Taxonomy and Phylogeny of Eight New Acrophialophora Species (Sordariales, Chaetomiaceae) from China
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling, Fungal Isolation, and Morphology
2.2. DNA Extraction, PCR Amplifications, and Sequencing
2.3. Phylogenetic Analyses
3. Results
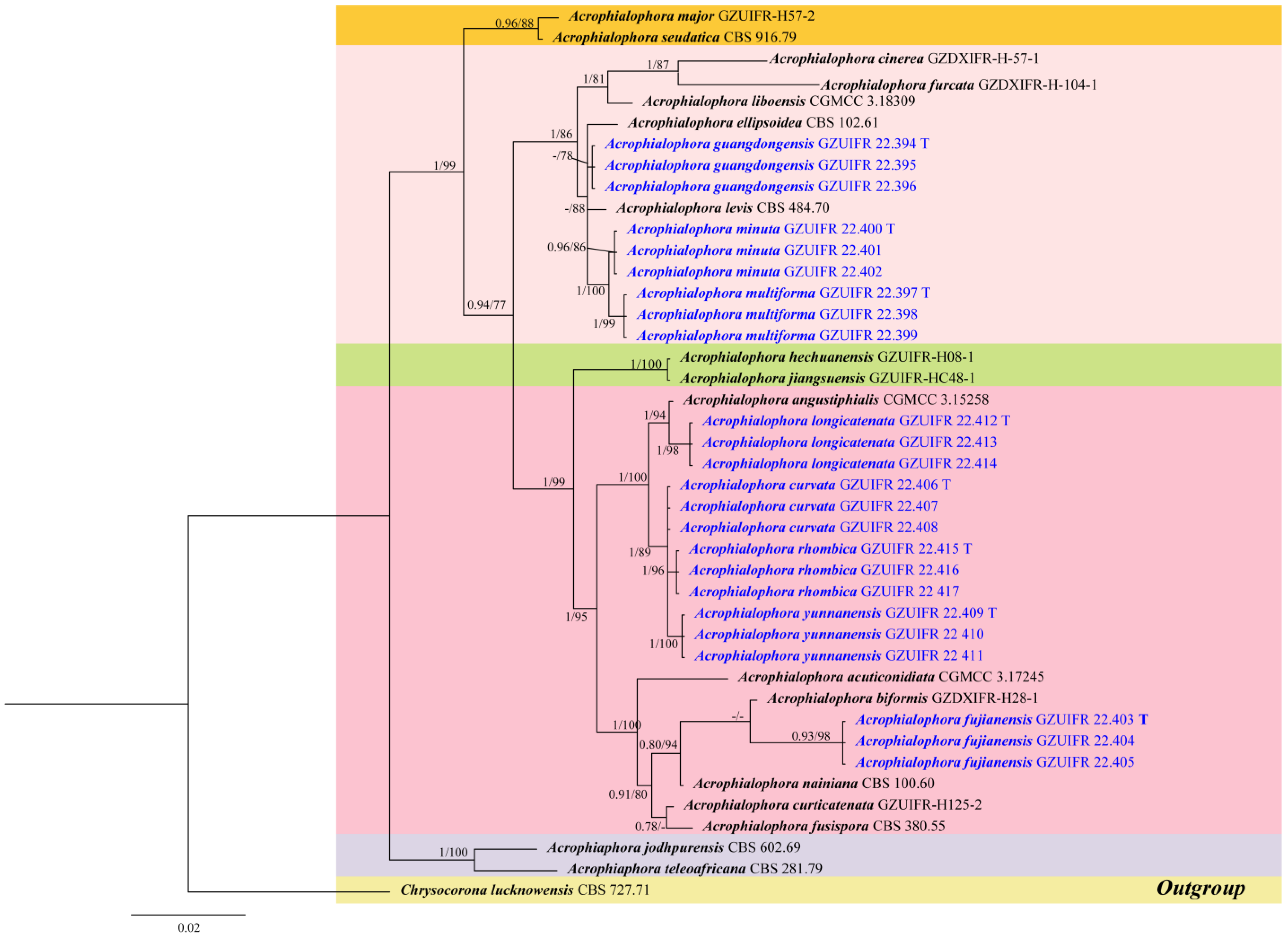
3.1. Phylogeny Analysis
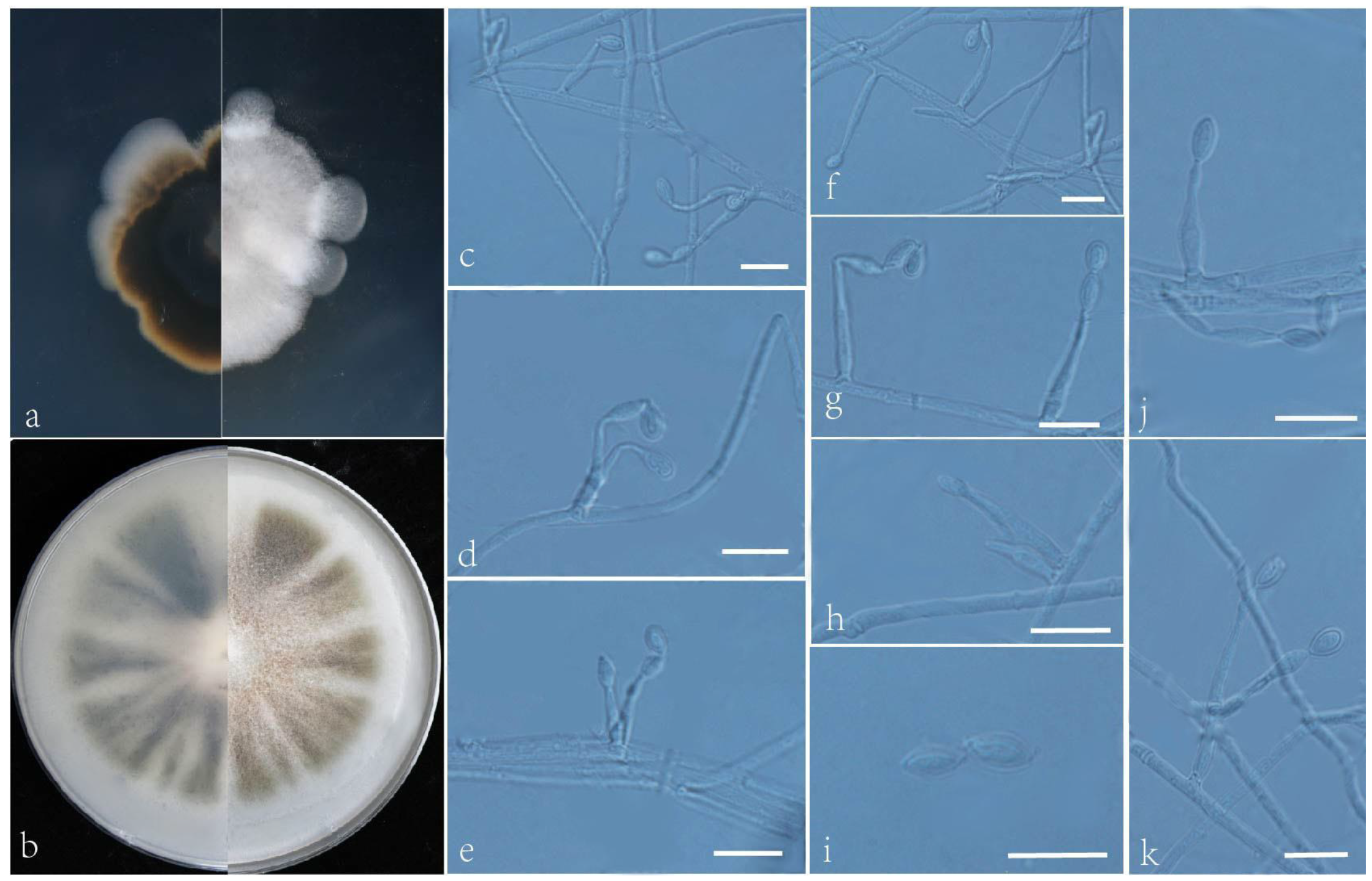
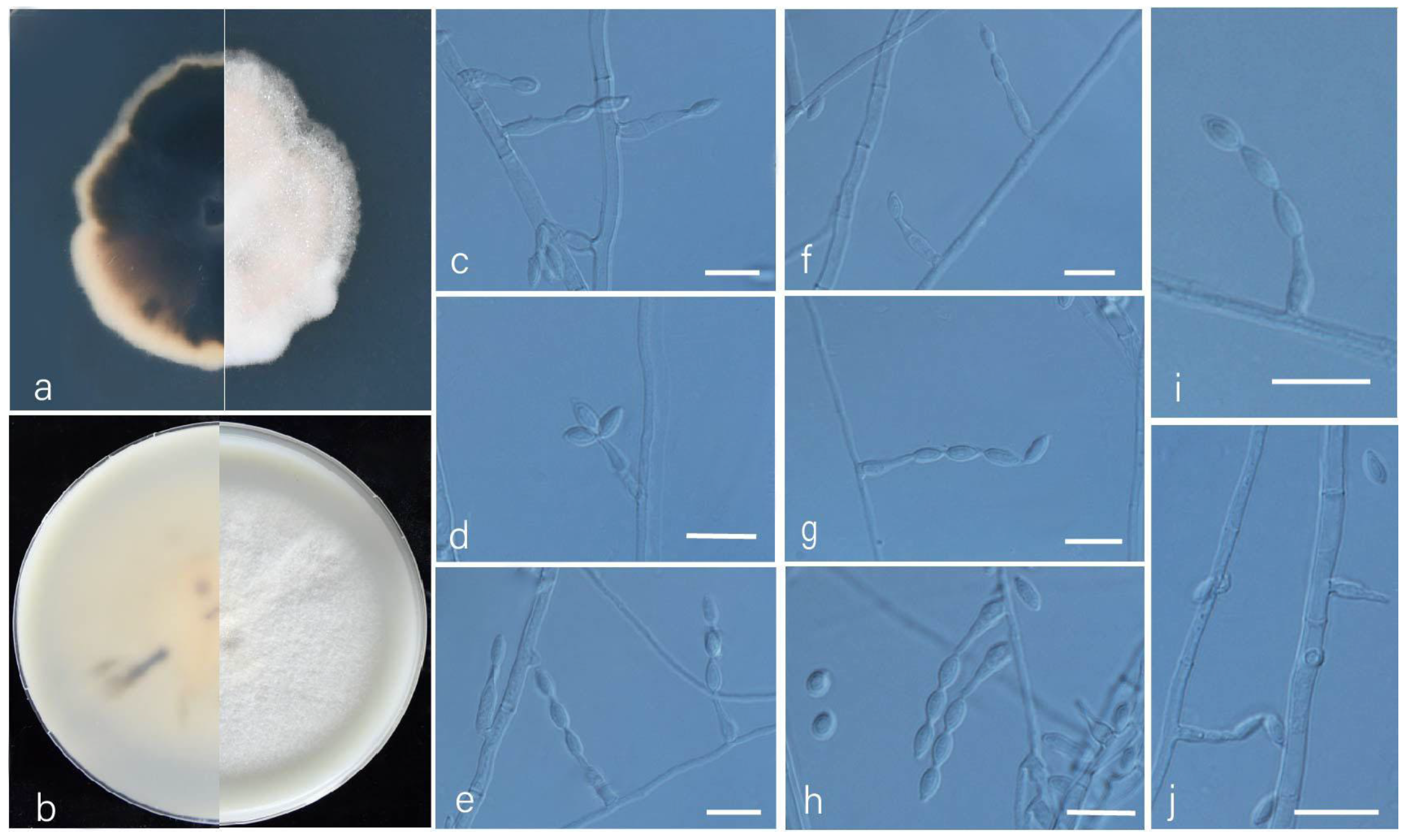
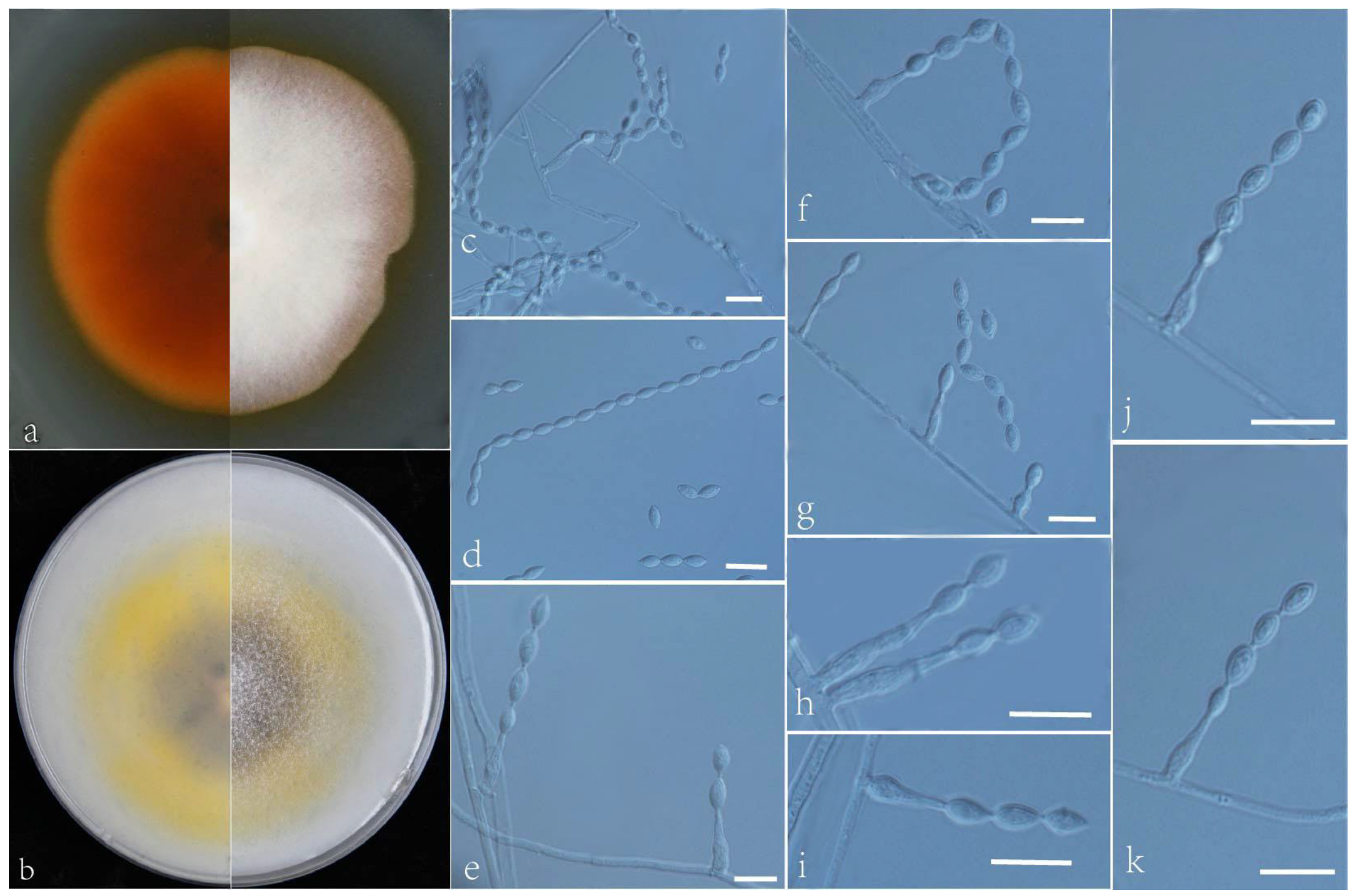
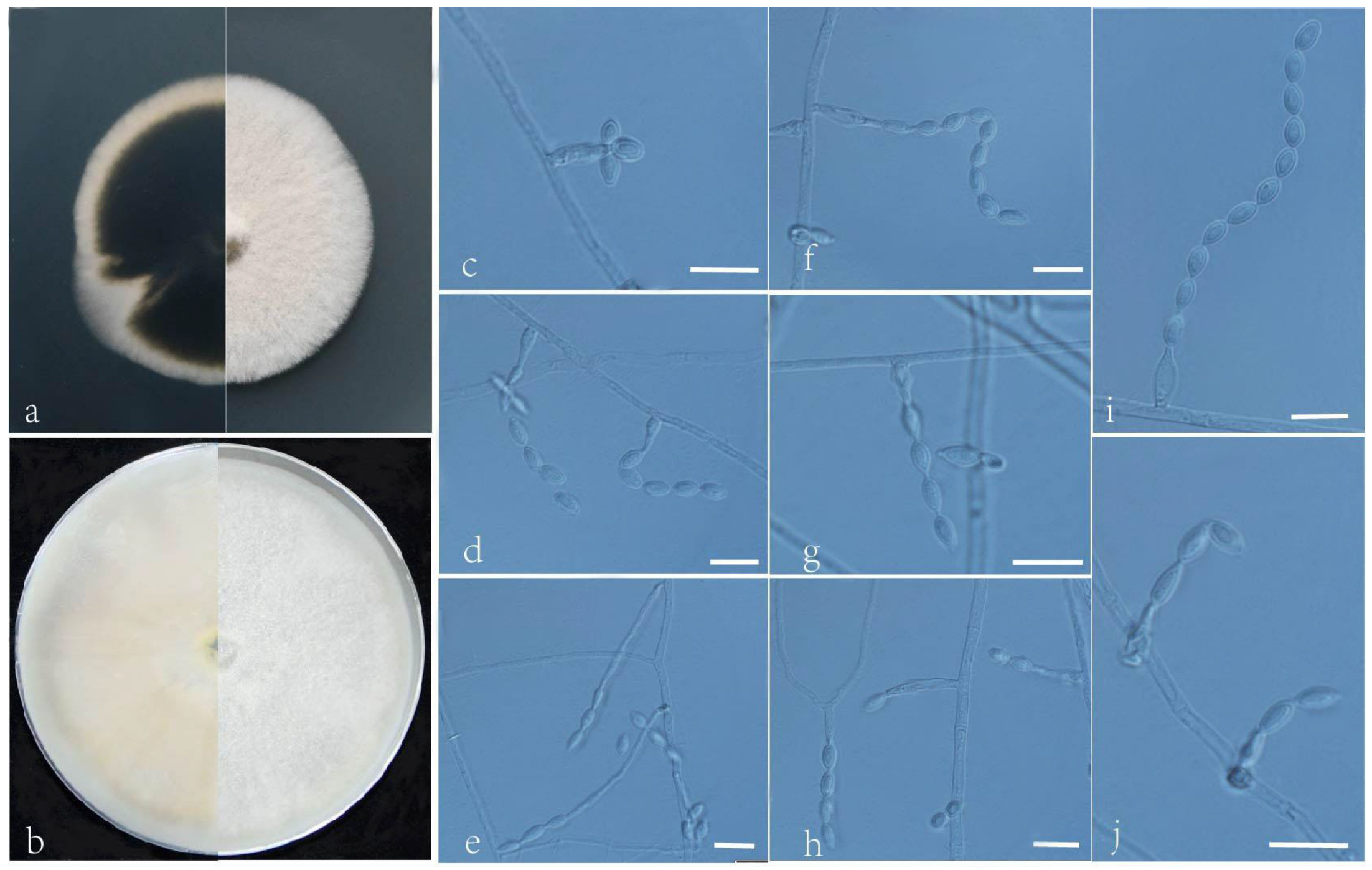
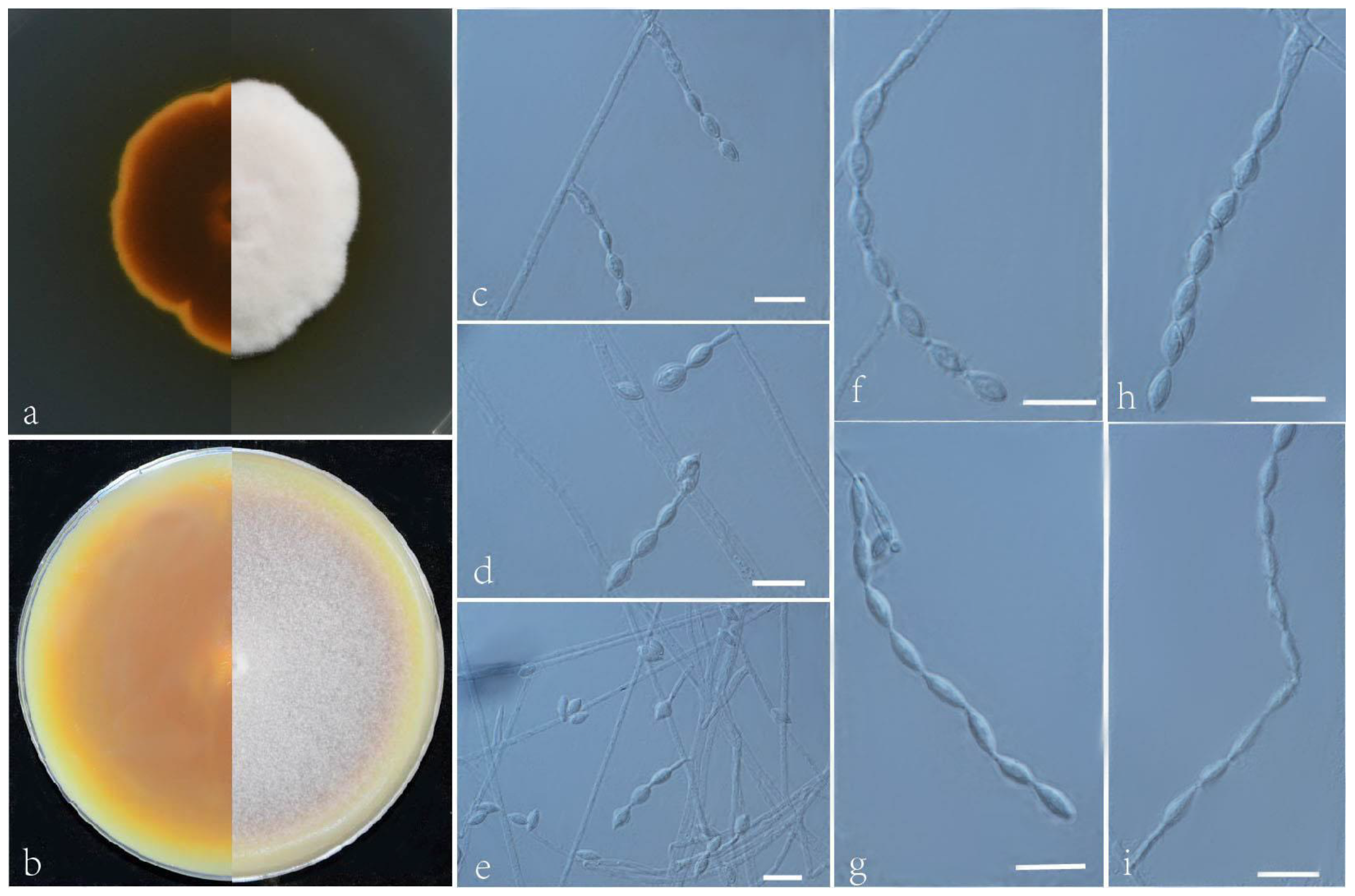
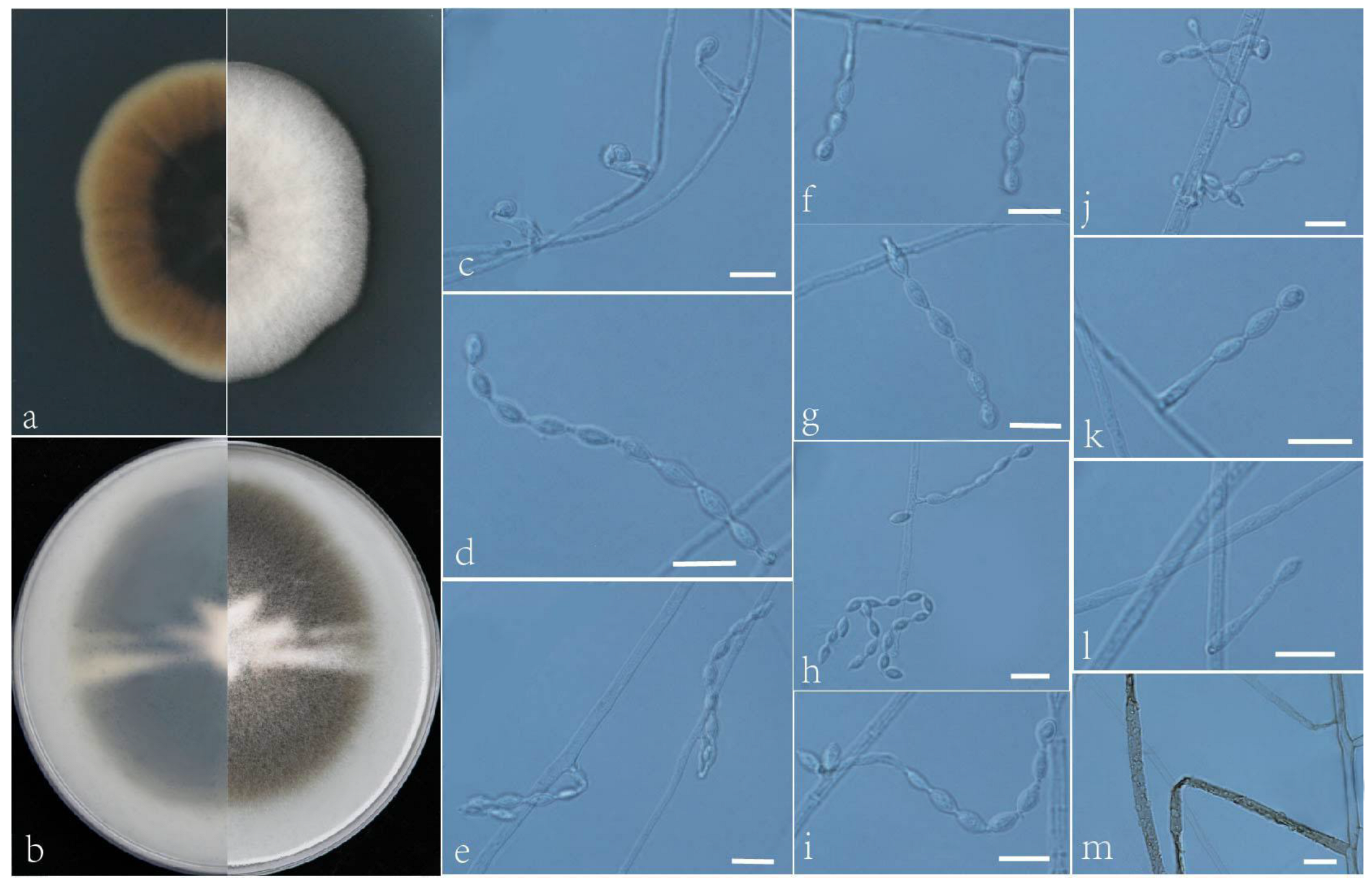
3.2. Taxonomy


4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Edward, J.C. A new genus of the Moniliaceae. Mycologia 1959, 51, 781–786. [Google Scholar] [CrossRef]
- Barron, G.L.; Peterson, J.L. The Genera of Hyphomycetes from Soil; Williams & Wilkins Co.: Baltimore, MD, USA, 1968; p. 477. [Google Scholar]
- Dal Vesco, G.; Peyronel, B. Funghi isolati dal suolo di due isole del Pacifico meridionale. Allionia 1968, 14, 31–49. [Google Scholar]
- Samson, R.A.; Mahmood, T. The genus Acrophialophora (fungi, moniliales). Acta Bot. Neerl. 1970, 19, 804–808. [Google Scholar] [CrossRef]
- Luangsa-Ard, J.J.; Hywel-Jones, N.L.; Samson, R.A. The polyphyletic nature of Paecilomyces sensu lato based on 18S-generated rDNA phylogeny. Mycologia 2004, 96, 773–780. [Google Scholar] [CrossRef]
- Brown, A.H.S.; Smith, G. The genus Paecilomyces Bainier and its perfect stage Byssochlamys Westling. Trans. Br. Mycol. Soc. 1957, 40, 17–89. [Google Scholar] [CrossRef]
- Onions, A.H.S.; Barron, G.L. Monophialidic species of Paecilomyces. Mycological 1967, 107, 1–25. [Google Scholar]
- Gams, W. Cephalosporium-Artige Schimmelpilze (Hyphomycetes); Gustav Fischer: Stuttgart, Germany, 1971; pp. 38–130. [Google Scholar]
- Liang, Z.Q.; Han, Y.F.; Chu, H.L. Studies on the genus Paecilomyces in China. IV. Two new species of Paecilomyces with monophialides. Mycotaxon 2006, 97, 13–20. [Google Scholar]
- Liang, Z.Q.; Han, Y.F.; Chu, H.L. A new thermotolerant Paecilomyces species which produces laccase and a biform sporogenous structure. Fungal Divers. 2007, 27, 95–102. [Google Scholar]
- Han, Y.F.; Liang, Z.Q.; Chu, H.L. A new thermophilic species of Paecilomyces, Paecilomyces curticatenatus. Mycosystema 2007, 26, 13–16. [Google Scholar]
- Chu, H.L.; Liang, Z.Q.; Han, Y.F. A thermotolerant Paecilomyces inflatus var. major Liang Z Q, Chu H L et Han Y F var. nov. Which Produces Laccase. J. Fungal Res. 2004, 2, 43–46. [Google Scholar]
- Liang, Z.Q.; Han, Y.F.; Chu, H.L.; Fox, R.T. Studies on the genus Paecilomyces in China, V. Taifanglania gen. nov. for some monophialidic species. Fungal Divers. 2009, 34, 69–77. [Google Scholar]
- Han, Y.F.; Liang, J.D.; Liang, Z.Q.; Zou, X.; Dong, X. Two new Taifanglania species identified through DELTA-assisted phenetic analysis. Mycotaxon 2010, 112, 325–333. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.R.; Han, Y.F.; Liang, Z.Q. A new thermotolerant species of Taifanglania. Mycosystem 2015, 34, 345–349. [Google Scholar]
- Perdomo, H.; García, D.; Gené, J.; Cano, J.; Sutton, D.A.; Summerbell, R.; Guarro, J. Phialemoniopsis, a new genus of Sordariomycetes, and new species of Phialemonium and Lecythophora. Mycologia 2013, 105, 398–421. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, F.; Wu, W.; Cai, L. A phylogenetic assessment and taxonomic revision of the thermotolerant hyphomycete genera Acrophialophora and Taifanglania. Mycologia 2015, 107, 768–779. [Google Scholar] [CrossRef]
- Sandoval-Denis, M.; Gené, J.; Sutton, D.A.; Wiederhold, N.P.; Guarro, J. Acrophialophora, a poorly known fungus with clinical significance. J. Clin. Microbiol. 2015, 53, 1549–1555. [Google Scholar] [CrossRef]
- Zhang, Y.W.; Wang, Y.; Zeng, G.P.; Chen, W.H.; Zou, X.; Han, Y.F.; Qiu, S.Y.; Liang, Z.Q. A new species of Acrophialophora from Guizhou Province, China. Phytotaxa 2017, 302, 266–272. [Google Scholar] [CrossRef]
- Wang, X.W.; Bai, F.Y.; Bensch, K.; Meijer, M.; Sun, B.D.; Han, Y.F.; Crous, P.W.; Samson, R.A.; Yang, F.Y.; Houbraken, J. Phylogenetic re-evaluation of Thielavia with the introduction of a new family Podosporaceae. Stud. Mycol. 2019, 93, 155–252. [Google Scholar] [CrossRef]
- Wang, X.W.; Han, P.J.; Bai, F.Y.; Luo, A.; Bensch, K.; Meijer, M.; Kraak, B.; Han, D.Y.; Sun, B.D.; Crous, P.W.; et al. Taxonomy, phylogeny and identification of Chaetomiaceae with emphasis on thermophilic species. Stud. Mycol. 2022, 101, 121–243. [Google Scholar] [CrossRef]
- Hendy, M.H.; Hashem, A.H.; Sulieman, W.B.; Sultan, M.H.; Abdelraof, M. Purification, characterization and anticancer activity of l-methionine γ-lyase from thermo-tolerant aspergillus fumigatus. Microb. Cell Fact. 2023, 22, 8. [Google Scholar] [CrossRef]
- Barros, R.R.O.; Oliveira, R.A.; Gottschalk, L.M.F.; Bon, E.P.S. Production of cellulolytic enzymes by fungi Acrophialophora nainiana and Ceratocystis paradoxa using different carbon sources. Appl. Biochem. Biotechnol. 2010, 161, 448–454. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef]
- Vilgalys, R.; Sun, B.L. Ancient and recent patterns of geographic speciation in the oyster mushroom Pleurotus revealed by phylogenetic analysis of ribosomal DNA sequences. Proc. Natl. Acad. Sci. USA 1994, 91, 4599–4603. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.L.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; Haeseler, A.V.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van, D.M.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Matushima, T. Microfungi of the Solomon Islands and Papua-New Guinea; Matushima: Kobe, Japan, 1971; p. 42. [Google Scholar]
- Matushima, T. Icones Microfungorum a Matushima Lectorum; Matushima: Kobe, Japan, 1975; p. 104. [Google Scholar]
- Arx, J.A. The Genera of Fungi Sporulating in Pure Culture; Cramer: Vaduz, Liechtenstein, 1981; pp. 283–331. [Google Scholar]
- Samson, R.A. Paecilomyces and some allied hyphomycetes. Stud. Mycol. 1974, 6, 1–119. [Google Scholar]
- Zhang, Y.W.; Chen, W.H.; Wang, Y.R.; Han, Y.F.; Liang, Z.Q. Screening on cellulase from thermophilic Taifanglania spp. Chin. Brew. 2013, 3, 45–47. [Google Scholar]
- Wang, Y.; Han, Y.F.; Liang, Z.Q. Rice straw degradation and enzyme production of two Taifanglania strains. Mycosystema 2017, 36, 598–603. [Google Scholar]
| Species | Strains | GenBank Accession Numbers | |||
|---|---|---|---|---|---|
| ITS | tub2 | LSU | RPB2 | ||
| Acrophialophora acuticonidiata | CGMCC 3.17245 | KJ026975 | KJ147441 | — | — |
| Acrophialophora angustiphialis | CGMCC 3.15258 | KJ026972 | KJ147438 | — | — |
| Acrophialophora biformis | GZDXIFR-H28-1 | DQ191963 | — | — | — |
| Acrophialophora cinerea | GZDXIFR-H-57-1 | DQ243694 | KP143110 | OP419973 | OP886702 |
| Acrophialophora curticatenata | GZUIFR-H125-2 | EU004811 | — | — | — |
| Acrophialophora curvata | GZUIFR 22.406 | OP454351 | OP547306 | OP454363 | OP802834 |
| Acrophialophora curvata | GZUIFR 22.407 | OP454352 | OP547307 | OP454364 | OP802835 |
| Acrophialophora curvata | GZUIFR 22.408 | OP454353 | OP547308 | OP454365 | OP802836 |
| Acrophialophora ellipsoidea | CBS 102.61 | MK926786 | MK926886 | MK926786 | MK876748 |
| Acrophialophora fujianensis | GZUIFR 22.403 | OP454345 | OP536984 | OP454372 | OP820579 |
| Acrophialophora fujianensis | GZUIFR 22.404 | OP454346 | OP536985 | OP454373 | OP820580 |
| Acrophialophora fujianensis | GZUIFR 22.405 | OP454347 | OP536986 | OP454374 | OP820581 |
| Acrophialophora furcata | GZDXIFR-H-104-1 | DQ243695 | KP143113 | OP456145 | OP886703 |
| Acrophialophora fusispora | CBS 380.55 | MK926788 | MK926888 | MK926788 | MK876750 |
| Acrophialophora guangdongensis | GZUIFR 22.394 T | OP454339 | OP547315 | OP454369 | OP491393 |
| Acrophialophora guangdongensis | GZUIFR 22.395 | OP454340 | OP547316 | OP454370 | OP491394 |
| Acrophialophora guangdongensis | GZUIFR 22.396 | OP454341 | OP547317 | OP454371 | OP491395 |
| Acrophialophora hechuanensis | GZUIFR-H08-1 | MK926789 | MK926889 | MK926789 | MK876751 |
| Acrophialophora jiangsuensis | GZUIFR HC48.1 | KF719171 | KP143112 | OP456146 | OP491392 |
| Acrophialophora jodhpurensis | CBS 602.69 | MK926790 | MK926890 | MK926790 | MK876752 |
| Acrophialophora levis | CBS 484.70 | KP233038 | KP233044 | KM995840 | — |
| Acrophialophora liboensis | CGMCC 3.18309 | KP192127 | KP999978 | OP456147 | OP886704 |
| Acrophialophora longicatenata | GZUIFR 22.412 T | OP454357 | OP547309 | OP454378 | OP834081 |
| Acrophialophora longicatenata | GZUIFR 22.413 | OP454358 | OP547310 | OP454379 | OP834082 |
| Acrophialophora longicatenata | GZUIFR 22.414 | OP454359 | OP547311 | OP454380 | OP834083 |
| Acrophialophora major | GZUIFR-H57-2 | MK926792 | MK926892 | MK926792 | MK876754 |
| Acrophialophora minuta | GZUIFR 22.400 T | OP454342 | OP547318 | OP454366 | OP880249 |
| Acrophialophora minuta | GZUIFR 22.401 | OP454343 | OP547319 | OP454367 | OP880250 |
| Acrophialophora minuta | GZUIFR 22.402 | OP454344 | OP547320 | OP454368 | OP880251 |
| Acrophialophora multiforma | GZUIFR 22.397 T | OP454336 | OP547321 | OP454360 | OP880243 |
| Acrophialophora multiforma | GZUIFR 22.398 | OP454337 | OP547322 | OP454361 | OP880244 |
| Acrophialophora multiforma | GZUIFR 22.399 | OP454338 | OP547323 | OP454362 | OP880245 |
| Acrophialophora nainiana | CBS 100.60 | MK926793 | MK926893 | MK926793 | MK876755 |
| Acrophialophora rhombica | GZUIFR 22.415 T | OP454354 | OP731571 | OP454381 | OP834084 |
| Acrophialophora rhombica | GZUIFR 22.416 | OP454355 | OP731572 | OP454382 | OP834085 |
| Acrophialophora rhombica | GZUIFR 22.417 | OP454356 | OP731573 | OP454383 | OP834086 |
| Acrophialophora seudatica | CBS 916.79 | LN736030 | LN736032 | LN736031 | — |
| Acrophialophora teleoafricana | CBS 281.79 | MK926795 | MK926895 | MK926795 | MK876757 |
| Acrophialophora yunnanensis | GZUIFR 22.409 T | OP454348 | OP547312 | OP454375 | OP880246 |
| Acrophialophora yunnanensis | GZUIFR 22.410 | OP454349 | OP547313 | OP454376 | OP880247 |
| Acrophialophora yunnanensis | GZUIFR 22.411 | OP454350 | OP547314 | OP454377 | OP880248 |
| Chrysocorona lucknowensis | CBS 727.71 T | MK926813 | MK926913 | MK926813 | MK876773 |
| ITS | LSU | tub2 | RPB2 | |
|---|---|---|---|---|
| ML analysis | K3P+G4 | F81+F+I | HKY+F+G4 | GTR+F+G4 |
| BI analysis | K2P+G4 | F81+F+I | HKY+F+G4 | GTR+F+G4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, L.; Zhang, Y.-W.; Wang, H.-Y.; Dong, C.-B.; Chen, W.-H.; Liang, J.-D.; Han, Y.-F. Taxonomy and Phylogeny of Eight New Acrophialophora Species (Sordariales, Chaetomiaceae) from China. J. Fungi 2023, 9, 645. https://doi.org/10.3390/jof9060645
Peng L, Zhang Y-W, Wang H-Y, Dong C-B, Chen W-H, Liang J-D, Han Y-F. Taxonomy and Phylogeny of Eight New Acrophialophora Species (Sordariales, Chaetomiaceae) from China. Journal of Fungi. 2023; 9(6):645. https://doi.org/10.3390/jof9060645
Chicago/Turabian StylePeng, Lan, Yan-Wei Zhang, Hai-Yan Wang, Chun-Bo Dong, Wan-Hao Chen, Jian-Dong Liang, and Yan-Feng Han. 2023. "Taxonomy and Phylogeny of Eight New Acrophialophora Species (Sordariales, Chaetomiaceae) from China" Journal of Fungi 9, no. 6: 645. https://doi.org/10.3390/jof9060645
APA StylePeng, L., Zhang, Y.-W., Wang, H.-Y., Dong, C.-B., Chen, W.-H., Liang, J.-D., & Han, Y.-F. (2023). Taxonomy and Phylogeny of Eight New Acrophialophora Species (Sordariales, Chaetomiaceae) from China. Journal of Fungi, 9(6), 645. https://doi.org/10.3390/jof9060645






