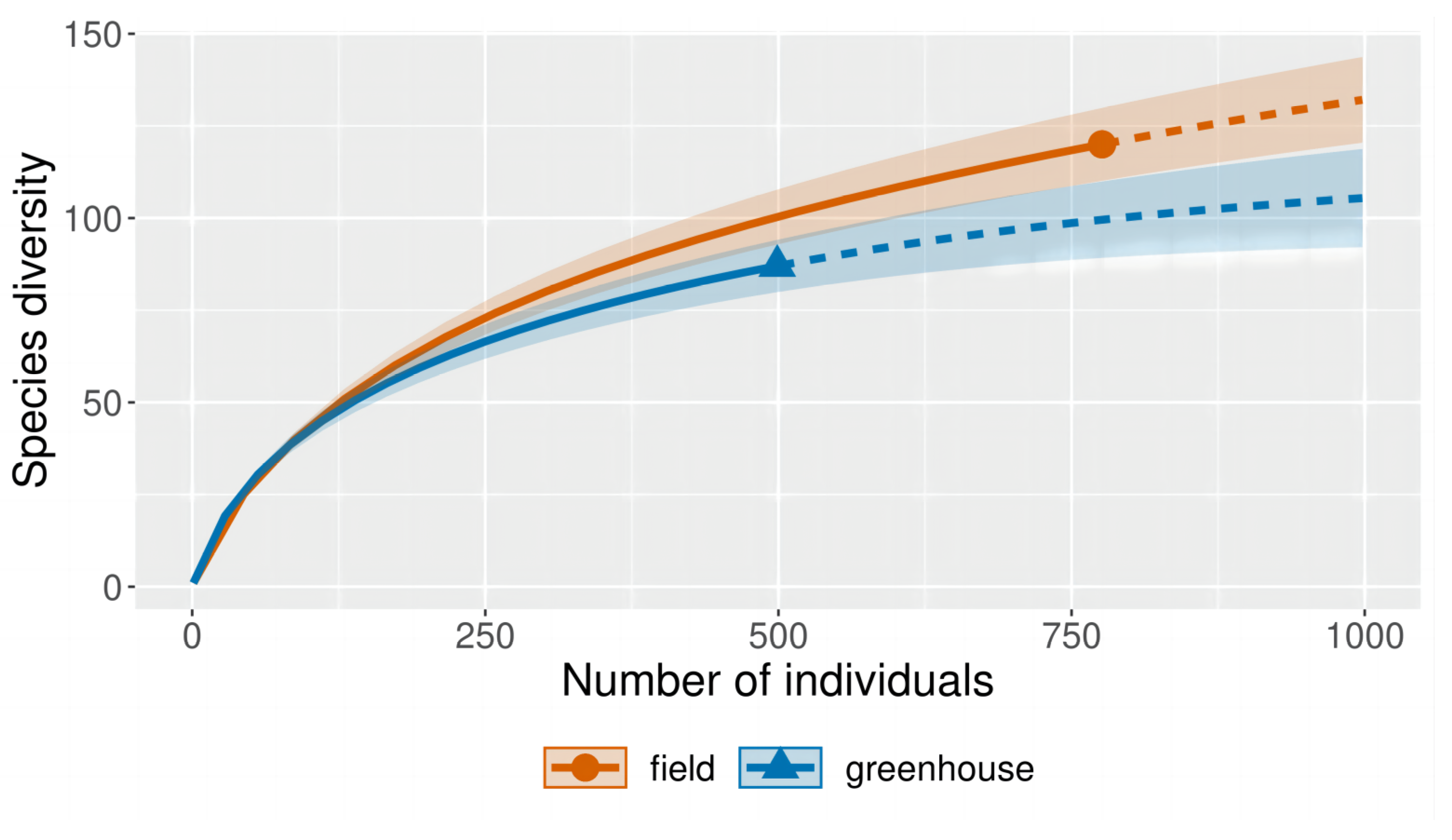
3.1. Community Composition of Endophytic Fungi and Rhizosphere Soil Fungi
3.2. Influence of Culture Medium on Fungal Isolation
3.3. Effects of Culture Medium on Endophytic Fungal Diversity
3.4. Effect of the Medium Type on Endophytic Fungal Diversity in Different Tissues of the Plant
3.5. Effects of Culture Medium on Soil Fungal Diversity
3.6. Effects of Culture Medium on Trichoderma and Fusarium with High Isolation Efficiency
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bamisile, B.S.; Dash, C.K.; Akutse, K.S.; Keppanan, R.; Afolabi, O.G.; Hussain, M.; Qasim, M.; Wang, L. Prospects of endophytic fungal entomopathogens as biocontrol and plant growth promoting agents: An insight on how artificial inoculation methods affect endophytic colonization of host plants. Microbiol. Res. 2018, 217, 34–50. [Google Scholar] [CrossRef] [PubMed]
- Vinale, F.; Sivasithamparam, K.; Ghisalberti, E.L.; Marra, R.; Woo, S.L.; Lorito, M. Trichoderma–plant–pathogen interactions. Soil Biol. Biochem. 2008, 40, 1–10. [Google Scholar] [CrossRef]
- Arenz, B.E.; Held, B.W.; Jurgens, J.A.; Farrell, R.L.; Blanchette, R.A. Fungal diversity in soils and historic wood from the Ross Sea Region of Antarctica. Soil Biol. Biochem. 2006, 38, 3057–3064. [Google Scholar] [CrossRef]
- Salazar-Cerezo, S.; Martinez-Montiel, N.; Cruz-Lopez, M.D.C.; Martinez-Contreras, R.D. Fungal diversity and community composition of culturable fungi in Stanhopea trigrina cast gibberellin producers. Front. Microbiol. 2018, 9, 612. [Google Scholar] [CrossRef]
- Nischitha, R.; Shivanna, M.B. Antimicrobial activity and metabolite profiling of endophytic fungi in Digitaria bicornis (Lam) Roem. and Schult. and Paspalidium flavidum (Retz.) A. Camus. 3 Biotech. 2021, 11, 53. [Google Scholar] [CrossRef]
- Muggia, L.; Kopun, T.; Grube, M. Effects of growth media on the diversity of culturable fungi from lichens. Molecules 2017, 22, 824. [Google Scholar] [CrossRef]
- Prior, R.; Görges, K.; Yurkov, A.; Begerow, D. New isolation method for endophytes based on enzyme digestion. Mycol. Prog. 2014, 13, 849–856. [Google Scholar] [CrossRef]
- Cosoveanu, A.; Cabrera, R. Fungi as endophytes in Artemisia thuscula: Juxtaposed elements of diversity and phylogeny. J. Fungi 2018, 4, 17. [Google Scholar] [CrossRef]
- Zeng, Z.H.; Yuan, Z.L.; Huang, X.N.; Liu, F. Isolation and diversity of mangrove endophytic fungi. Prot. For. Sci. Technol. 2022, 39–42, 45. [Google Scholar] [CrossRef]
- Smith, H.; Wingfield, M.J.; Petrini, O. Botryosphaeria dothidea endophytic in Eucalyptus grandis and Eucalyptus nitens in South Africa. Forest Ecol. Manag. 1996, 89, 189–195. [Google Scholar] [CrossRef]
- Blodgett, J.T.; Swart, W.J.; Weeks, L.W.J. Species composition of endophytic fungi in Amaranthus hybridus leaves, petioles, stems, and roots. Mycologia 2000, 92, 853–859. [Google Scholar] [CrossRef]
- Bettucci, L.; Simeto, S.; Alonso, R.; Lupo, S. Endophytic fungi of twigs and leaves of three native species of Myrtaceae in Uruguay. Sydowia 2004, 56, 8–23. [Google Scholar]
- Blodgett, J.T.; Swart, W.J.; Louw, S.V.; Weeks, W.J. Soil amendments and watering influence the incidence of endophytic fungi in Amaranthus hybridus in South Africa. Appl. Soil. Ecol. 2007, 35, 311–318. [Google Scholar] [CrossRef]
- Xue, Q.W.; Niu, Y.C.; Deng, H. Diversity of endophytic fungi in common cucurbit plants. Mycosystema 2015, 34, 196–203. [Google Scholar] [CrossRef]
- Belfiori, B.; Rubini, A.; Riccioni, C. Diversity of endophytic and pathogenic fungi of Saffron (Crocus sativus) plants from cultivation sites in Italy. Diversity 2021, 13, 535. [Google Scholar] [CrossRef]
- Wang, Z.B.; Zhou, X.; Ma, X.B.; Wang, J.L. Isolation of endophytic fungi from medicinal plants and screening and identification of antagonism. Chin. Agric. Sci. Bull. 2022, 38, 75–81. [Google Scholar]
- Evans, H.C.; Holmes, K.A.; Thomas, S.E. Endophytes and mycoparasites associated with an indigenous forest tree, Theobroma gileri, in Ecuador and a preliminary assessment of their potential as biocontrol agents of cocoa diseases. Mycol. Prog. 2003, 2, 149–160. [Google Scholar] [CrossRef]
- Khan, A.R.; Waqas, M.; Ullah, I.; Khan, A.L.; Khan, M.A.; Lee, I.; Shin, J. Culturable endophytic fungal diversity in the cadmium hyperaccumulator Solanum nigrum L. and their role in enhancing phytoremediation. Environ. Exp. Bot. 2017, 135, 126–135. [Google Scholar] [CrossRef]
- Li, W.Z.; Duan, W.J.; Zhou, X.; Chen, J.; Jiang, X.Z.; Yang, Y.; Cai, L. Diversity of fungi in wheat seeds. Mycosystema 2021, 40, 487–501. [Google Scholar] [CrossRef]
- Guo, L.D.; Hyde, K.D.; Liew, E. Identification of endophytic fungi from Livistona chinensis based on morphology and rDNA sequences. New Phytol. 2000, 147, 617–630. [Google Scholar] [CrossRef]
- Xiang, L.B.; Gong, S.J.; Yang, L.J.; Hao, J.; Xue, M.F.; Zeng, F.S.; Zhang, X.J.; Shi, W.Q.; Wang, H.; Yu, D.Z. Biocontrol potential of endophytic fungi in medicinal plants from Wuhan Botanical Garden in China. Biol. Control 2016, 94, 47–55. [Google Scholar] [CrossRef]
- Wang, H.X.; Yu, Y.R.; Huang, B.K. Diversity of culturable endophytic fungi from Broussonetia papyrifera. Mycosystema 2020, 39, 2399–2408. [Google Scholar] [CrossRef]
- Figueredo, H.M.; Gonçalves, V.N.; Godinho, V.M.; Lopes, D.V.; Oliveira, F.S.; Rosa, L.H. Diversity and ecology of cultivable fungi isolated from the thermal soil gradients in Deception Island, Antarctica. Extremophiles 2020, 24, 219–225. [Google Scholar] [CrossRef]
- Xu, Y.L.; Liu, M.; Huang, H.Q.; Zhu, J.; Bao, S.X. Diversity of soil culturable fungi in Bamen Bay Mangrove Forests. Chin. J. Trop. Crops 2013, 34, 181–187. [Google Scholar]
- Zhang, J.Z.; Chen, X.R.; Yang, C.D.; Xue, L. A study on the diversity of soil cultured fungi in the alpine grassland of Eastern Qilian Mountains. Acta Prataculturae Sin. 2010, 19, 124–132. [Google Scholar]
- Vasanthakumari, M.M.; Shivanna, M.B. Fungal assemblages in the rhizosphere and rhizoplane of grasses of the subfamily Panicoideae in the Lakkavalli region of Karnataka, India. Microbes Environ. 2011, 26, 228–236. [Google Scholar] [CrossRef]
- Jiang, H.; Li, Y.C.; Jiang, Y. Seasonal variation of soil fungal diversity in the rhizosphere of Phragmites australis. J. Liaoning Norm. Univ. (Nat. Sci. Ed.) 2017, 40, 89–94. [Google Scholar]
- Ayob, Z.; Kusai, N.A.; Ali, S.R.A. Sequence-based identification and characterisation of cultivated filamentous fungi in the Alan Bunga peat ecosystems of Sarawak, Malaysia. Mires Peat 2018, 21, 1–19. [Google Scholar] [CrossRef]
- Zhou, J.; Miao, Y.F.; Fang, K.; Chen, L.; Yang, Z.P.; Dong, X.F.; Zhang, H.B. Diversity of the endophytic and rhizosphere soil fungi of Ageratina adenophora. Ecol. Sci. 2019, 38, 1–7. [Google Scholar] [CrossRef]
- Guo, M.J.; Zhao, X.Y.; Huang, J.H.; Wei, S.Z.; Xu, X.L. Isolation and activity analysis of fungi from the rhizosphere of mangroves in Xinglin Bay, Xiamen. Microbiology 2021, 48, 1496–1503. [Google Scholar] [CrossRef]
- Vieira, F.C.S.; Nahas, E. Comparison of microbial numbers in soils by using various culture media and temperatures. Microbiol. Res. 2005, 160, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Grishkan, I.; Beharav, A.; Kirzhner, V.; Nevo, E. Adaptive spatiotemporal distribution of soil microfungi in ‘Evolution Canyon’ III, Nahal Shaharut, extreme southern Negev Desert, Israel. Biol. J. Linn. Soc. 2007, 90, 263–277. [Google Scholar] [CrossRef]
- Watrud, L.S.; Martin, K.; Donegan, K.K.; Stone, J.K.; Coleman, C.G. Comparison of taxonomic, colony morphotype and PCR-RFLP methods to characterize microfungal diversity. Mycologia 2006, 98, 384–392. [Google Scholar] [CrossRef]
- Huang, L.Q.; Niu, Y.C.; Su, L.; Deng, H.; Lyu, H. The potential of endophytic fungi isolated from cucurbit plants for biocontrol of soilborne fungal diseases of cucumber. Microbiol. Res. 2020, 231, 126369. [Google Scholar] [CrossRef]
- Zimmerman, N.B.; Vitousek, P.M. Fungal endophyte communities reflect environmental structuring across a Hawaiian landscape. Proc. Natl. Acad. Sci. USA 2012, 109, 13022–13027. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Deng, H.; Niu, Y.C. Phialemoniopsis endophytica sp. nov., a new species of endophytic fungi from Luffa cylindrica in Henan, China. Mycol. Prog. 2016, 15, 48. [Google Scholar] [CrossRef]
- Lin, X.G. Principles and Methods of Soil Microbiology Research, 1st ed.; Higher Education Press: Beijing, China, 2010; pp. 31, 37, 38. [Google Scholar]
- Zhu, J.; Ren, X.; Liu, H.; Liang, C. Effect of irrigation with microcystins-contaminated water on growth and fruit quality of Cucumis sativus L. and the health risk. Agr. Water Manage 2018, 204, 91–99. [Google Scholar] [CrossRef]
- Arnold, A.E.; Maynard, Z.; Gilbert, G.S. Fungal endophytes in dicotyledonous neotropical trees: Patterns of abundance and diversity. Mycol. Res. 2001, 105, 1502–1507. [Google Scholar] [CrossRef]
- Qin, W.T.; Zhuang, W.Y. Seven new species of Trichoderma (Hypocreales) in the Harzianum and Strictipile clades. Phytotaxa 2017, 305, 121–139. [Google Scholar] [CrossRef]
- Fang, Z.D. Research Methods of Plant Disease, 3rd ed.; Chinese Agricultural Press: Beijing, China, 1998; pp. 125, 133. [Google Scholar]
- Hohmann, P.; Jones, E.E.; Hill, R.A.; Stewart, A. Understanding Trichoderma in the root system of Pinus radiata: Associations between rhizosphere colonisation and growth promotion for commercially grown seedlings. Fungal Biol. 2011, 115, 759–767. [Google Scholar] [CrossRef]
- de Hoog, G.S.; van den Ende, A. Molecular diagnostics of clinical strains of filamentous Basidiomycetes. Mycoses 1998, 41, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Li, D.W.; Huang, Q.T.; Huang, L.; Wen, J.K.; Luo, J.; Li, Q.; Peng, Y.; Zhang, Y.L. Baiting out a full length sequence from unmapped RNA-seq data. BMC Genom. 2021, 22, 857. [Google Scholar] [CrossRef] [PubMed]
- Tedersoo, L.; Sánchez-Ramírez, S.; Kõljalg, U.; Bahram, M.; Döring, M.; Schigel, D.; May, T.; Ryberg, M.; Abarenkov, K. High-level classification of the Fungi and a tool for evolutionary ecological analyses. Fungal Divers. 2018, 90, 135–159. [Google Scholar] [CrossRef]
- Lücking, R.; Aime, M.C.; Robbertse, B.; Miller, A.N.; Aoki, T.; Ariyawansa, H.A.; Cardinali, G.; Crous, P.W.; Druzhinina, I.S.; Geiser, D.M.; et al. Fungal taxonomy and sequence-based nomenclature. Nat. Microbiol. 2021, 6, 540–548. [Google Scholar] [CrossRef]
- Hsieh, T.C.; Ma, K.H.; Chao, A. iNEXT: An R package for rarefaction and extrapolation of species diversity (Hill numbers). Methods Ecol. Evol. 2016, 7, 1451–1456. [Google Scholar] [CrossRef]
- Chen, T.; Liu, Y.X.; Huang, L. ImageGP: An easy-to-use data visualization web server for scientific researchers. iMeta 2022, 1, e5. [Google Scholar] [CrossRef]
- Shannon, C.E.; Weaver, W. The Mathematical Theory of Communication; The University of Illinois Press: Urbana, IL, USA, 1949; pp. 3–24. [Google Scholar]
- Simpson, E.H. Measurement of Diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Armstrong, R.A. When to use the Bonferroni correction. Ophthalmic Physiol. Opt. J. Br. Coll. Ophthalmic Opt. 2014, 34, 502–508. [Google Scholar] [CrossRef]
- Gehan, E.A. A generalized Wilcoxon test for comparing arbitrarily singly-censored samples. Biometrika 1965, 52, 203–224. [Google Scholar] [CrossRef] [PubMed]
- Mu, L.X.; Niu, Y.C.; Deng, H. The endophytic mycobiota in summer growing cucumber in Beijing. Mycosystema 2010, 29, 214–221. [Google Scholar] [CrossRef]
- Yan, X.N.; Sikora, R.A.; Zheng, J.W. Potential use of cucumber (Cucumis sativus L.) endophytic fungi as seed treatment agents against root-knot nematode Meloidogyne incognita. J. Zhejiang Univ. Sci. B 2011, 12, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Deng, H.; Niu, Y.C. Phylogenetic analysis of Plectosphaerella species based on multi-locus DNA sequences and description of P. sinensis sp. nov. Mycol. Prog. 2017, 16, 823–829. [Google Scholar] [CrossRef]
- Su, L.; Niu, Y.C. Multilocus phylogenetic analysis of Talaromyces species isolated from cucurbit plants in China and description of two new species, T. cucurbitiradicus and T. endophyticus. Mycologia 2018, 110, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Girlanda, M.; Perotto, S.; Moenne-Loccoz, Y.; Bergero, R.; Lazzari, A.; Defago, G.; Bonfante, P.; Luppi, A.M. Impact of Biocontrol Pseudomonas fluorescens CHA0 and a Genetically Modified Derivative on the Diversity of Culturable Fungi in the Cucumber Rhizosphere. Appl. Environ. Microb. 2001, 67, 1851–1864. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.Y.; Wu, F.Z. Soil microbial community structure in cucumber rhizosphere of different resistance cultivars to fusarium wilt. Fems Microbiol. Ecol. 2010, 72, 456–463. [Google Scholar] [CrossRef]
- Vu, D.; Groenewald, M.; de Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J.; et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019, 92, 135–154. [Google Scholar] [CrossRef]
- Kiyuna, T.; An, K.; Kigawa, R.; Sano, C.; Miura, S.; Sugiyama, J. Molecular assessment of fungi in “black spots” that deface murals in the Takamatsuzuka and Kitora Tumuli in Japan: Acremonium sect. Gliomastix including Acremonium tumulicola sp. nov. and Acremonium felinum comb. nov. Mycoscience 2011, 52, 1–17. [Google Scholar] [CrossRef]
- Li, B.H.; Wang, C.C.; Dong, X.L.; Zhang, Z.F.; Wang, C.X. Acremonium Brown Spot, a new disease caused by Acremonium sclerotigenum on bagged apple fruit in China. Plant Dis. 2014, 98, 1012. [Google Scholar] [CrossRef]
- Luo, J.; Walsh, E.; Naik, A.; Zhuang, W.; Zhang, K.; Cai, L.; Zhang, N. Temperate pine barrens and tropical rain forests are both rich in undescribed fungi. PLoS ONE 2014, 9, e103753. [Google Scholar] [CrossRef]
- Wagner, L.; Stielow, B.; Hoffmann, K.; Petkovits, T.; Papp, T.; Vágvölgyi, C.; de Hoog, G.S.; Verkley, G.; Voigt, K. A comprehensive molecular phylogeny of the Mortierellales (Mortierellomycotina) based on nuclear ribosomal DNA. Pers. Int. Mycol. J. 2013, 30, 77–93. [Google Scholar] [CrossRef]
- Lombard, L.; Houbraken, J.; Decock, C.; Samson, R.A.; Meijer, M.; Réblová, M.; Groenewald, J.Z.; Crous, P.W. Generic hyper-diversity in Stachybotriaceae. Pers. Int. Mycol. J. 2016, 36, 156–246. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, J.; Nakashima, C. Japanese species of Alternaria and their species boundaries based on host range. Fungal Syst. Evol. 2020, 5, 197–281. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.W.; Groenewald, J.Z. A phylogenetic re-evaluation of Arthrinium. Ima Fungus 2013, 4, 133–154. [Google Scholar] [CrossRef] [PubMed]
- Pyrri, I.; Tripyla, E.; Zalachori, A.; Chrysopoulou, M.; Parmakelis, A.; Kapsanaki-Gotsi, E. Fungal contaminants of indoor air in the National Library of Greece. Aerobiologia 2020, 36, 387–400. [Google Scholar] [CrossRef]
- Schoch, C.L.; Robbertse, B.; Robert, V.; Vu, D.; Cardinali, G.; Irinyi, L.; Meyer, W.; Nilsson, R.H.; Hughes, K.; Miller, A.N.; et al. Finding needles in haystacks: Linking scientific names, reference specimens and molecular data for Fungi. Database-Oxford 2014, 2014, bau61. [Google Scholar] [CrossRef] [PubMed]
- Vesth, T.C.; Nybo, J.L.; Theobald, S.; Frisvad, J.C.; Larsen, T.O.; Nielsen, K.F.; Hoof, J.B.; Brandl, J.; Salamov, A.; Riley, R.; et al. Investigation of inter- and intraspecies variation through genome sequencing of Aspergillus section Nigri. Nat. Genet. 2018, 50, 1688–1695. [Google Scholar] [CrossRef] [PubMed]
- Frisvad, J.C.; Hubka, V.; Ezekiel, C.N.; Hong, S.B.; Nováková, A.; Chen, A.J.; Arzanlou, M.; Larsen, T.O.; Sklenář, F.; Mahakarnchanakul, W.; et al. Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Stud. Mycol. 2019, 93, 1–63. [Google Scholar] [CrossRef]
- Visagie, C.M.; Houbraken, J. Updating the taxonomy of Aspergillus in South Africa. Stud. Mycol. 2020, 95, 253–292. [Google Scholar] [CrossRef]
- Irinyi, L.; Serena, C.; Garcia-Hermoso, D.; Arabatzis, M.; Desnos-Ollivier, M.; Vu, D.; Cardinali, G.; Arthur, I.; Normand, A.; Giraldo, A.; et al. International Society of Human and Animal Mycology (ISHAM)-ITS reference DNA barcoding database—The quality controlled standard tool for routine identification of human and animal pathogenic fungi. Med. Mycol. 2015, 53, 313–337. [Google Scholar] [CrossRef]
- Simon, U.K.; Weiss, M. Intragenomic variation of fungal ribosomal genes is higher than previously thought. Mol. Biol. Evol. 2008, 25, 2251–2254. [Google Scholar] [CrossRef]
- Hou, L.W.; Groenewald, J.Z.; Pfenning, L.H.; Yarden, O.; Crous, P.W.; Cai, L. The phoma-like dilemma. Stud. Mycol. 2020, 96, 309–396. [Google Scholar] [CrossRef] [PubMed]
- Yaguchi, T.; Sano, A.; Yarita, K.; Suh, M.K.; Nishimura, K.; Udagawa, S. A new species of Cephalotheca isolated from a Korean patient. Mycotaxon 2006, 96, 309–322. [Google Scholar]
- De Melo, M.P.; Matos, K.S.; Moreira, S.I.; Silva, F.F.; Conceição, G.H.; Nechet, K.L.; Halfeld-Vieira, B.A.; Beserra Júnior, J.E.A.; Ventura, J.A.; Costa, H.; et al. Two new Ceratobasidium species causing white thread blight on tropical plants in Brazil. Trop. Plant Pathol. 2018, 43, 559–571. [Google Scholar] [CrossRef]
- Sugiyama, K.; Sano, A.; Murakami, M.; Ogawa, T.; Mishima, H.; Otake, H.; Kamei, K.; Sugiyama, S. Three isolations of Chaetomium globosum from erythematous epilation of canine skin. Med. Mycol. 2008, 46, 505–510. [Google Scholar] [CrossRef]
- Bensch, K.; Groenewald, J.Z.; Meijer, M.; Dijksterhuis, J.; Jurjević, Ž.; Andersen, B.; Houbraken, J.; Crous, P.W.; Samson, R.A. Cladosporium species in indoor environments. Stud. Mycol. 2018, 89, 177–301. [Google Scholar] [CrossRef]
- Liu, C.H.; Huang, X.; Xie, T.-N.; Duan, N.; Xue, Y.-R.; Zhao, T.-X.; Lever, M.A.; Hinrichs, K.; Inagaki, F. Exploration of cultivable fungal communities in deep coal-bearing sediments from ∼1.3 to 2.5 km below the ocean floor. Environ. Microbiol. 2017, 19, 803–818. [Google Scholar] [CrossRef]
- Nirenberg, H.I.; Feiler, U.; Hagedorn, G. Description of Colletotrichum lupini comb. nov. in modern terms. Mycologia 2002, 94, 307–320. [Google Scholar] [CrossRef]
- Gherbawy, Y.; Kesselboth, C.; Elhariry, H.; Hoffmann, K. Molecular Barcoding of Microscopic Fungi with Emphasis on the Mucoralean Genera Mucor and Rhizopus. In Molecular Identification of Fungi; Springer: Berlin/Heidelberg, Germany, 2010; pp. 213–250. [Google Scholar] [CrossRef]
- Da Cunha, K.C.; Sutton, D.A.; Fothergill, A.W.; Gené, J.; Cano, J.; Madrid, H.; Hoog, S.D.; Crous, P.W.; Guarro, J. In vitro antifungal susceptibility and molecular identity of 99 clinical isolates of the opportunistic fungal genus Curvularia. Diagn. Micr. Infec. Dis. 2013, 76, 168–174. [Google Scholar] [CrossRef]
- Pearce, T.L.; Scott, J.B.; Crous, P.W.; Pethybridge, S.J.; Hay, F.S. Tan spot of pyrethrum is caused by a Didymella species complex. Plant Pathol. 2016, 65, 1170–1184. [Google Scholar] [CrossRef]
- Crous, P.W.; Braun, U.; Wingfield, M.J.; Wood, A.R.; Shin, H.D.; Summerell, B.A.; Alfenas, A.C.; Cumagun, C.J.R.; Groenewald, J.Z. Phylogeny and taxonomy of obscure genera of microfungi. Pers. Int. Mycol. J. 2009, 22, 139–161. [Google Scholar] [CrossRef]
- Balasingham, S.; Chalkias, S.; Balasingham, A.; Saul, Z.; Wickes, B.L.; Sutton, D.A. A case of bovine valve endocarditis caused by Engyodontium album. Med. Mycol. 2011, 49, 430–434. [Google Scholar] [CrossRef] [PubMed]
- Ezekiel, C.N.; Kraak, B.; Sandoval-Denis, M.; Sulyok, M.; Oyedele, O.A.; Ayeni, K.I.; Makinde, O.M.; Akinyemi, O.M.; Krska, R.; Crous, P.W.; et al. Diversity and toxigenicity of fungi and description of Fusarium madaense sp. nov. from cereals, legumes and soils in north-central Nigeria. MycoKeys 2020, 67, 95–124. [Google Scholar] [CrossRef] [PubMed]
- Wiemann, P.; Sieber, C.M.K.; von Bargen, K.W.; Studt, L.; Niehaus, E.; Espino, J.J.; Huß, K.; Michielse, C.B.; Albermann, S.; Wagner, D.; et al. Deciphering the cryptic genome: Genome-wide analyses of the rice pathogen Fusarium fujikuroi reveal complex regulation of secondary metabolism and novel metabolites. PLoS Pathog. 2013, 9, e1003475. [Google Scholar] [CrossRef] [PubMed]
- King, R.; Urban, M.; Hammond-Kosack, M.C.U.; Hassani-Pak, K.; Hammond-Kosack, K.E. The completed genome sequence of the pathogenic ascomycete fungus Fusarium graminearum. BMC Genom. 2015, 16, 544. [Google Scholar] [CrossRef] [PubMed]
- Surovy, M.Z.; Kabir, M.K.; Gupta, D.R.; Hassan, O.; Mahmud, N.U.; Sabir, A.A.; Rahman, M.M.; Chang, T.; Panaccione, D.G.; Islam, M.T. First report of Fusarium wilt caused by Fusarium oxysporum on strawberry in Bangladesh. Plant Dis. 2019, 103, 367. [Google Scholar] [CrossRef]
- Schlegel, M.; Queloz, V.; Sieber, T.N. The endophytic mycobiome of European ash and sycamore maple leaves—Geographic patterns, host specificity and influence of ash dieback. Front. Microbiol. 2018, 9, 2345. [Google Scholar] [CrossRef]
- Bills, G.F.; Platas, G.; Overy, D.P.; Collado, J.; Fillola, A.; Jiménez, M.R.; Martín, J.; Del Val, A.G.; Vicente, F.; Tormo, J.R.; et al. Discovery of the parnafungins, antifungal metabolites that inhibit mRNA polyadenylation, from the Fusarium larvarum complex and other Hypocrealean fungi. Mycologia 2017, 101, 449–472. [Google Scholar] [CrossRef]
- Tran, V.T.; Braus-Stromeyer, S.A.; Timpner, C.; Braus, G.H. Molecular diagnosis to discriminate pathogen and apathogen species of the hybrid Verticillium longisporum on the oilseed crop Brassica napus. Appl. Microbiol. Biot. 2013, 97, 4467–4483. [Google Scholar] [CrossRef]
- Hyde, K.D.; Nilsson, R.H.; Alias, S.A.; Ariyawansa, H.A.; Blair, J.E.; Cai, L.; de Cock, A.W.A.M.; Dissanayake, A.J.; Glockling, S.L.; Goonasekara, I.D.; et al. One stop shop: Backbones trees for important phytopathogenic genera: I (2014). Fungal Divers. 2014, 67, 21–125. [Google Scholar] [CrossRef]
- Kumar, D.; Sigler, L.; Gibas, C.F.C.; Mohan, S.; Schuh, A.; Medeiros, B.C.; Peckham, K.; Humar, A. Graphium basitruncatum fungemia in a patient with acute leukemia. J. Clin. Microbiol. 2007, 45, 1644–1647. [Google Scholar] [CrossRef]
- Hage, H.; Miyauchi, S.; Virágh, M.; Drula, E.; Min, B.; Chaduli, D.; Navarro, D.; Favel, A.; Norest, M.; Lesage Meessen, L.; et al. Gene family expansions and transcriptome signatures uncover fungal adaptations to wood decay. Environ. Microbiol. 2021, 23, 5716–5732. [Google Scholar] [CrossRef]
- Tan, D.H.S.; Sigler, L.; Gibas, C.F.C.; Fong, I.W. Disseminated fungal infection in a renal transplant recipient involving Macrophomina phaseolina and Scytalidium dimidiatum: Case report and review of taxonomic changes among medically important members of the Botryosphaeriaceae. Med. Mycol. 2008, 46, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Alaei, H.; De Backer, M.; Nuytinck, J.; Maes, M.; Höfte, M.; Heungens, K. Phylogenetic relationships of Puccinia horiana and other rust pathogens of Chrysanthemum×morifolium based on rDNA ITS sequence analysis. Mycol. Res. 2009, 113, 668–683. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, E.; Arakawa, M.; Yamagishi, K.; Hara, A. Phylogenetic and structural analyses of the mating-type loci in Clavicipitaceae. Fems. Microbiol. Lett. 2006, 264, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Freed, S.; Jin, F.; Ren, S. Determination of genetic variability among the isolates of Metarhizium anisopliae var. anisopliae from different geographical origins. World J. Microb. Biot. 2011, 27, 359–370. [Google Scholar] [CrossRef]
- Stavrou, A.A.; Lackner, M.; Lass-Flörl, C.; Boekhout, T. The changing spectrum of Saccharomycotina yeasts causing candidemia: Phylogeny mirrors antifungal susceptibility patterns for azole drugs and amphothericin B. Fems. Yeast Res. 2019, 19, foz037. [Google Scholar] [CrossRef]
- Błońska, E.; Lasota, J.; Jankowiak, R.; Michalcewicz, J.; Wojas, T.; Zbyryt, A.; Ciach, M. Biological and physicochemical properties of the nests of White Stork Ciconia ciconia reveal soil entirely formed, modified and maintained by birds. Sci. Total Environ. 2021, 763, 143020. [Google Scholar] [CrossRef]
- Crous, P.W.; Schumacher, R.K.; Wingfield, M.J.; Akulov, A.; Denman, S.; Roux, J.; Braun, U.; Burgess, T.I.; Carnegie, A.J.; Váczy, K.Z.; et al. New and interesting fungi. 1. Fungal Syst. Evol. 2018, 1, 169–215. [Google Scholar] [CrossRef]
- De Carvalho Parahym, A.M.R.; Da Silva, C.M.; Domingos, I.D.F.; Gonçalves, S.S.; de Melo Rodrigues, M.; de Morais, V.L.L.; Neves, R.P. Pulmonary infection due to Pseudozyma aphidis in a patient with burkitt lymphoma: First case report. Diagn. Microbiol. Infect. Dis. 2013, 75, 104–106. [Google Scholar] [CrossRef]
- Salem, I.B.; Correia, K.C.; Boughalleb, N.; Michereff, S.J.; León, M.; Abad-Campos, P.; García-Jiménez, J.; Armengol, J. Monosporascus eutypoides, a cause of root rot and vine decline in Tunisia, and evidence that M. cannonballus and M. eutypoides are distinct species. Plant Dis. 2013, 97, 737–743. [Google Scholar] [CrossRef]
- Mackenzie, D.A.; Wongwathanarat, P.; Carter, A.T.; Archer, D.B. Isolation and use of a homologous histone H4 promoter and a ribosomal DNA region in a transformation vector for the oil-producing fungus Mortierella alpina. Appl. Environ. Microb. 2000, 66, 4655–4661. [Google Scholar] [CrossRef] [PubMed]
- Hurdeal, V.G.; Gentekaki, E.; Hyde, K.D.; Nguyen, T.T.T.; Lee, H.B. Novel Mucor species (Mucoromycetes, Mucoraceae) from northern Thailand. MycoKeys 2021, 84, 57–78. [Google Scholar] [CrossRef] [PubMed]
- Bukovská, P.; Jelínková, M.; Hršelová, H.; Sýkorová, Z.; Gryndler, M. Terminal restriction fragment length measurement errors are affected mainly by fragment length, G + C nucleotide content and secondary structure melting point. J. Microbiol. Meth. 2010, 82, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Jankowiak, R.; Ciach, M.; Bilański, P.; Linnakoski, R. Diversity of wood-inhabiting fungi in woodpecker nest cavities in southern Poland. Acta Mycol. 2019, 54, 1126. [Google Scholar] [CrossRef]
- Ligoxigakis, E.K.; Papaioannou, I.A.; Markakis, E.A.; Typas, M.A. First report of pink rot of Phoenix and Washingtonia Species Caused by Nalanthamala vermoesenii in Greece. Plant Dis. 2013, 97, 285. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, N.; Hattori, M.; Yokoyama, E.; Isomura, S.; Ujita, M.; Hara, A. Entomogenous fungi that produce 2,6-pyridine dicarboxylic acid (dipicolinic acid). J. Biosci. Bioeng. 2006, 102, 365–368. [Google Scholar] [CrossRef]
- Gorfer, M.; Blumhoff, M.; Klaubauf, S.; Urban, A.; Inselsbacher, E.; Bandian, D.; Mitter, B.; Sessitsch, A.; Wanek, W.; Strauss, J. Community profiling and gene expression of fungal assimilatory nitrate reductases in agricultural soil. ISME J. 2011, 5, 1771–1783. [Google Scholar] [CrossRef]
- Andrade, K.C.R.; Fernandes, R.A.; Pinho, D.B.; de Freitas, M.M.; Filho, E.X.F.; Pessoa, A.; Silva, J.I.; Magalhães, P.O. Sequencing and characterization of an L-asparaginase gene from a new species of Penicillium section Citrina isolated from Cerrado. Sci. Rep. 2021, 11, 17861. [Google Scholar] [CrossRef]
- Arteau, M.; Labrie, S.; Roy, D. Terminal-restriction fragment length polymorphism and automated ribosomal intergenic spacer analysis profiling of fungal communities in Camembert cheese. Int. Dairy J. 2010, 20, 545–554. [Google Scholar] [CrossRef]
- Pornsuriya, C.; Chitphithak, I. Blue mold caused by Penicillium oxalicum on muskmelon (Cucumis melo) in Thailand. Australasian Plant Dis. Notes 2018, 13, 46. [Google Scholar] [CrossRef]
- Faisal, M.; Elsayed, E.; Fitzgerald, S.D.; Silva, V.; Mendoza, L. Outbreaks of phaeohyphomycosis in the chinook salmon (Oncorhynchus tshawytscha) caused by Phoma herbarum. Mycopathologia 2007, 163, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Gómez Luciano, L.B.; Tsai, I.J.; Chuma, I.; Tosa, Y.; Chen, Y.; Li, J.; Li, M.; Lu, M.J.; Nakayashiki, H.; Li, W. Blast fungal genomes show frequent chromosomal changes, gene gains and losses, and effector gene turnover. Mol. Biol. Evol. 2019, 36, 1148–1161. [Google Scholar] [CrossRef] [PubMed]
- Robideau, G.P.; De Cock, A.W.A.M.; Coffey, M.D.; Voglmayr, H.; Brouwer, H.; Bala, K.; Chitty, D.W.; Désaulniers, N.; Eggertson, Q.A.; Gachon, C.M.M.; et al. DNA barcoding of oomycetes with cytochrome oxidase subunit I and internal transcribed spacer. Mol. Ecol. Resour. 2011, 11, 1002–1011. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.W.; Braun, U.; Schubert, K.; Groenewald, J.Z. Delimiting Cladosporium from morphologically similar genera. Stud. Mycol. 2007, 58, 33–56. [Google Scholar] [CrossRef]
- Ogawa, Y.; Tokumasu, S.; Tubaki, K. An original habitat of tempeh molds. Mycoscience 2004, 45, 271–276. [Google Scholar] [CrossRef]
- Ponizovskaya, V.B.; Rebrikova, N.L.; Kachalkin, A.V.; Antropova, A.B.; Bilanenko, E.N.; Mokeeva, V.L. Micromycetes as colonizers of mineral building materials in historic monuments and museums. Fungal Biol. 2019, 123, 290–306. [Google Scholar] [CrossRef]
- Buzina, W.; Lang-Loidolt, D.; Braun, H.; Freudenschuss, K.; Stammberger, H. Development of molecular methods for identification of Schizophyllum commune from clinical samples. J. Clin. Microbiol. 2001, 39, 2391–2396. [Google Scholar] [CrossRef]
- Simon, U.K.; Groenewald, J.Z.; Crous, P.W. Cymadothea trifolii, an obligate biotrophic leaf parasite of Trifolium, belongs to Mycosphaerellaceae as shown by nuclear ribosomal DNA analyses. Pers. Int. Mycol. J. 2009, 22, 49–55. [Google Scholar] [CrossRef]
- Pyrri, I.; Visagie, C.M.; Soccio, P.; Houbraken, J. Re-evaluation of the taxonomy of Talaromyces minioluteus. J. Fungi 2021, 7, 993. [Google Scholar] [CrossRef]
- Li, C.; Zhao, S.; Zhang, T.; Xian, L.; Liao, L.; Liu, J.; Feng, J. Genome sequencing and analysis of Talaromyces pinophilus provide insights into biotechnological applications. Sci. Rep. 2017, 7, 490. [Google Scholar] [CrossRef]
- Xue, J.; Wu, P.; Xu, L.; Wei, X. Penicillitone, a potent in vitro anti-inflammatory and cytotoxic rearranged sterol with an unusual tetracycle core produced by Penicillium purpurogenum. Org. Lett. 2014, 16, 1518–1521. [Google Scholar] [CrossRef] [PubMed]
- Krueger, D. Monographic Studies in the Genus Polyporus (Basidiomycotina); The University of Tennessee: Knoxville, TN, USA, 2023; Volume 5, p. 23. [Google Scholar]
- Madrid, H.; Hernández-Restrepo, M.; Gené, J.; Cano, J.; Guarro, J.; Silva, V. New and interesting chaetothyrialean fungi from Spain. Mycol. Progress 2016, 15, 1179–1201. [Google Scholar] [CrossRef]
- Chen, L.L.; Liu, L.J.; Shi, M.M.; Song, X.; Zheng, C.; Chen, X.; Zhang, Y. Characterization and gene cloning of a novel serine protease with nematicidal activity from Trichoderma pseudokoningii SMF2. Fems. Microbiol. Lett. 2009, 299, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Chung, D.; Kwon, Y.M.; Yang, Y. Telomere-to-telomere genome assembly of asparaginase-producing Trichoderma simmonsii. BMC Genom. 2021, 22, 830. [Google Scholar] [CrossRef]
- Jankowiak, R.; Stępniewska, H.; Bilański, P.; Taerum, S.J. Fungi as potential factors limiting natural regeneration of pedunculate oak (Quercus robur) in mixed-species forest stands in Poland. Plant Pathol. 2022, 71, 805–817. [Google Scholar] [CrossRef]




| Collection Time | Location | Growth Stage [38] | Number of Plants | Soil Number | Treatment Row/Pot Number |
|---|---|---|---|---|---|
| 20 July 2022 | Field | Seedling | 16 | 16 | CC 1-4; CT-4; CF-4; TF-4 |
| 5 August 2022 | Greenhouse | Seedling | 16 | 16 | |
| 14 August 2022 | Field | Flowering | 16 | 16 | |
| 21 August 2022 | Greenhouse | Vine growth | 16 | 16 | |
| Total | 64 | 64 |
| Fungal Type | Media | Number of Genera | Strain Number | Specific Genera Compared to PDA/Specific Genera with PDA |
|---|---|---|---|---|
| Endophytic fungi (Total 44 genera, 504 strains) | CMA | 26 | 138 | Acrocalymma 1; Boeremia; Clonostachys; Graphium; Moesziomyces 2; Mucor; Paramyrothecium |
| MEA | 26 | 121 | Ceratobasidium; Curvularia; Mucor; Paramyrothecium; Preussia | |
| Rhizosphere soil fungi (Total 61 genera, 771 strains) | CZA | 45 | 290 | Antarctomyces; Bisifusarium; Edenia; Geomyces; Marquandomyces; Meyerozyma; Microascus; Monosporascus; Nalanthamala; Ochroconis; Pyricularia; Sarocladium; Striaticonidium; Trichocladium |
| Endophytic fungi Rhizosphere soil fungi Total fungi (74 genera, 1275 strains) | PDA | 32 | 131 | Acremonium; Aureobasidium; Cephalotheca; Humicola; Pythium |
| 45 | 331 | Boeremia; Cephalotheca; Engyodontium; Glomerella; Linnemannia; Metarhizium; Paraisaria; Schizophyllum | ||
| 55 | 445 | |||
| Endophytic fungi | MRBA | 27 | 114 | Globisporangium; Graphium; Harknessia; Mucor; Trametes |
| Rhizosphere soil fungi | 33 | 150 | Chaetomidium; Irpex; Monosporascus | |
| Total fungi | 45 | 281 |
| Genera | Isolated from All 5 Cultural Media | Isolated from All 3 Media Used to Isolate Rhizosphere Fungi | Isolated from All 4 Media Used to Isolate Endophytic Fungi | Isolated from Roots, Stems and Leaves of Cucumber Plants | |
|---|---|---|---|---|---|
| 1 | Acremonium | + | |||
| 2 | Acrocalymma | + | |||
| 3 | Albifimbria | + | |||
| 4 | Alternaria * | + | + | + | + |
| 5 | Apiospora | + | |||
| 6 | Aspergillus | + | + | + | + |
| 7 | Chaetomium | + | + | + | |
| 8 | Cladosporium | + | + | + | + |
| 9 | Clonostachys | + | |||
| 10 | Colletotrichum | + | + | + | + |
| 11 | Coniochaeta | + | + | ||
| 12 | Edenia | + | + | ||
| 13 | Fusarium | + | + | + | + |
| 14 | Humicola | + | |||
| 15 | Lectera | + | |||
| 16 | Malassezia | + | |||
| 17 | Microdochium | + | + | ||
| 18 | Monosporascus | + | + | + | |
| 19 | Mortierella | + | |||
| 20 | Mucor | + | + | ||
| 21 | Paramyrothecium | + | + | ||
| 22 | Penicillium | + | + | + | + |
| 23 | Phoma | + | |||
| 24 | Plectosphaerella | + | + | + | + |
| 25 | Pseudogymnoascus | + | + | + | |
| 26 | Stagonosporopsis | + | + | + | + |
| 27 | Talaromyces | + | + | + | + |
| 28 | Trichoderma | + | + | + | + |
| Total | 17 | 24 | 16 | 12 |
| Plant Part/Cultural Medium | CMA | MEA | PDA | MRBA |
|---|---|---|---|---|
| Root | 5.50 a AB 1 | 2.50 b B | 3.00 b AB | 6.50 a A |
| Stem | 7.75 a AB | 6.25 a AB | 8.75 a B | 4.50 a A |
| Leaf | 4.25 a A | 3.75 ab A | 4.75 b A | 3.25 a A |
| Media/Isolates | CMA | CZA | MEA | MRBA | PDA |
|---|---|---|---|---|---|
| Fusarium (E)1 | 10.00 ± 1.73 a 3 | - | 8.50 ± 4.04 a | 9.25 ± 5.80 a | 6.50 ± 3.70 a |
| Fusarium (R)2 | - | 7.00 ± 3.37 ab | - | 5.75 ± 4.78 ab | 11.00 ± 7.35 ab |
| Trichoderma (E)1 | 21.21 ± 3.69 ab | - | 24.50 ± 4.65 a | 15.75 ± 5.25 b | 20.75 ± 5.32 ab |
| Trichoderma (R)2 | - | 93.50 ± 8.06 a | - | 128.50 ± 53.98 a | 100.00 ± 34.69 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, C.-Y.; Zhang, M.-Y.; Niu, Y.-C.; Zhang, M.; Geng, Y.-H.; Deng, H. Comparison of Fungal Genera Isolated from Cucumber Plants and Rhizosphere Soil by Using Various Cultural Media. J. Fungi 2023, 9, 934. https://doi.org/10.3390/jof9090934
Cheng C-Y, Zhang M-Y, Niu Y-C, Zhang M, Geng Y-H, Deng H. Comparison of Fungal Genera Isolated from Cucumber Plants and Rhizosphere Soil by Using Various Cultural Media. Journal of Fungi. 2023; 9(9):934. https://doi.org/10.3390/jof9090934
Chicago/Turabian StyleCheng, Chong-Yang, Ming-Yuan Zhang, Yong-Chun Niu, Meng Zhang, Yue-Hua Geng, and Hui Deng. 2023. "Comparison of Fungal Genera Isolated from Cucumber Plants and Rhizosphere Soil by Using Various Cultural Media" Journal of Fungi 9, no. 9: 934. https://doi.org/10.3390/jof9090934
APA StyleCheng, C.-Y., Zhang, M.-Y., Niu, Y.-C., Zhang, M., Geng, Y.-H., & Deng, H. (2023). Comparison of Fungal Genera Isolated from Cucumber Plants and Rhizosphere Soil by Using Various Cultural Media. Journal of Fungi, 9(9), 934. https://doi.org/10.3390/jof9090934





