Spectrometric Characterization of Clinical and Environmental Isolates of Aspergillus Series Versicolores
Abstract
:1. Introduction
2. Materials and Methods
2.1. Clinical and Environmental Isolates
2.2. Molecular Identification
2.3. Generation of the Local Database
2.3.1. Culture
2.3.2. Protein Extraction
2.3.3. Spectral Acquisition
2.3.4. Quality Control
2.3.5. Creation of MSP
2.4. Evaluation of MSP Library Performance
3. Results
3.1. Interest of Culture Media
3.2. Characteristics of the Local Database
3.3. Evaluation of the Local Database Performance and Comparison with the Bruker Database
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Domsch, K.H.; Gams, W.; Anderson, T.H. Compendium of Soil Fungi; Academic Press: London, UK; New York, NY, USA, 1980; ISBN 978-0-12-220401-2. [Google Scholar]
- Pitt, J.I.; Hocking, A.D. Fungi and Food Spoilage; Springer: New York, NY, USA, 2009; ISBN 978-0-387-92207-2. [Google Scholar]
- Delanoë, A.; Heutte, N.; Gente, S.; Séguin, V.; Garon, D. Relationships between Exposure to Bioaerosols, Moldy Surface and Symptoms in French Mold-Damaged Homes. Atmosphere 2020, 11, 223. [Google Scholar] [CrossRef]
- Géry, A.; Lepetit, C.; Heutte, N.; Séguin, V.; Bonhomme, J.; Garon, D. Cellular Cytotoxicity and Oxidative Potential of Recurrent Molds of the Genus Aspergillus Series Versicolores. Microorganisms 2022, 10, 228. [Google Scholar] [CrossRef] [PubMed]
- De, H.; Azad, S.M.; Giri, P.P.; Pal, P.; Ghosh, A.; Maitra, A. Two Cases of Non-Cystic Fibrosis (CF) Bronchiectasis with Allergic Bronchopulmonary Aspergillosis. Respir. Med. Case Rep. 2017, 20, 68–71. [Google Scholar] [CrossRef]
- Benndorf, D.; Müller, A.; Bock, K.; Manuwald, O.; Herbarth, O.; Von Bergen, M. Identification of Spore Allergens from the Indoor Mould Aspergillus Versicolor. Allergy 2008, 63, 454–460. [Google Scholar] [CrossRef] [PubMed]
- Charles, M.P.; Noyal, M.J.; Easow, J.M.; Ravishankar, M. Invasive Pulmonary Aspergillosis Caused by Aspergillus Versicolor in a Patient on Mechanical Ventilation. Australas. Med. J. 2011, 4, 632–634. [Google Scholar] [CrossRef]
- Gomes, C.C.; Pinto, L.C.C.; Victor, F.L.; da Silva, E.A.B.; Ribeiro, A.d.A.; Sarquis, M.I.d.M.; Camões, I.C.G. Aspergillus in Endodontic Infection near the Maxillary Sinus. Braz. J. Otorhinolaryngol. 2015, 81, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Moreno, G.; Arenas, R. Other Fungi Causing Onychomycosis. Clin. Dermatol. 2010, 28, 160–163. [Google Scholar] [CrossRef]
- Perri, P.; Campa, C.; Incorvaia, C.; Parmeggiani, F.; Lamberti, G.; Costagliola, C.; Sebastiani, A. Endogenous Aspergillus Versicolor Endophthalmitis in an Immuno-Competent HIV-Positive Patient. Mycopathologia 2005, 160, 259–261. [Google Scholar] [CrossRef]
- Baddley, J.W.; Marr, K.A.; Andes, D.R.; Walsh, T.J.; Kauffman, C.A.; Kontoyiannis, D.P.; Ito, J.I.; Balajee, S.A.; Pappas, P.G.; Moser, S.A. Patterns of Susceptibility of Aspergillus Isolates Recovered from Patients Enrolled in the Transplant-Associated Infection Surveillance Network. J. Clin. Microbiol. 2009, 47, 3271–3275. [Google Scholar] [CrossRef]
- Géry, A.; Séguin, V.; Eldin de Pécoulas, P.; Bonhomme, J.; Garon, D. Aspergilli Series Versicolores: Importance of Species Identification in the Clinical Setting. Crit. Rev. Microbiol. 2023, 49, 485–498. [Google Scholar] [CrossRef]
- Smith, G. A Manual of the Aspergilli. Nature 1946, 157, 462. [Google Scholar] [CrossRef]
- Peterson, S.W. Phylogenetic Analysis of Aspergillus Species Using DNA Sequences from Four Loci. Mycologia 2008, 100, 205–226. [Google Scholar] [CrossRef] [PubMed]
- Houbraken, J.; Kocsubé, S.; Visagie, C.M.; Yilmaz, N.; Wang, X.-C.; Meijer, M.; Kraak, B.; Hubka, V.; Bensch, K.; Samson, R.A.; et al. Classification of Aspergillus, Penicillium, Talaromyces and Related Genera (Eurotiales): An Overview of Families, Genera, Subgenera, Sections, Series and Species. Stud. Mycol. 2020, 95, 5–169. [Google Scholar] [CrossRef] [PubMed]
- Jurjevic, Z.; Peterson, S.W.; Horn, B.W. Aspergillus Section Versicolores: Nine New Species and Multilocus DNA Sequence Based Phylogeny. IMA Fungus Glob. Mycol. J. 2012, 3, 59–79. [Google Scholar] [CrossRef] [PubMed]
- Sklenář, F.; Glässnerová, K.; Jurjević, Ž.; Houbraken, J.; Samson, R.A.; Visagie, C.M.; Yilmaz, N.; Gené, J.; Cano, J.; Chen, A.J.; et al. Taxonomy of Aspergillus Series Versicolores: Species Reduction and Lessons Learned about Intraspecific Variability. Stud. Mycol. 2022, 102, 53–93. [Google Scholar] [CrossRef] [PubMed]
- Géry, A.; Rioult, J.-P.; Heutte, N.; Séguin, V.; Bonhomme, J.; Garon, D. First Characterization and Description of Aspergillus Series Versicolores in French Bioaerosols. J. Fungi 2021, 7, 676. [Google Scholar] [CrossRef] [PubMed]
- Patel, R. A Moldy Application of MALDI: MALDI-ToF Mass Spectrometry for Fungal Identification. J. Fungi 2019, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Li, T.-Y.; Liu, B.-H.; Chie, C. Characterization of Aspergillus Spores by Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry. Rapid Commun. Mass Spectrom. 2000, 14, 2393–2400. [Google Scholar] [CrossRef]
- Barker, K.R.; Kus, J.V.; Normand, A.-C.; Gharabaghi, F.; McTaggart, L.; Rotstein, C.; Richardson, S.E.; Campigotto, A.; Tadros, M. A Practical Workflow for the Identification of Aspergillus, Fusarium, Mucorales by MALDI-TOF MS: Database, Medium, and Incubation Optimization. J. Clin. Microbiol. 2022, 60, e01032-22. [Google Scholar] [CrossRef]
- Vidal-Acuña, M.R.; Ruiz-Pérez de Pipaón, M.; Torres-Sánchez, M.J.; Aznar, J. Identification of Clinical Isolates of Aspergillus, Including Cryptic Species, by Matrix Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS). Med. Mycol. 2018, 56, 838–846. [Google Scholar] [CrossRef]
- Imbert, S.; Normand, A.C.; Gabriel, F.; Cassaing, S.; Bonnal, C.; Costa, D.; Lachaud, L.; Hasseine, L.; Kristensen, L.; Schuttler, C.; et al. Multi-Centric Evaluation of the Online MSI Platform for the Identification of Cryptic and Rare Species of Aspergillus by MALDI-TOF. Med. Mycol. 2019, 57, 962–968. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.; Wang, Q.; Wei, L.; Wan, Z.; Li, R.; Yu, J. Limitations of Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry for the Identification of Aspergillus Species. Med. Mycol. 2022, 60, myab084. [Google Scholar] [CrossRef]
- Garrigos, T.; Neuwirth, C.; Chapuis, A.; Bador, J.; Amoureux, L.; Andre, E.; Barbier, E.; Caillon, J.; Cardot-Martin, E.; Cattoir, V.; et al. Development of a Database for the Rapid and Accurate Routine Identification of Achromobacter Species by Matrix-Assisted Laser Desorption/Ionization–Time-of-Flight Mass Spectrometry (MALDI-TOF MS). Clin. Microbiol. Infect. 2021, 27, e1–e126. [Google Scholar] [CrossRef]
- Asare, P.T.; Lee, C.-H.; Hürlimann, V.; Teo, Y.; Cuénod, A.; Akduman, N.; Gekeler, C.; Afrizal, A.; Corthesy, M.; Kohout, C.; et al. A MALDI-TOF MS Library for Rapid Identification of Human Commensal Gut Bacteria from the Class Clostridia. Front. Microbiol. 2023, 14, 1104707. [Google Scholar] [CrossRef]
- Ouarti, B.; Laroche, M.; Righi, S.; Meguini, M.N.; Benakhla, A.; Raoult, D.; Parola, P. Development of MALDI-TOF Mass Spectrometry for the Identification of Lice Isolated from Farm Animals. Parasite 2020, 27, 28. [Google Scholar] [CrossRef] [PubMed]
- Reeve, M.A.; Buddie, A.G.; Pollard, K.M.; Varia, S.; Seier, M.K.; Offord, L.C.; Cock, M.J.W. A Highly-Simplified and Inexpensive MALDI-TOF Mass Spectrometry Sample-Preparation Method with Broad Applicability to Microorganisms, Plants, and Insects. J. Biol. Methods 2018, 5, e103. [Google Scholar] [CrossRef] [PubMed]
- Reeve, M.A.; Caine, T.S.; Buddie, A.G. Spectral Grouping of Nominally Aspergillus Versicolor Microbial-Collection Deposits by MALDI-TOF MS. Microorganisms 2019, 7, 235. [Google Scholar] [CrossRef]
- Masih, A.; Singh, P.K.; Kathuria, S.; Agarwal, K.; Meis, J.F.; Chowdhary, A. Identification by Molecular Methods and Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry and Antifungal Susceptibility Profiles of Clinically Significant Rare Aspergillus Species in a Referral Chest Hospital in Delhi, India. J. Clin. Microbiol. 2016, 54, 2354–2364. [Google Scholar] [CrossRef]
- Alanio, A.; Beretti, J.-L.; Dauphin, B.; Mellado, E.; Quesne, G.; Lacroix, C.; Amara, A.; Berche, P.; Nassif, X.; Bougnoux, M.-E. Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry for Fast and Accurate Identification of Clinically Relevant Aspergillus Species. Clin. Microbiol. Infect. 2011, 17, 750–755. [Google Scholar] [CrossRef]
- Becker, P.T.; de Bel, A.; Martiny, D.; Ranque, S.; Piarroux, R.; Cassagne, C.; Detandt, M.; Hendrickx, M. Identification of Filamentous Fungi Isolates by MALDI-TOF Mass Spectrometry: Clinical Evaluation of an Extended Reference Spectra Library. Med. Mycol. 2014, 52, 826–834. [Google Scholar] [CrossRef]
- Cassagne, C.; Ranque, S.; Normand, A.C.; Fourquet, P.; Thiebault, S.; Planard, C.; Hendrickx, M.; Piarroux, R. Mould Routine Identification in the Clinical Laboratory by Matrix-Assisted Laser Desorption Ionization Time-Of-Flight Mass Spectrometry. PLoS ONE 2011, 6, e28425. [Google Scholar] [CrossRef] [PubMed]
- Normand, A.C.; Gabriel, F.; Riat, A.; Cassagne, C.; Bourgeois, N.; Huguenin, A.; Chauvin, P.; Geyter, D.; Bexkens, M.; Rubio Garcia, E.; et al. Optimization of MALDI-ToF Mass Spectrometry for Yeast Identification: A Multicenter Study. Med. Mycol. 2019, 58, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Valentine, N.; Wunschel, S.; Wunschel, D.; Petersen, C.; Wahl, K. Effect of Culture Conditions on Microorganism Identification by Matrix-Assisted Laser Desorption Ionization Mass Spectrometry. Appl. Environ. Microbiol. 2005, 71, 58–64. [Google Scholar] [CrossRef] [PubMed]
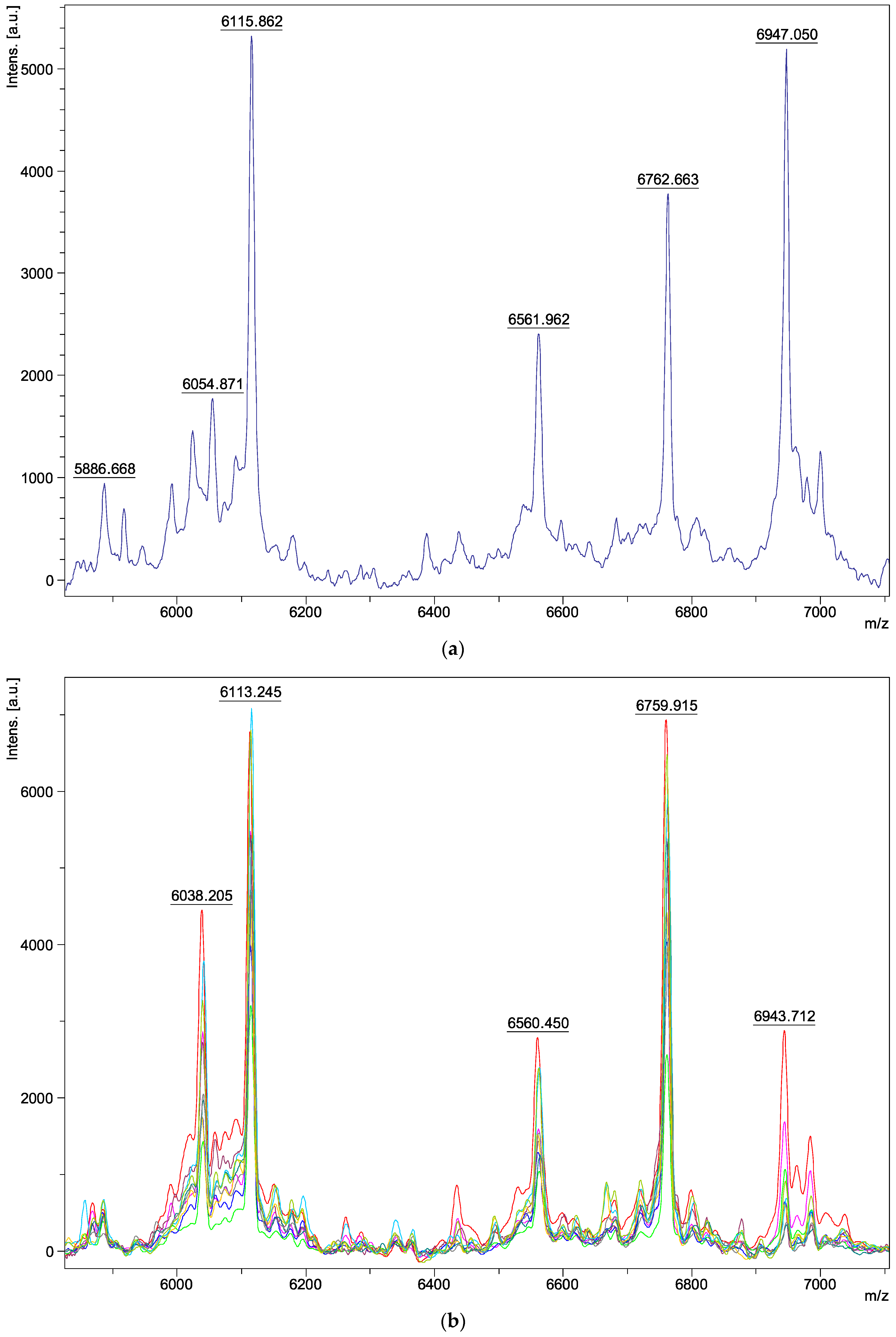
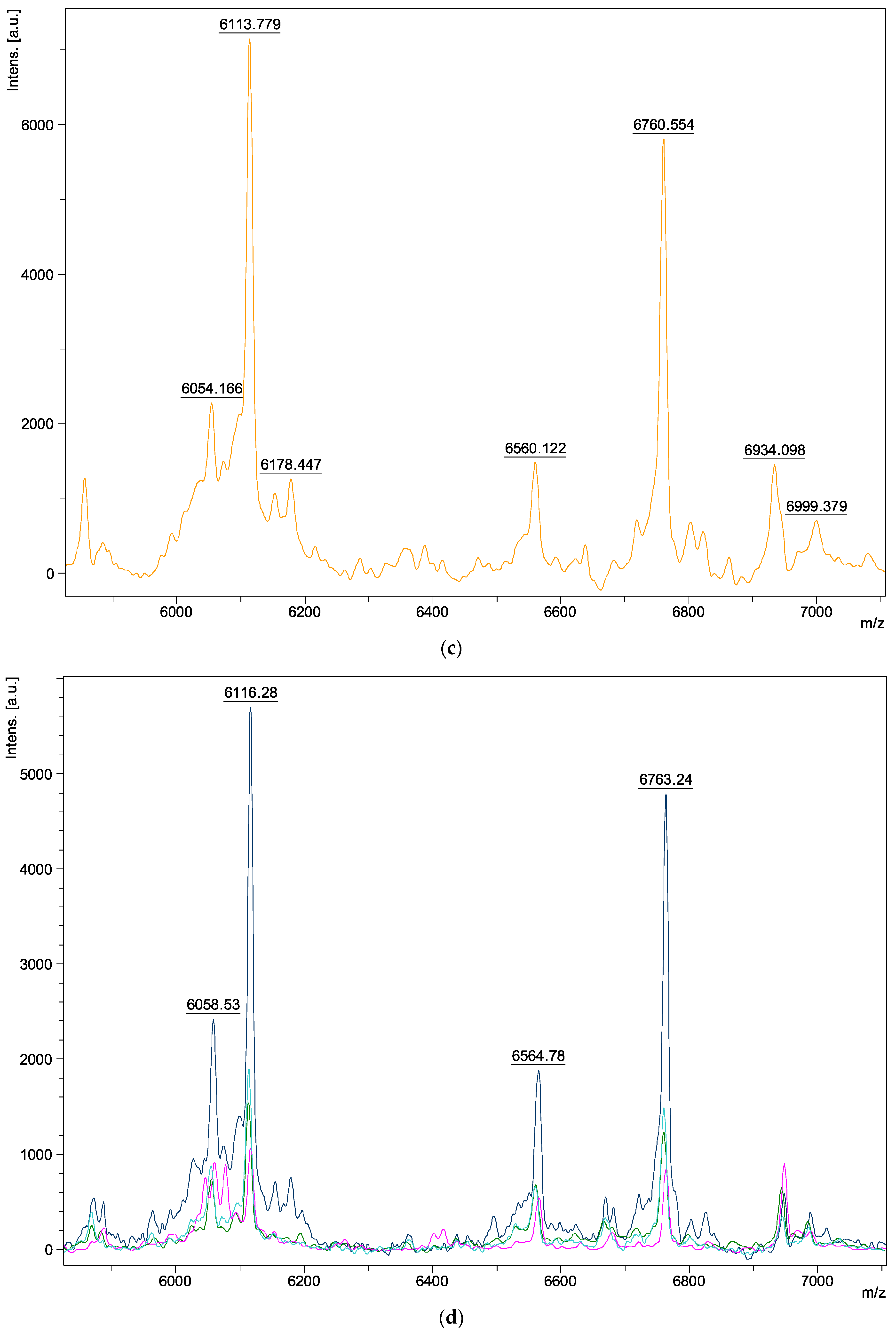
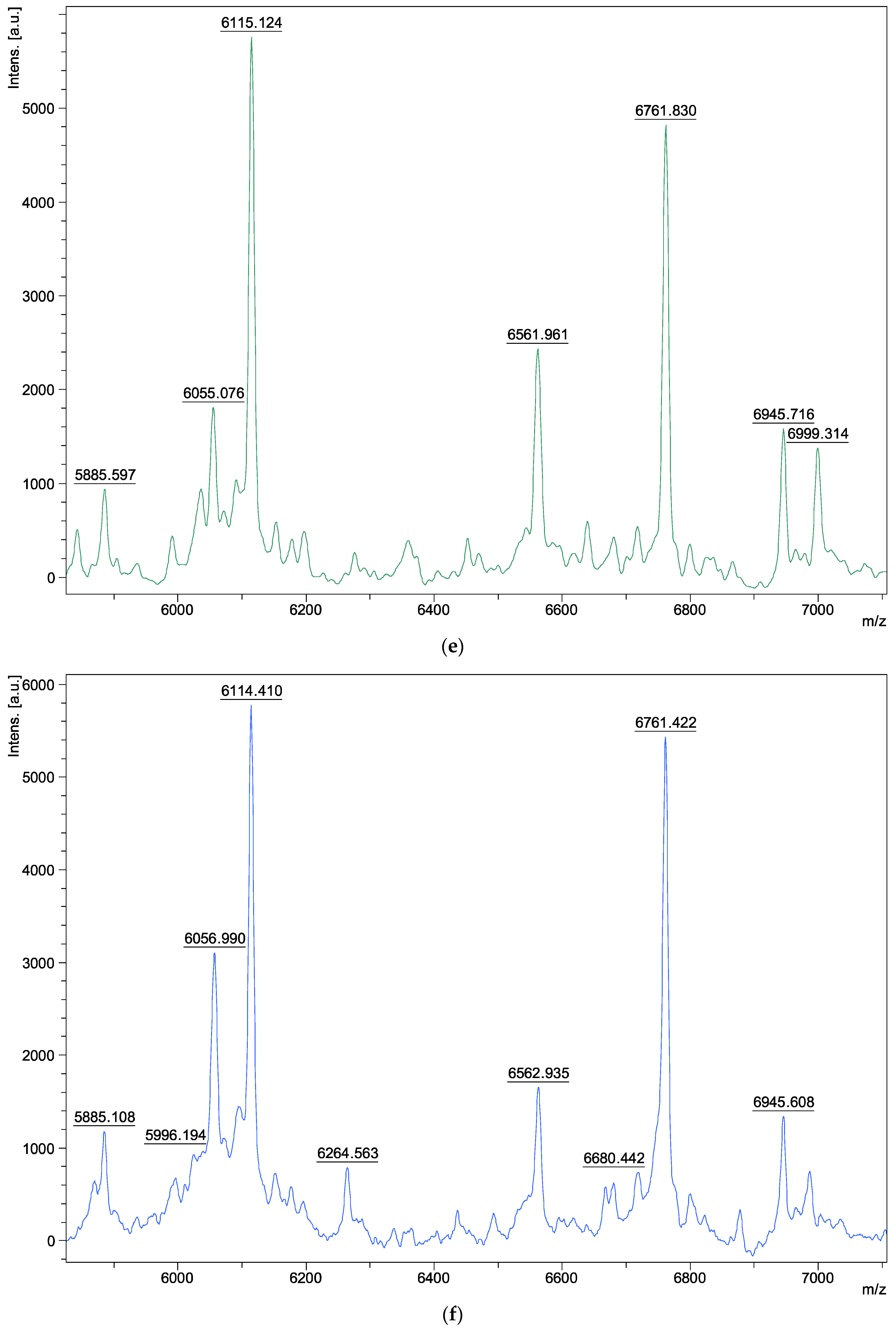

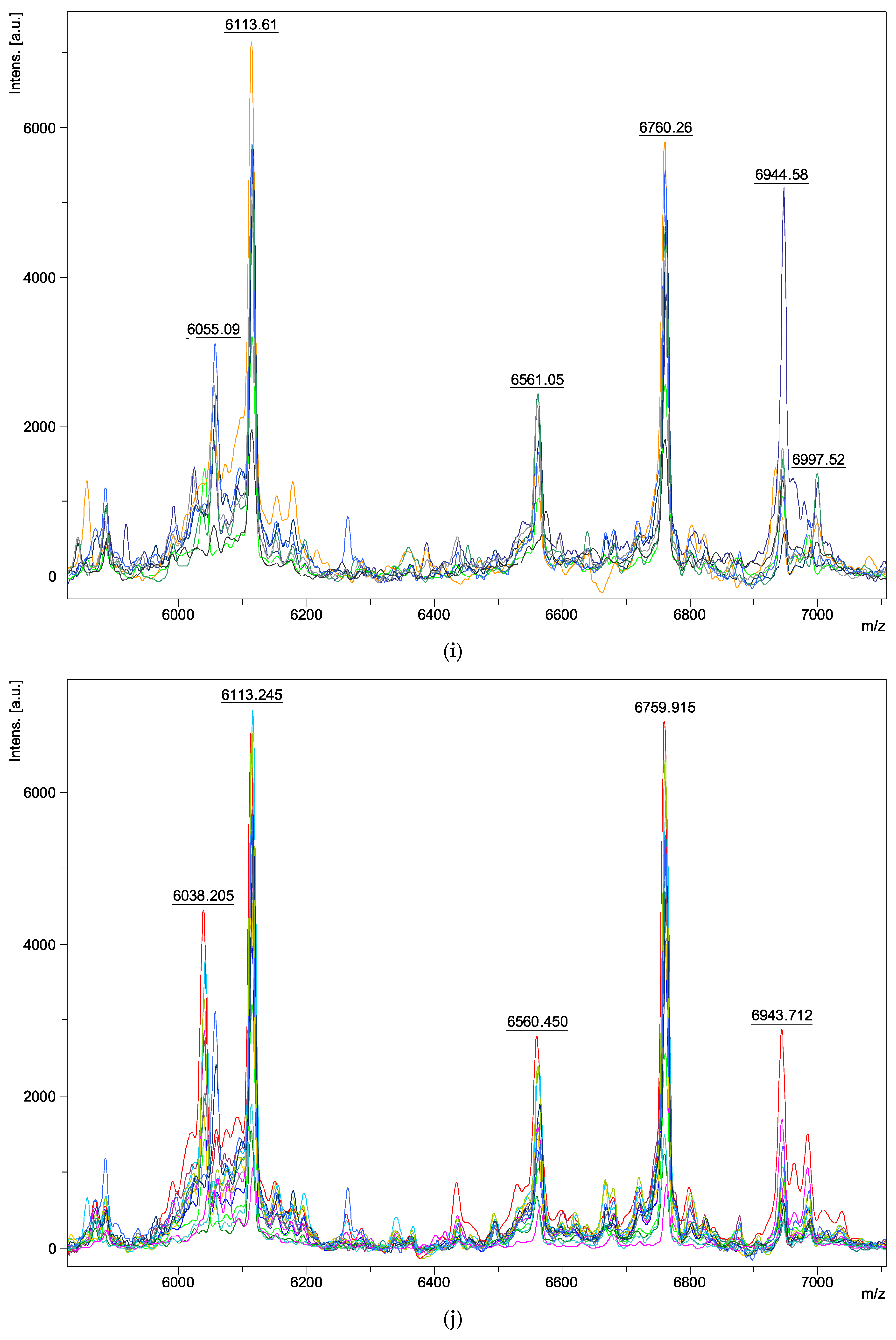
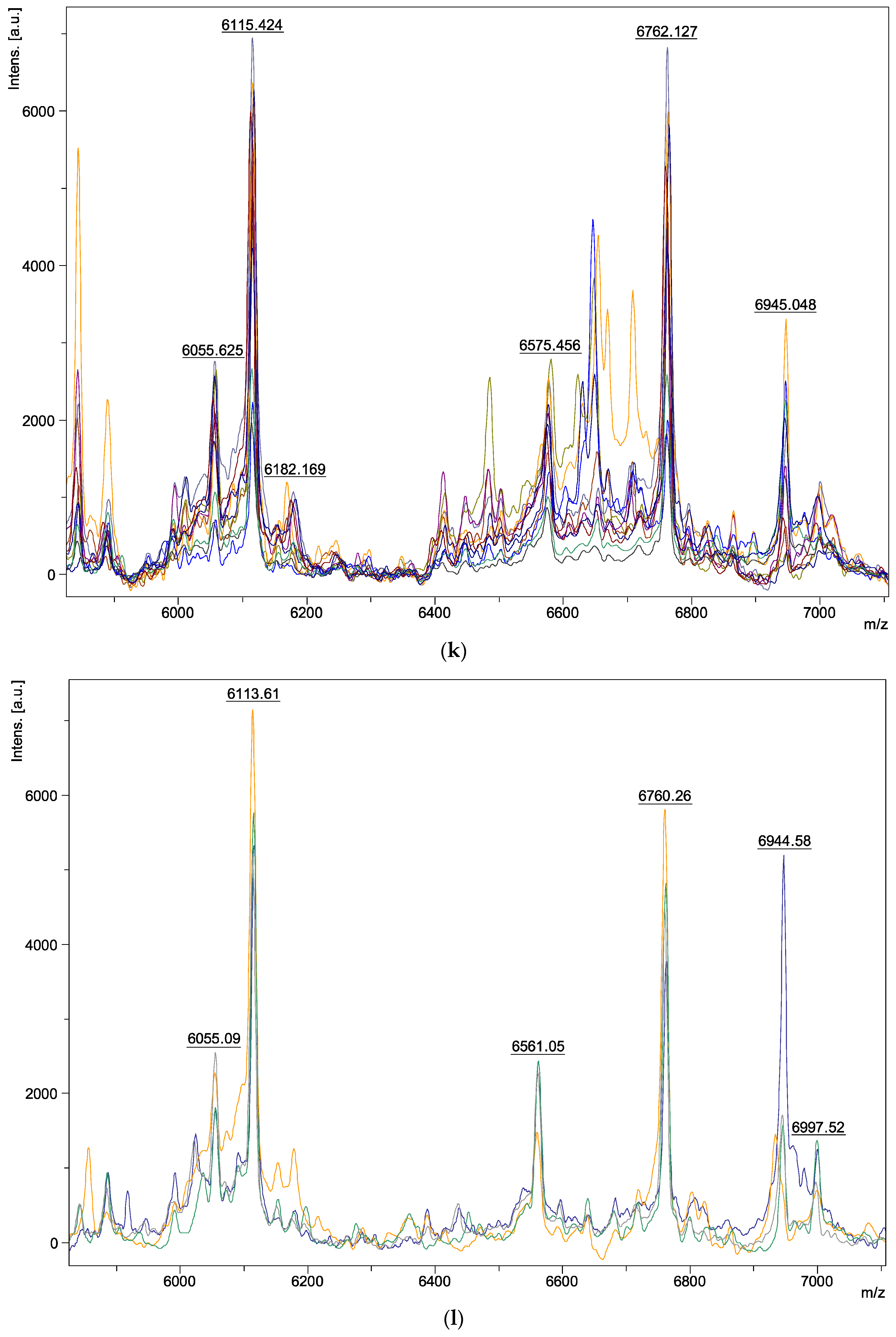
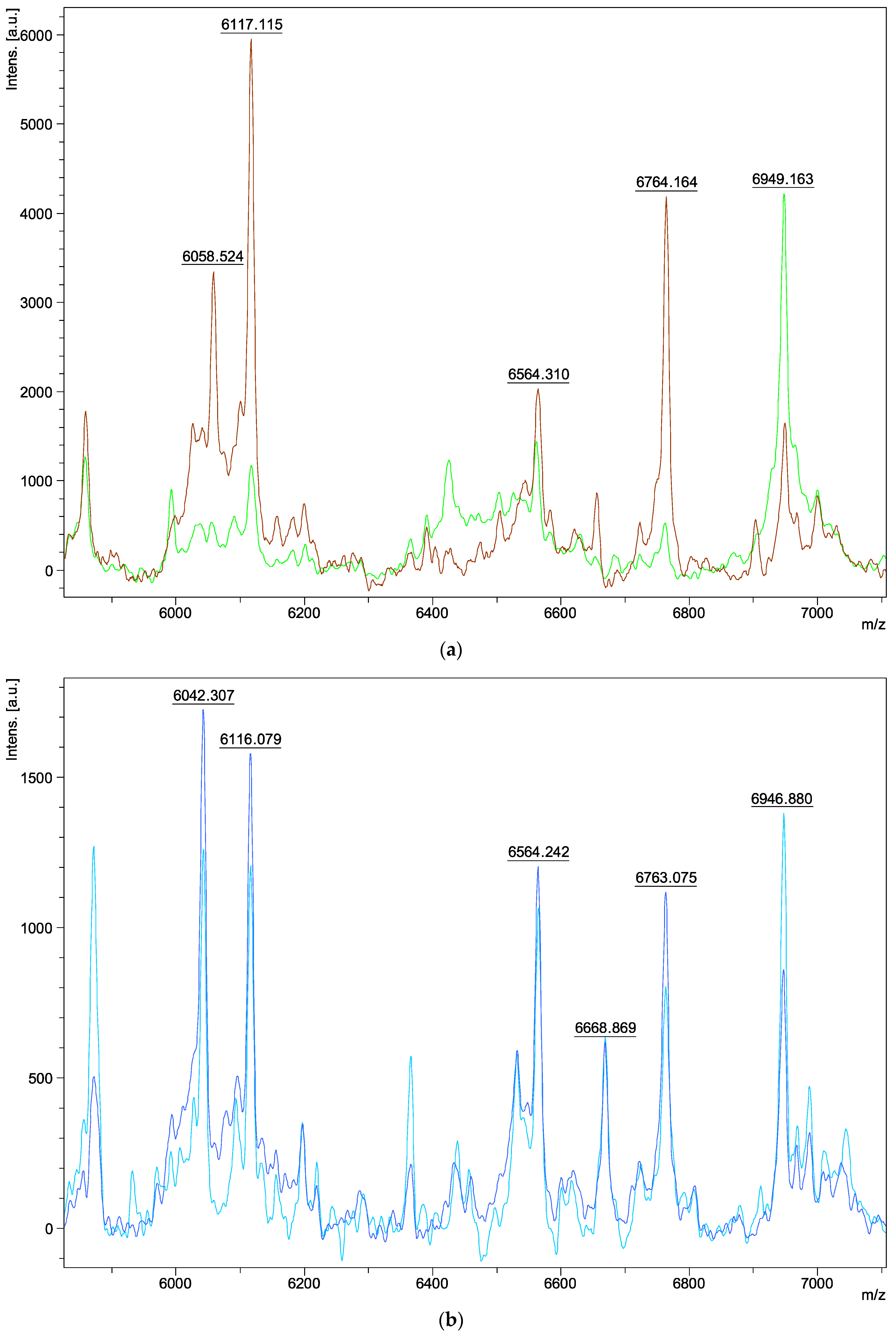
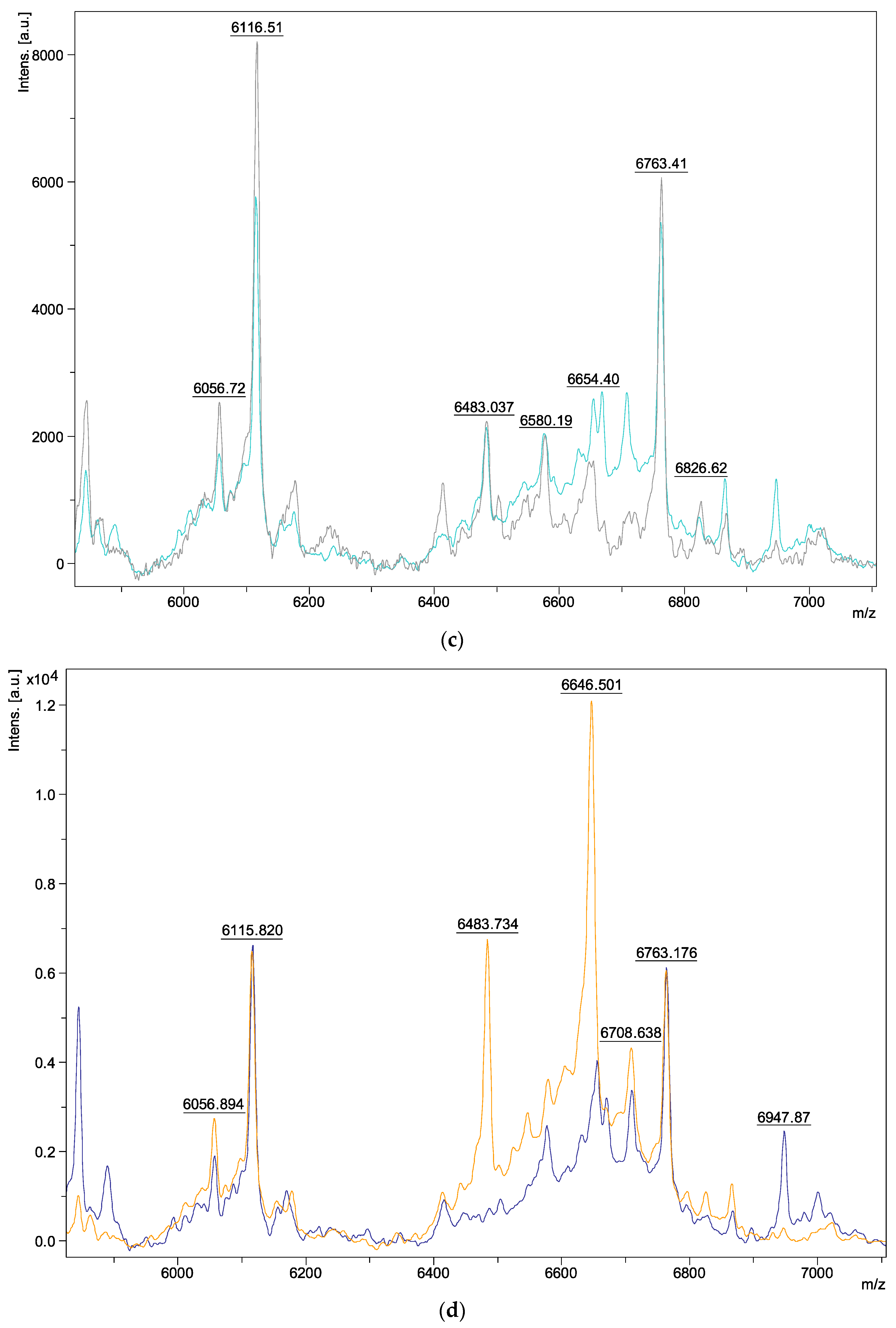
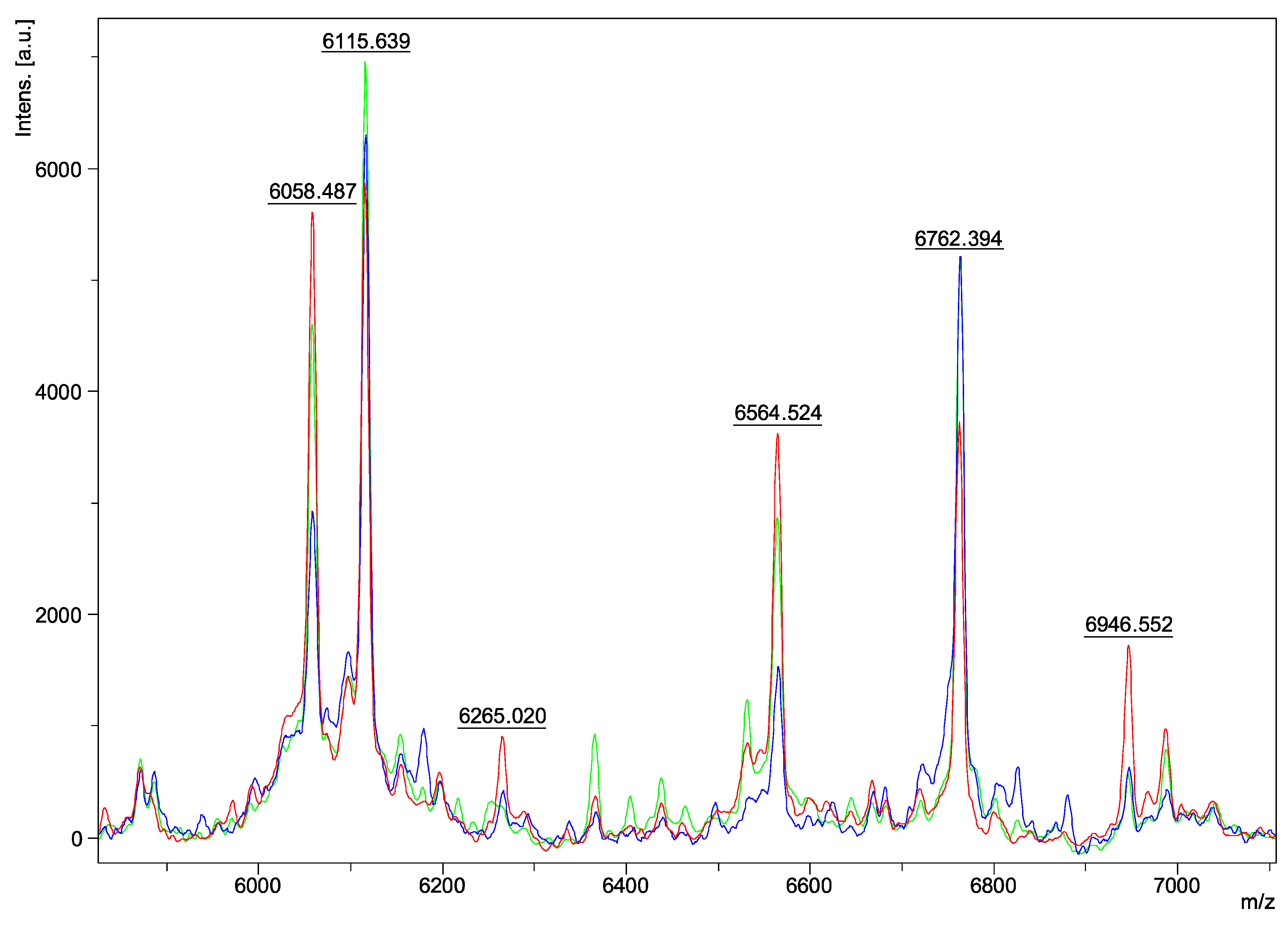
| Species | Isolate Number | Origins |
|---|---|---|
| Aspergillus amoenus | 1 (E) | Mold-damaged home |
| Aspergillus creber | 2 (E) | Cancer treatment center |
| 3 (E) | Cancer treatment center | |
| 4 (E) | Serpula lacrymans-damaged home | |
| 5 (C) | Skin scales of right foot | |
| 6 (C) | Skin scales of right foot | |
| 7 (C) | Sputum | |
| 8 (E) | Mold-damaged home | |
| 9 (E) | Mold-damaged home | |
| 10 (E) | Mold-damaged home | |
| 11 (E) | Mold-damaged home | |
| Aspergillus fructus | 12 (C) | Nail of big toe |
| Aspergillus jensenii | 13 (C) | Scalp |
| 14 (C) | Bronchoalveolar lavage fluid | |
| 15 (C) | Bronchoalveolar lavage fluid | |
| 16 (E) | Mold-damaged home | |
| Aspergillus protuberus | 17 (E) | Cancer treatment center |
| Aspergillus puulaauensis | 18 (C) | Armpit skin |
| Aspergillus sydowii | 19 (C) | Bronchoalveolar lavage fluid |
| 20 (C) | Bronchoalveolar lavage fluid | |
| 21 (C) | External auditory canal | |
| 22 (C) | Bronchoalveolar lavage fluid | |
| 23 (C) | Bronchoalveolar lavage fluid | |
| 24 (C) | Bronchoalveolar lavage fluid | |
| 25 (C) | Bronchoalveolar lavage fluid | |
| 26 (C) | Nail | |
| 27 (C) | Bronchoalveolar lavage fluid | |
| 28 (C) | Bronchoalveolar lavage fluid | |
| 29 (C) | Bronchoalveolar lavage fluid | |
| Aspergillus tabacinus | 30 (C) | Sputum (cystic fibrosis) |
| Species | Isolate Number | Culture Medium | Number of MSPs Obtained | ||||||
|---|---|---|---|---|---|---|---|---|---|
| SAB | MEA | CZA | LIQUID SAB | ||||||
| First Attempt | Second Attempt | Third Attempt | First Attempt | Second Attempt | First Attempt | First Attempt | |||
| Aspergillus amoenus | 1 I | 1 MSP | / | / | 2 MSPs | / | 0 MSP | / | 3 |
| Aspergillus creber | 2 (E) | 0 MSP | 0 MSP | 1 MSP | 0 MSP | 0 MSP | 0 MSP | 1 MSP | 2 |
| 3 (E) | 0 MSP | 0 MSP | 1 MSP | 1 MSP | / | / | / | 2 | |
| 4 (E) | 0 MSP | 0 MSP | 1 MSP | 0 MSP | 0 MSP | 0 MSP | 1 MSP | 2 | |
| 5 (C) | 0 MSP | 0 MSP | 1 MSP | 1 MSP | / | / | / | 2 | |
| 6 (C) | 1 MSP | / | / | 0 MSP | 0 MSP | 0 MSP | 0 MSP | 1 | |
| 7 (C) | 0 MSP | 0 MSP | 1 MSP | 0 MSP | 1 MSP | 0 MSP | / | 2 | |
| 8 (E) | 1 MSP | / | / | 0 MSP | 0 MSP | 0 MSP | 0 MSP | 1 | |
| 9 (E) | 0 MSP | 0 MSP | 1 MSP | 1 MSP | / | / | / | 2 | |
| 10 (E) | 1 MSP | / | / | 2 MSPs | / | / | / | 3 | |
| 11 (E) | 0 MSP | 0 MSP | 1 MSP | 1 MSP | / | / | / | 2 | |
| Aspergillus fructus | 12 (C) | 1 MSP | / | / | 1 MSP | / | 0 MSP | / | 2 |
| Aspergillus jensenii | 13 (C) | 1 MSP | / | / | 1 MSP | / | / | / | 2 |
| 14 (C) | 0 MSP | 0 MSP | 1 MSP | 0 MSP | 0 MSP | 0 MSP | 0 MSP | 1 | |
| 15 (C) | 1 MSP | / | / | 1 MSP | / | / | / | 2 | |
| 16 (E) | 0 MSP | 0 MSP | 1 MSP | 0 MSP | 0 MSP | 0 MSP | 0 MSP | 1 | |
| Aspergillus protuberus | 17 (E) | 0 MSP | 0 MSP | 1 MSP | 0 MSP | 0 MSP | 0 MSP | 0 MSP | 1 |
| Aspergillus puulaauensis | 18 (C) | 0 MSP | 0 MSP | 1 MSP | 1 MSP | / | 0 MSP | 1 MSP | 3 |
| Aspergillus sydowii | 19 (C) | 2 MSPs | / | / | 1 MSP | / | / | / | 3 |
| 20 (C) | 1 MSP | / | / | 1 MSP | / | / | / | 2 | |
| 21 (C) | 1 MSP | / | / | 1 MSP | / | / | / | 2 | |
| 22 (C) | 1 MSP | / | / | 1 MSP | / | / | / | 2 | |
| 23 (C) | 2 MSPs | / | / | 0 MSP | 0 MSP | 0 MSP | / | 2 | |
| 24 (C) | 1 MSP | / | / | 1 MSP | / | / | / | 2 | |
| 25 (C) | 1 MSP | / | / | 1 MSP | / | / | / | 2 | |
| 26 (C) | 1 MSP | / | / | 1 MSP | / | / | / | 2 | |
| 27 (C) | 1 MSP | / | / | 1 MSP | / | / | / | 2 | |
| 28 (C) | 1 MSP | / | / | 1 MSP | / | / | / | 2 | |
| 29 (C) | 1 MSP | / | / | 1 MSP | / | / | / | 2 | |
| Aspergillus tabacinus | 30 (C) | 1 MSP | / | / | 1 MSP | / | 0 MSP | / | 2 |
| Species | Isolates Number | Bruker Database | Local Database | Total Number of Profiles (/10) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Log Score ≥ 2.00 | Number of Profiles | 1.70 ≤ Log Score < 2.00 | Number of Profiles | Log Score ≥ 2.00 | Number of Profiles | 1,70 ≤ Log Score < 2.00 | Number of Profiles | ||||
| Aspergillus amoenus | 1 (E) | Aspergillus versicolor 120227_14 ETL | 1 | 0 | Aspergillus amoenus | 2 | 0 | 10 | |||
| Aspergillus versicolor 392 UGB | 1 | Aspergillus tabacinus | 1 | ||||||||
| Aspergillus versicolor D_16_256_8_1 LLH | 1 | Aspergillus fructus | 2 | ||||||||
| Aspergillus versicolor 1343 MPA | 1 | Aspergillus protuberus | 1 | ||||||||
| Aspergillus creber | 2 (E) | Aspergillus versicolor F51 LLH | 1 | 0 | Aspergillus creber | 7 | 0 | 10 | |||
| Aspergillus versicolor DSM 63292 DSM | 1 | Aspergillus puulaauensis | 1 | ||||||||
| 3 (E) | 0 | 0 | Aspergillus creber | 9 | 0 | 10 | |||||
| Aspergillus puulaauensis | 1 | ||||||||||
| 4 (E) | Aspergillus versicolor F51 LLH | 1 | 0 | Aspergillus creber | 8 | 0 | 10 | ||||
| Aspergillus puulaauensis | 1 | ||||||||||
| 5 (C) | 0 | 0 | Aspergillus creber | 9 | 0 | 10 | |||||
| Aspergillus puulaauensis | 1 | ||||||||||
| 6 (C) | Aspergillus versicolor F51 LLH | 1 | 0 | Aspergillus creber | 8 | 0 | 10 | ||||
| Aspergillus puulaauensis | 1 | ||||||||||
| 7 (C) | 0 | 0 | Aspergillus creber | 10 | 0 | 10 | |||||
| 8 (E) | Aspergillus versicolor F51 LLH | 1 | 0 | Aspergillus creber | 9 | 0 | 10 | ||||
| 9 (E) | Aspergillus versicolor F51 LLH | 1 | 0 | Aspergillus creber | 9 | 0 | 10 | ||||
| 10 (E) | 0 | 0 | Aspergillus creber | 10 | 0 | 10 | |||||
| 11 (E) | Aspergillus versicolor F51 LLH | 1 | 0 | Aspergillus creber | 8 | 0 | 10 | ||||
| Aspergillus puulaauensis | 1 | ||||||||||
| Aspergillus fructus | 12 (C) | Aspergillus versicolor 120227_14 ETL | 1 | Aspergillus versicolor 1343 MPA | 1 | Aspergillus fructus | 2 | Aspergillus protuberus | 1 | 10 | |
| Aspergillus versicolor 392 UGB | 1 | Aspergillus amoenus | 1 | Aspergillus puulaauensis | 1 | ||||||
| Aspergillus versicolor D_16_256_8_1 LLH | 1 | Aspergillus tabacinus | 1 | ||||||||
| Aspergillus jensenii | 13 (C) | 0 | 0 | Aspergillus jensenii | 4 | 0 | 10 | ||||
| Aspergillus creber | 3 | ||||||||||
| Aspergillus puulaauensis | 3 | ||||||||||
| 14 (C) | 0 | 0 | Aspergillus jensenii | 3 | Aspergillus jensenii | 1 | 10 | ||||
| Aspergillus creber | 2 | Aspergillus creber | 3 | ||||||||
| Aspergillus puulaauensis | 1 | ||||||||||
| 15 (C) | 0 | 0 | Aspergillus jensenii | 5 | 0 | 10 | |||||
| Aspergillus creber | 4 | ||||||||||
| Aspergillus puulaauensis | 1 | ||||||||||
| 16 (E) | 0 | 0 | Aspergillus jensenii | 3 | Aspergillus jensenii | 2 | 10 | ||||
| Aspergillus creber | 2 | Aspergillus creber | 2 | ||||||||
| Aspergillus puulaauensis | 1 | ||||||||||
| Aspergillus protuberus | 17 (E) | 0 | Aspergillus versicolor 120227_14 ETL | 1 | Aspergillus protuberus | 1 | Aspergillus fructus | 2 | 10 | ||
| Aspergillus tabacinus | 1 | Aspergillus sydowii | 3 | ||||||||
| Aspergillus amoenus | 1 | Aspergillus puulaauensis | 1 | ||||||||
| Aspergillus puulaauensis | 18 (C) | Aspergillus versicolor F51 LLH | 1 | 0 | Aspergillus puulaauensis | 3 | 0 | 10 | |||
| Aspergillus jensenii | 2 | ||||||||||
| Aspergillus creber | 4 | ||||||||||
| Aspergillus sydowii | 19 (C) | Aspergillus versicolor 1343 MPA | 1 | 0 | Aspergillus sydowii | 8 | Aspergillus sydowii | 1 | 10 | ||
| 20 (C) | 0 | 0 | Aspergillus sydowii | 9 | Aspergillus sydowii | 1 | 10 | ||||
| 21 (C) | 0 | 0 | Aspergillus sydowii | 9 | Aspergillus sydowii | 1 | 10 | ||||
| 22 (C) | Aspergillus versicolor 1343 MPA | 1 | 0 | Aspergillus sydowii | 6 | Aspergillus sydowii | 3 | 10 | |||
| 23 (C) | 0 | Aspergillus sydowii 2008_141783 MUZ | 1 | Aspergillus sydowii | 2 | Aspergillus sydowii | 7 | 10 | |||
| 24 (C) | 0 | Aspergillus versicolor 1343 MPA | 1 | Aspergillus sydowii | 5 | Aspergillus sydowii | 4 | 10 | |||
| 25 (C) | 0 | Aspergillus sydowii 2008_141783 MUZ | 1 | Aspergillus sydowii | 8 | Aspergillus sydowii | 1 | 10 | |||
| 26 (C) | 0 | 0 | Aspergillus sydowii | 10 | 0 | 10 | |||||
| 27 (C) | Aspergillus versicolor 1343 MPA | 1 | 0 | Aspergillus sydowii | 8 | 0 | 10 | ||||
| Aspergillus amoenus | 1 | ||||||||||
| 28 (C) | 0 | Aspergillus versicolor 1343 MPA | 1 | 0 | Aspergillus sydowii | 8 | 10 | ||||
| Aspergillus versicolor 2009_137364 MUZ | 1 | ||||||||||
| 29 (C) | Aspergillus versicolor 1343 MPA | 1 | 0 | Aspergillus sydowii | 9 | 0 | 10 | ||||
| Aspergillus tabacinus | 30 (C) | Aspergillus versicolor 120227_14 ETL | 1 | 0 | Aspergillus tabacinus | 2 | Aspergillus fructus | 1 | 10 | ||
| Aspergillus amoenus | 1 | Aspergillus jensenii | 2 | ||||||||
| Aspergillus protuberus | 1 | Aspergillus amoenus | 1 | ||||||||
| Aspergillus fructus | 1 | ||||||||||
| Total number of profiles | 18 | 9 | 225 | 48 | 300 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jomat, O.; Géry, A.; Leudet, A.; Capitaine, A.; Garon, D.; Bonhomme, J. Spectrometric Characterization of Clinical and Environmental Isolates of Aspergillus Series Versicolores. J. Fungi 2023, 9, 868. https://doi.org/10.3390/jof9090868
Jomat O, Géry A, Leudet A, Capitaine A, Garon D, Bonhomme J. Spectrometric Characterization of Clinical and Environmental Isolates of Aspergillus Series Versicolores. Journal of Fungi. 2023; 9(9):868. https://doi.org/10.3390/jof9090868
Chicago/Turabian StyleJomat, Océane, Antoine Géry, Astrid Leudet, Agathe Capitaine, David Garon, and Julie Bonhomme. 2023. "Spectrometric Characterization of Clinical and Environmental Isolates of Aspergillus Series Versicolores" Journal of Fungi 9, no. 9: 868. https://doi.org/10.3390/jof9090868
APA StyleJomat, O., Géry, A., Leudet, A., Capitaine, A., Garon, D., & Bonhomme, J. (2023). Spectrometric Characterization of Clinical and Environmental Isolates of Aspergillus Series Versicolores. Journal of Fungi, 9(9), 868. https://doi.org/10.3390/jof9090868






