Biogeographic Patterns of Fungal Sub-Communities under Different Land-Use Types in Subtropical China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site and Soil Sampling
2.2. Determination of Soil Physicochemical Properties
2.3. Soil DNA Extraction and 16S rRNA Gene Sequencing
2.4. Processing of Sequence Analysis
2.5. Statistical and Bioinformatic Analyses
3. Results
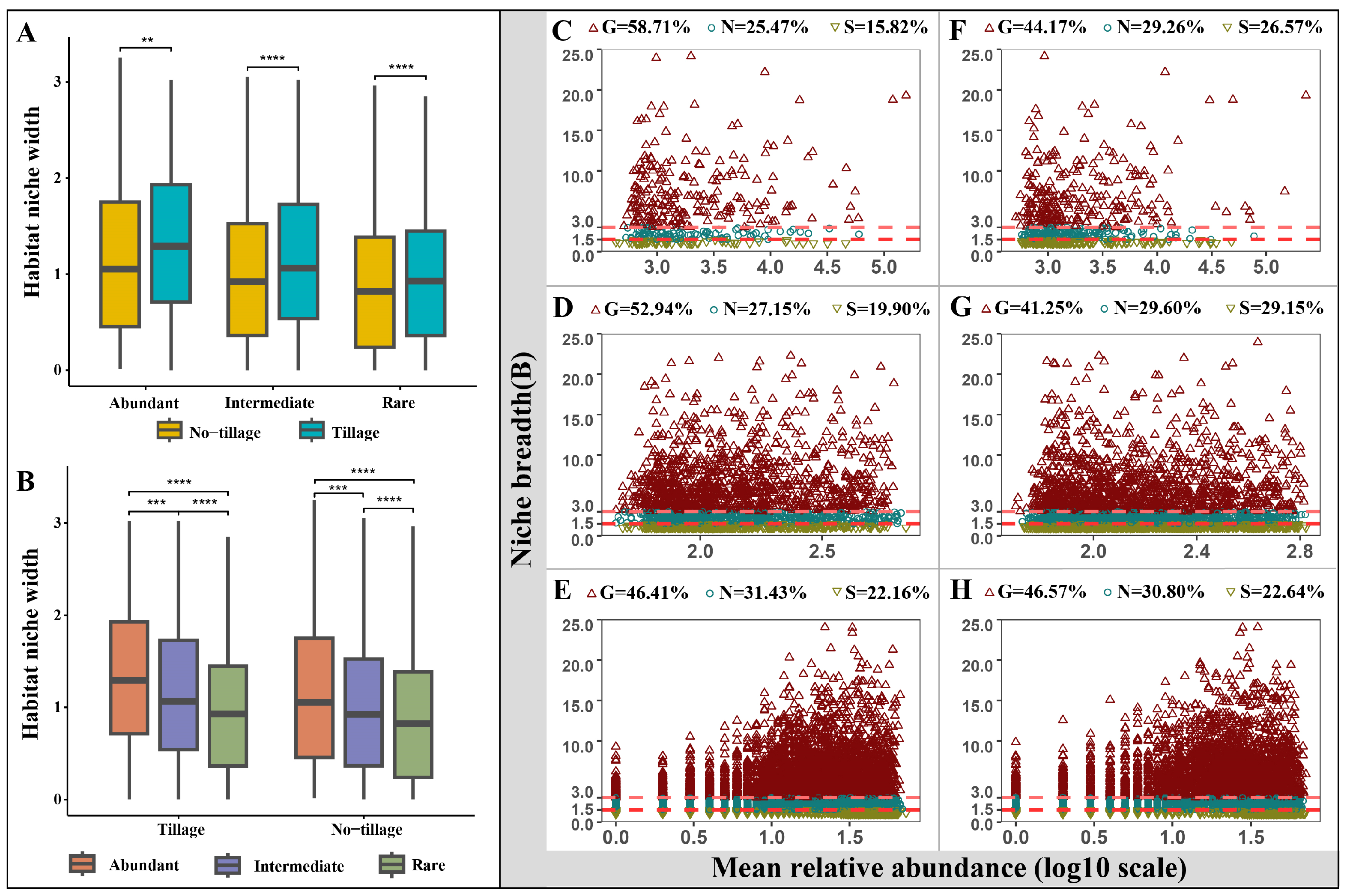
3.1. Diversity and Homogeneity of Fungal Sub-Communities in Tilled and Untilled Soils
3.2. Niche Width and Generalists/Specialists for Abundant, Intermediate, and Rare Taxa
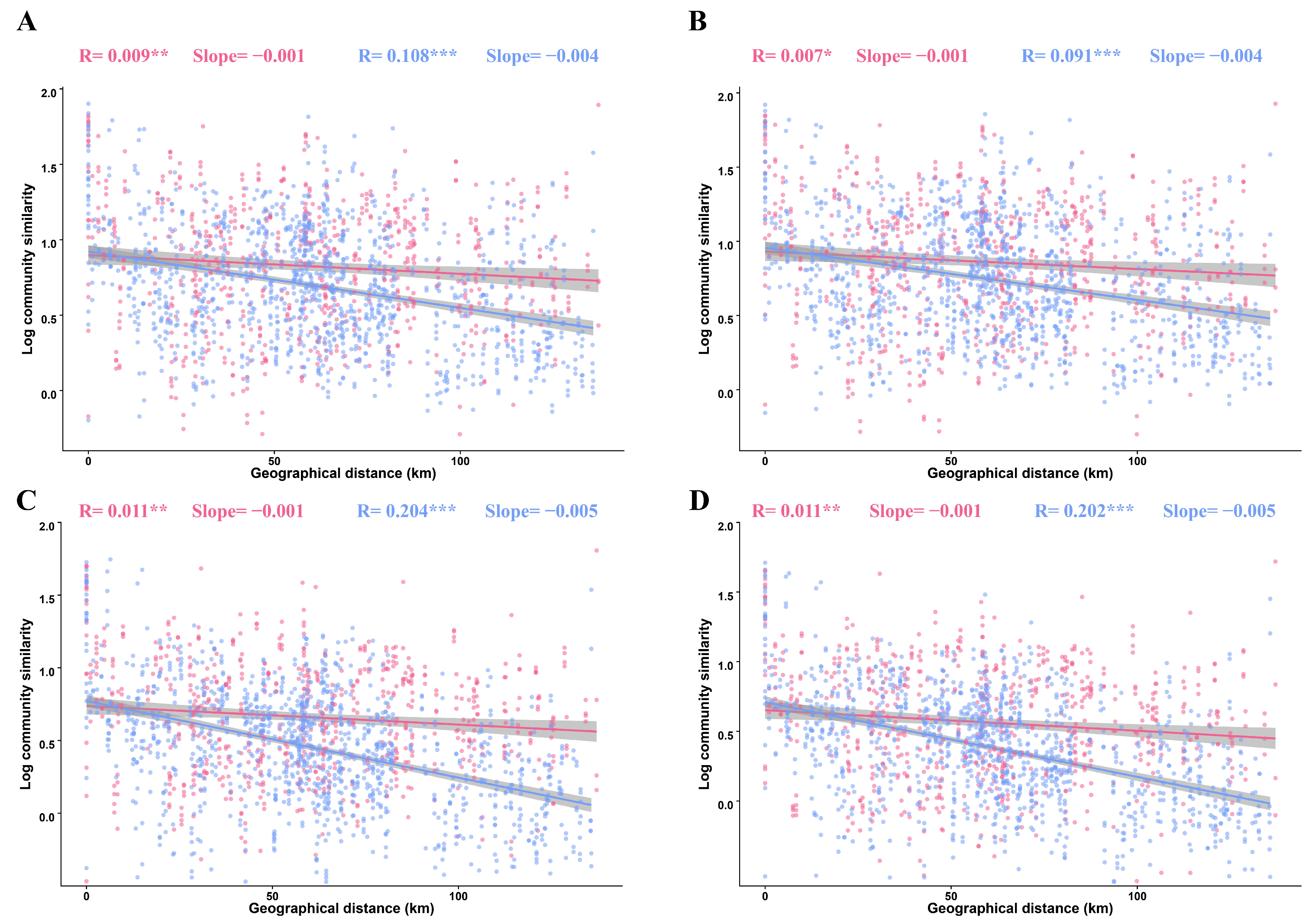
3.3. Distance–Decay Patterns of Fungal Sub-Communities in Tilled and Untilled Soils
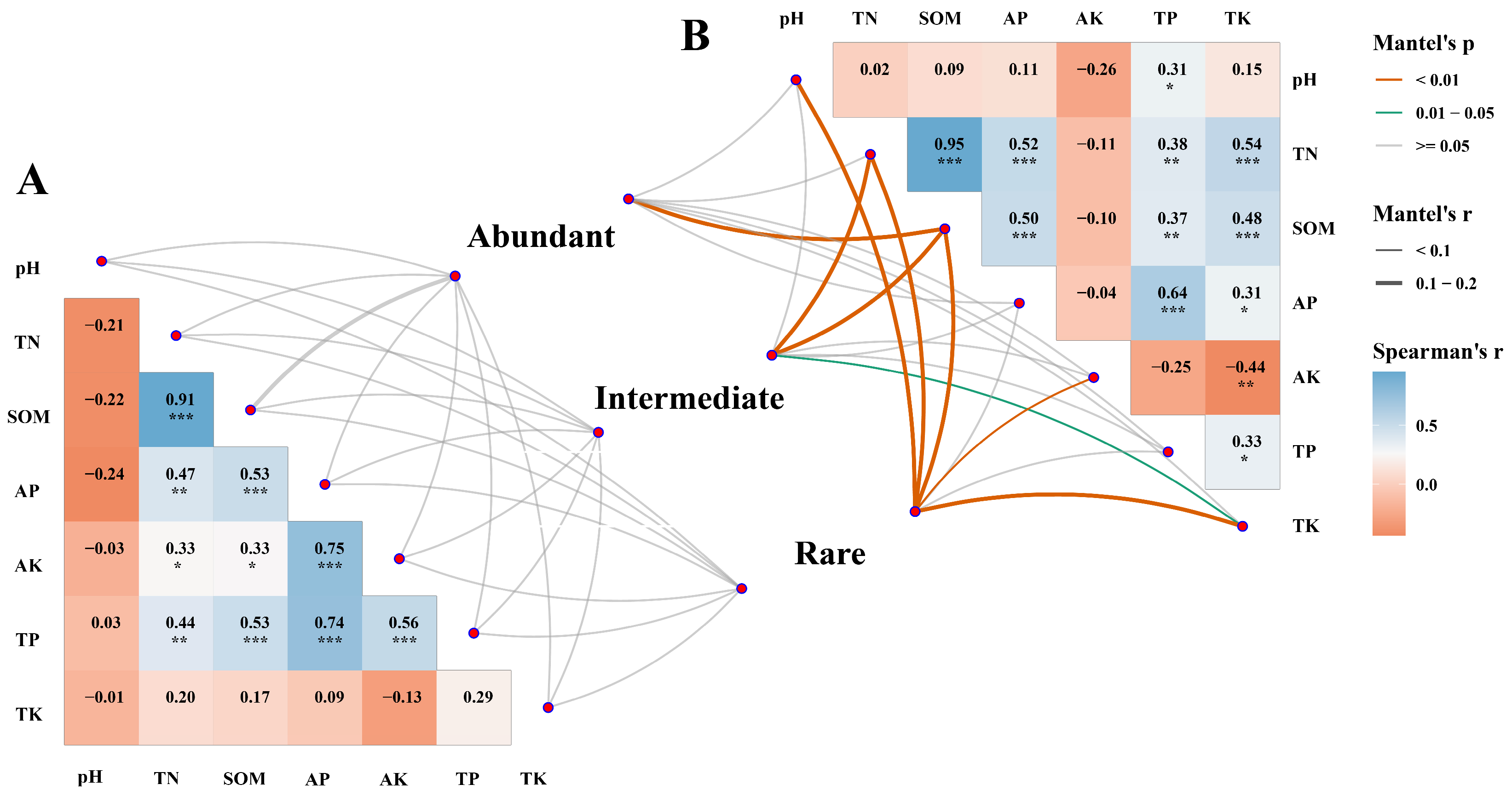
3.4. The Effect of Environmental Factors on Fungal Sub-Communities
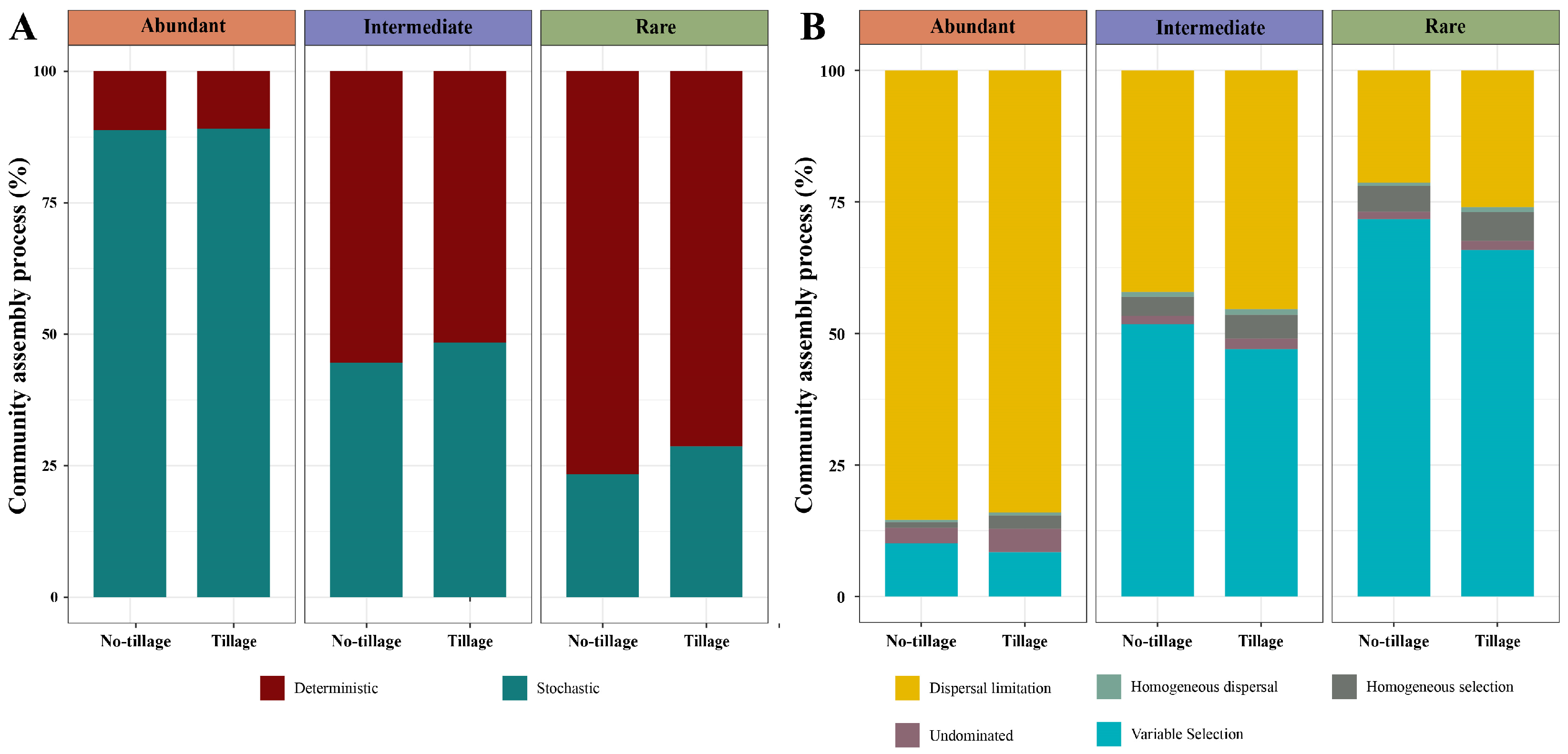
3.5. Assembly Processes in Abundant, Intermediate, and Rare Fungal Sub-Communities in the Tilled and Untilled Soils
4. Discussion
4.1. Distinct Fungal Sub-Community Structures between Tilled and Untilled Soils
4.2. Fungal Sub-Communities in Tilled and Untilled Soils Exhibited Different Biogeographic Patterns
4.3. Relative Contributions of Stochastic and Deterministic Processes in Tilled and Untilled Soils
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wagg, C.; Bender, S.F.; Widmer, F.; Van Der Heijden, M.G. Soil biodiversity and soil community composition determine ecosystem multifunctionality. Proc. Natl. Acad. Sci. USA 2014, 111, 5266–5270. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, S.; Walder, F.; Büchi, L.; Meyer, M.; Held, A.Y.; Gattinger, A.; Keller, T.; Charles, R.; van der Heijden, M.G. Agricultural intensification reduces microbial network complexity and the abundance of keystone taxa in roots. ISME J. 2019, 13, 1722–1736. [Google Scholar] [CrossRef] [Green Version]
- Allan, E.; Manning, P.; Alt, F.; Binkenstein, J.; Blaser, S.; Blüthgen, N.; Böhm, S.; Grassein, F.; Hölzel, N.; Klaus, V.H. Land use intensification alters ecosystem multifunctionality via loss of biodiversity and changes to functional composition. Ecol. Lett. 2015, 18, 834–843. [Google Scholar] [CrossRef] [Green Version]
- Foley, J.A.; DeFries, R.; Asner, G.P.; Barford, C.; Bonan, G.; Carpenter, S.R.; Chapin, F.S.; Coe, M.T.; Daily, G.C.; Gibbs, H.K. Global consequences of land use. Science 2005, 309, 570–574. [Google Scholar] [CrossRef] [Green Version]
- Hu, W.; Zhang, Y.; Huang, B.; Teng, Y. Soil environmental quality in greenhouse vegetable production systems in eastern China: Current status and management strategies. Chemosphere 2017, 170, 183–195. [Google Scholar] [CrossRef]
- Hottenstein, J.D.; Neilson, J.W.; Gil-Loaiza, J.; Root, R.A.; White, S.A.; Chorover, J.; Maier, R.M. Soil microbiome dynamics during pyritic mine tailing phytostabilization: Understanding microbial bioindicators of soil acidification. Front. Microbiol. 2019, 10, 1211. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Nan, J.; Xu, D.; Mo, L.; Zheng, Y.; Chao, L.; Qu, H.; Guo, Y.; Li, F.; Bao, Y. Response differences between soil fungal and bacterial communities under opencast coal mining disturbance conditions. Catena 2020, 194, 104779. [Google Scholar] [CrossRef]
- Chen, R.; Zhong, L.; Jing, Z.; Guo, Z.; Li, Z.; Lin, X.; Feng, Y. Fertilization decreases compositional variation of paddy bacterial community across geographical gradient. Soil Biol. Biochem. 2017, 114, 181–188. [Google Scholar] [CrossRef]
- Rodrigues, J.L.; Pellizari, V.H.; Mueller, R.; Baek, K.; Jesus, E.d.C.; Paula, F.S.; Mirza, B.; Hamaoui Jr, G.S.; Tsai, S.M.; Feigl, B. Conversion of the Amazon rainforest to agriculture results in biotic homogenization of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2013, 110, 988–993. [Google Scholar] [CrossRef] [Green Version]
- Gossner, M.M.; Lewinsohn, T.M.; Kahl, T.; Grassein, F.; Boch, S.; Prati, D.; Birkhofer, K.; Renner, S.C.; Sikorski, J.; Wubet, T. Land-use intensification causes multitrophic homogenization of grassland communities. Nature 2016, 540, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Madsen, E.L. Microorganisms and their roles in fundamental biogeochemical cycles. Curr. Opin. Biotechnol. 2011, 22, 456–464. [Google Scholar] [CrossRef] [PubMed]
- Pedrós-Alió, C. The rare bacterial biosphere. Annu. Rev. Mar. Sci. 2012, 4, 449–466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lynch, M.D.; Neufeld, J.D. Ecology and exploration of the rare biosphere. Nat. Rev. Microbiol. 2015, 13, 217–229. [Google Scholar] [CrossRef] [PubMed]
- Elshahed, M.S.; Youssef, N.H.; Spain, A.M.; Sheik, C.; Najar, F.Z.; Sukharnikov, L.O.; Roe, B.A.; Davis, J.P.; Schloss, P.D.; Bailey, V.L. Novelty and uniqueness patterns of rare members of the soil biosphere. Appl. Environ. Microbiol. 2008, 74, 5422–5428. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Xue, Y.; Shi, J.; Pan, A.; Tang, X.; Ming, F. The response of dominant and rare taxa for fungal diversity within different root environments to the cultivation of Bt and conventional cotton varieties. Microbiome 2018, 6, 184. [Google Scholar] [CrossRef]
- Jiao, S.; Lu, Y. Abundant fungi adapt to broader environmental gradients than rare fungi in agricultural fields. Glob. Chang. Biol. 2020, 26, 4506–4520. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, H.; Liu, S.; Hussain, S.; Zhang, L.; Yu, X.; Cao, K.; Xin, X.; Cao, H.; Zhu, A. Response of Fungal Sub-Communities in a Maize-Wheat Rotation Field Subjected to Long-Term Conservation Tillage Management. Front. Microbiol. 2022, 13, 565. [Google Scholar] [CrossRef]
- Ma, B.; Wang, H.; Dsouza, M.; Lou, J.; He, Y.; Dai, Z.; Brookes, P.C.; Xu, J.; Gilbert, J.A. Geographic patterns of co-occurrence network topological features for soil microbiota at continental scale in eastern China. ISME J. 2016, 10, 1891–1901. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Liebner, S.; Alawi, M.; Ebenhöh, O.; Wagner, D. Taxonomic database and cut-off value for processing mcrA gene 454 pyrosequencing data by MOTHUR. J. Microbiol. Methods 2014, 103, 3–5. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Liu, L.; Yang, J.; Yu, Z.; Wilkinson, D.M. The biogeography of abundant and rare bacterioplankton in the lakes and reservoirs of China. ISME J. 2015, 9, 2068–2077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Song, H.; Lei, Y.; Korpelainen, H.; Li, C. Distinct co-occurrence patterns and driving forces of rare and abundant bacterial subcommunities following a glacial retreat in the eastern Tibetan Plateau. Biol. Fertil. Soils 2019, 55, 351–364. [Google Scholar] [CrossRef] [Green Version]
- Lam, F.; Longnecker, M. A modified Wilcoxon rank sum test for paired data. Biometrika 1983, 70, 510–513. [Google Scholar] [CrossRef]
- Jari Oksanen, F.; Blanchet, G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Wagner, H. Vegan: Community Ecology Package, R package version2.5–3. 2018. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 27 April 2023).
- Anderson, M.J.; Willis, T.J. Canonical analysis of principal coordinates: A useful method of constrained ordination for ecology. Ecology 2003, 84, 511–525. [Google Scholar] [CrossRef]
- Anderson, M.J. Permutational multivariate analysis of variance (PERMANOVA). In Wiley Statsref: Statistics Reference Online; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2014; pp. 1–15. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.; O’Hara, R.; Simpson, G.; Solymos, P. Vegan: Community Ecology Package, R package version 2.4–6. 2018. 2019. Available online: https://CRAN.R-project.org/package=vegan(accessed on 27 April 2023).
- Roughgarden, J. Evolution of niche width. Am. Nat. 1972, 106, 683–718. [Google Scholar] [CrossRef]
- Pandit, S.N.; Kolasa, J.; Cottenie, K. Contrasts between habitat generalists and specialists: An empirical extension to the basic metacommunity framework. Ecology 2009, 90, 2253–2262. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.; Logares, R.; Huang, B.; Hsieh, C.H. Abundant and rare picoeukaryotic sub-communities present contrasting patterns in the epipelagic waters of marginal seas in the northwestern P acific O cean. Environ. Microbiol. 2017, 19, 287–300. [Google Scholar] [CrossRef]
- Xiong, J.; Li, X.; Yan, M.; Lu, J.; Qiu, Q.; Chen, J. Comparable ecological processes govern the temporal succession of gut bacteria and microeukaryotes as shrimp aged. Microb. Ecol. 2020, 80, 935–945. [Google Scholar] [CrossRef]
- Nekola, J.C.; White, P.S. The distance decay of similarity in biogeography and ecology. J. Biogeogr. 1999, 26, 867–878. [Google Scholar] [CrossRef] [Green Version]
- Soininen, J.; McDonald, R.; Hillebrand, H. The distance decay of similarity in ecological communities. Ecography 2007, 30, 3–12. [Google Scholar] [CrossRef]
- Jackson, D.A.; Somers, K.M. Are probability estimates from the permutation model of Mantel’s test stable? Can. J. Zool. 1989, 67, 766–769. [Google Scholar] [CrossRef] [Green Version]
- Chave, J. Neutral theory and community ecology. Ecol. Lett. 2004, 7, 241–253. [Google Scholar] [CrossRef]
- Chase, J.M.; Kraft, N.J.; Smith, K.G.; Vellend, M.; Inouye, B.D. Using null models to disentangle variation in community dissimilarity from variation in α-diversity. Ecosphere 2011, 2, 1–11. [Google Scholar] [CrossRef]
- Chase, J.M. Ecological niche theory. Theory Ecol. 2011, 93–107. [Google Scholar]
- Stegen, J.C.; Lin, X.; Konopka, A.E.; Fredrickson, J.K. Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 2012, 6, 1653–1664. [Google Scholar] [CrossRef] [Green Version]
- Stegen, J.C.; Lin, X.; Fredrickson, J.K.; Chen, X.; Kennedy, D.W.; Murray, C.J.; Rockhold, M.L.; Konopka, A. Quantifying community assembly processes and identifying features that impose them. ISME J. 2013, 7, 2069–2079. [Google Scholar] [CrossRef]
- Olden, J.D.; Poff, N.L. Toward a mechanistic understanding and prediction of biotic homogenization. Am. Nat. 2003, 162, 442–460. [Google Scholar] [CrossRef] [Green Version]
- Xiong, C.; He, J.Z.; Singh, B.K.; Zhu, Y.G.; Wang, J.T.; Li, P.P.; Zhang, Q.B.; Han, L.L.; Shen, J.P.; Ge, A.H. Rare taxa maintain the stability of crop mycobiomes and ecosystem functions. Environ. Microbiol. 2021, 23, 1907–1924. [Google Scholar] [CrossRef]
- Zhou, Z.; Zhang, Y.; Zhang, F. Abundant and rare bacteria possess different diversity and function in crop monoculture and rotation systems across regional farmland. Soil Biol. Biochem. 2022, 171, 108742. [Google Scholar] [CrossRef]
- Dias, T.; Dukes, A.; Antunes, P.M. Accounting for soil biotic effects on soil health and crop productivity in the design of crop rotations. J. Sci. Food Agric. 2015, 95, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Venter, Z.S.; Jacobs, K.; Hawkins, H.-J. The impact of crop rotation on soil microbial diversity: A meta-analysis. Pedobiologia 2016, 59, 215–223. [Google Scholar] [CrossRef]
- Garbeva, P.v.; Van Veen, J.A.; Van Elsas, J.D. Microbial diversity in soil: Selection of microbial populations by plant and soil type and implications for disease suppressiveness. Annu. Rev. Phytopathol. 2004, 42, 243–270. [Google Scholar] [CrossRef]
- Berg, G.; Smalla, K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 2009, 68, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Benitez, M.-S.; Ewing, P.M.; Osborne, S.L.; Lehman, R.M. Rhizosphere microbial communities explain positive effects of diverse crop rotations on maize and soybean performance. Soil Biol. Biochem. 2021, 159, 108309. [Google Scholar] [CrossRef]
- Kuerban, M.; Cong, W.-F.; Jing, J.; Bezemer, T.M. Microbial soil legacies of crops under different water and nitrogen levels determine succeeding crop performance. Plant Soil 2022, 1–14. [Google Scholar] [CrossRef]
- Graco-Roza, C.; Aarnio, S.; Abrego, N.; Acosta, A.T.; Alahuhta, J.; Altman, J.; Angiolini, C.; Aroviita, J.; Attorre, F.; Baastrup-Spohr, L. Distance decay 2.0–a global synthesis of taxonomic and functional turnover in ecological communities. Glob. Ecol. Biogeogr. 2022, 31, 1399–1421. [Google Scholar] [CrossRef]
- Hazard, C.; Gosling, P.; Van Der Gast, C.J.; Mitchell, D.T.; Doohan, F.M.; Bending, G.D. The role of local environment and geographical distance in determining community composition of arbuscular mycorrhizal fungi at the landscape scale. ISME J. 2013, 7, 498–508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kivlin, S.N.; Winston, G.C.; Goulden, M.L.; Treseder, K.K. Environmental filtering affects soil fungal community composition more than dispersal limitation at regional scales. Fungal Ecol. 2014, 12, 14–25. [Google Scholar] [CrossRef] [Green Version]
- Clark, D.R.; Mathieu, M.; Mourot, L.; Dufosse, L.; Underwood, G.J.; Dumbrell, A.J.; McGenity, T.J. Biogeography at the limits of life: Do extremophilic microbial communities show biogeographical regionalization? Glob. Ecol. Biogeogr. 2017, 26, 1435–1446. [Google Scholar] [CrossRef] [Green Version]
- Bissett, A.; Richardson, A.E.; Baker, G.; Wakelin, S.; Thrall, P.H. Life history determines biogeographical patterns of soil bacterial communities over multiple spatial scales. Mol. Ecol. 2010, 19, 4315–4327. [Google Scholar] [CrossRef] [PubMed]
- Soininen, J.; Korhonen, J.J.; Karhu, J.; Vetterli, A. Disentangling the spatial patterns in community composition of prokaryotic and eukaryotic lake plankton. Limnol. Oceanogr. 2011, 56, 508–520. [Google Scholar] [CrossRef]
- Martiny, J.B.; Eisen, J.A.; Penn, K.; Allison, S.D.; Horner-Devine, M.C. Drivers of bacterial β-diversity depend on spatial scale. Proc. Natl. Acad. Sci. USA 2011, 108, 7850–7854. [Google Scholar] [CrossRef] [Green Version]
- Clark, D.R.; Underwood, G.J.; McGenity, T.J.; Dumbrell, A.J. What drives study-dependent differences in distance–decay relationships of microbial communities? Glob. Ecol. Biogeogr. 2021, 30, 811–825. [Google Scholar] [CrossRef]
- Li, P.; Li, W.; Dumbrell, A.J.; Liu, M.; Li, G.; Wu, M.; Jiang, C.; Li, Z. Spatial variation in soil fungal communities across paddy fields in subtropical China. Msystems 2020, 5, e00704–e00719. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.; Lu, H.-P.; Sastri, A.; Yeh, Y.-C.; Gong, G.-C.; Chou, W.-C.; Hsieh, C.-H. Contrasting the relative importance of species sorting and dispersal limitation in shaping marine bacterial versus protist communities. ISME J. 2018, 12, 485–494. [Google Scholar] [CrossRef] [Green Version]
- Meyer, K.M.; Memiaghe, H.; Korte, L.; Kenfack, D.; Alonso, A.; Bohannan, B.J. Why do microbes exhibit weak biogeographic patterns? ISME J. 2018, 12, 1404–1413. [Google Scholar] [CrossRef]
- Feng, M.; Adams, J.M.; Fan, K.; Shi, Y.; Sun, R.; Wang, D.; Guo, X.; Chu, H. Long-term fertilization influences community assembly processes of soil diazotrophs. Soil Biol. Biochem. 2018, 126, 151–158. [Google Scholar] [CrossRef]
- Wang, Z.; Li, Y.; Li, T.; Zhao, D.; Liao, Y. Conservation tillage decreases selection pressure on community assembly in the rhizosphere of arbuscular mycorrhizal fungi. Sci. Total Environ. 2020, 710, 136326. [Google Scholar] [CrossRef]
- Nuccio, E.E.; Anderson-Furgeson, J.; Estera, K.Y.; Pett-Ridge, J.; De Valpine, P.; Brodie, E.L.; Firestone, M.K. Climate and edaphic controllers influence rhizosphere community assembly for a wild annual grass. Ecology 2016, 97, 1307–1318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fierer, N.; Jackson, R.B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lozupone, C.A.; Knight, R. Global patterns in bacterial diversity. Proc. Natl. Acad. Sci. USA 2007, 104, 11436–11440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xun, W.; Li, W.; Xiong, W.; Ren, Y.; Liu, Y.; Miao, Y.; Xu, Z.; Zhang, N.; Shen, Q.; Zhang, R. Diversity-triggered deterministic bacterial assembly constrains community functions. Nat. Commun. 2019, 10, 3833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, Z.; Luo, W.; Zhang, C. Rare bacterial biosphere is more environmental controlled and deterministically governed than abundant one in sediment of thermokarst lakes across the Qinghai-Tibet Plateau. Front. Microbiol. 2022, 13, 944646. [Google Scholar] [CrossRef]
- Wilson, B.; Hayek, L.-A.C. Distinguishing relative specialist and generalist species in the fossil record. Mar. Micropaleontol. 2015, 119, 7–16. [Google Scholar] [CrossRef]
- Kurm, V.; Van Der Putten, W.H.; De Boer, W.; Naus-Wiezer, S.; Hol, W.G. Low abundant soil bacteria can be metabolically versatile and fast growing. Ecology 2017, 98, 555–564. [Google Scholar] [CrossRef] [Green Version]
- Jia, X.; Dini-Andreote, F.; Salles, J.F. Community assembly processes of the microbial rare biosphere. Trends Microbiol. 2018, 26, 738–747. [Google Scholar] [CrossRef]
- Wan, W.; Gadd, G.M.; Yang, Y.; Yuan, W.; Gu, J.; Ye, L.; Liu, W. Environmental adaptation is stronger for abundant rather than rare microorganisms in wetland soils from the Qinghai-Tibet Plateau. Mol. Ecol. 2021, 30, 2390–2403. [Google Scholar] [CrossRef]
- Shi, Y.; Dang, K.; Dong, Y.; Feng, M.; Wang, B.; Li, J.; Chu, H. Soil fungal community assembly processes under long-term fertilization. Eur. J. Soil Sci. 2020, 71, 716–726. [Google Scholar] [CrossRef]
- Dini-Andreote, F.; Stegen, J.C.; Van Elsas, J.D.; Salles, J.F. Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. Proc. Natl. Acad. Sci. USA 2015, 112, E1326–E1332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vellend, M. Conceptual synthesis in community ecology. Q. Rev. Biol. 2010, 85, 183–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Wu, F. Land-use conversion from open field to greenhouse cultivation differently affected the diversities and assembly processes of soil abundant and rare fungal communities. Sci. Total Environ. 2021, 788, 147751. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Han, H.; Zhang, R.; Xu, W.; Wang, Y.; Zhang, B.; Yin, Y.; Cao, H. Biogeographic Patterns of Fungal Sub-Communities under Different Land-Use Types in Subtropical China. J. Fungi 2023, 9, 646. https://doi.org/10.3390/jof9060646
Liu H, Han H, Zhang R, Xu W, Wang Y, Zhang B, Yin Y, Cao H. Biogeographic Patterns of Fungal Sub-Communities under Different Land-Use Types in Subtropical China. Journal of Fungi. 2023; 9(6):646. https://doi.org/10.3390/jof9060646
Chicago/Turabian StyleLiu, Hao, Heming Han, Ruoling Zhang, Weidong Xu, Yuwei Wang, Bo Zhang, Yifan Yin, and Hui Cao. 2023. "Biogeographic Patterns of Fungal Sub-Communities under Different Land-Use Types in Subtropical China" Journal of Fungi 9, no. 6: 646. https://doi.org/10.3390/jof9060646
APA StyleLiu, H., Han, H., Zhang, R., Xu, W., Wang, Y., Zhang, B., Yin, Y., & Cao, H. (2023). Biogeographic Patterns of Fungal Sub-Communities under Different Land-Use Types in Subtropical China. Journal of Fungi, 9(6), 646. https://doi.org/10.3390/jof9060646





