Molecular Phylogeny and Morphology Reveal Cryptic Species in the Cordyceps militaris Complex from Vietnam
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimen Collection and Fungus Isolation
2.2. Morphological Observations
2.3. DNA Extraction, Polymerase Chain Reaction (PCR), and Sequencing
2.4. Phylogenetic Analyses
3. Results
3.1. Sequencing and Phylogenetic Analyses
| Species List | Voucher Information | Host/Substrate | GenBank Accession Number | Reference | ||||
|---|---|---|---|---|---|---|---|---|
| nrSSU | nrLSU | TEF | RPB1 | RPB2 | ||||
| Cordyceps albocitrinus | spat 07-174 | MF416575 | MF416467 | MF416629 | [27] | |||
| Cordyceps amoene-rosea | CBS 107.73 | Coleoptera | AY526464 | MF416550 | MF416494 | MF416651 | MF416445 | [27,28] |
| Cordyceps amoene-rosea | CBS 729.73 | Coleoptera | MF416604 | MF416551 | MF416495 | MF416652 | MF416446 | [27,28] |
| Cordyceps araneae | BCC 85065 | Arachnid | MT003037 | MT017850 | MT017810 | MT017828 | [29] | |
| Cordyceps araneae | BCC 85066 | Arachnid | MT003038 | MT017851 | MT017811 | MT017829 | [29] | |
| Cordyceps araneae | BCC 88291 | Arachnid | MT003039 | MT017852 | MT017812 | MT017830 | [29] | |
| Cordyceps bifusispora | spat 08-129 | Lepidoptera | MF416576 | MF416523 | MF416468 | MF416630 | [27] | |
| Cordyceps bifusispora | spat 08-133.1 | Lepidoptera | MF416577 | MF416524 | MF416469 | MF416631 | MF416434 | [27] |
| Cordyceps bifusispora | EFCC 5690 | Lepidoptera | EF468952 | EF468806 | EF468746 | EF468854 | EF468909 | [12] |
| Cordyceps bifusispora | EFCC 8260 | Lepidoptera | EF468953 | EF468807 | EF468747 | EF468855 | EF468910 | [12] |
| Cordyceps blackwelliae | TBRC 7255 | Lepidoptera | MF140703 | MF140823 | MF140772 | MF140796 | [30] | |
| Cordyceps blackwelliae | TBRC 7256 | Coleoptera | MF140702 | MF140822 | MF140771 | MF140795 | [30] | |
| Cordyceps blackwelliae | YFCC 856 | Lepidoptera | MW181780 | MW173992 | MW168233 | MW168199 | MW168216 | [31] |
| Cordyceps brevistroma | BCC 78209 | Lepidoptera | MT003044 | MT017855 | MT017817 | MT017835 | [29] | |
| Cordyceps brevistroma | BCC 79253 | Lepidoptera | MT003045 | MT017856 | – | MT017836 | [29] | |
| Cordyceps buttonspora | YFCC 8400 | Lepidoptera | OL468555 | OL468575 | OL473523 | OL739569 | OL473534 | [25] |
| Cordyceps buttonspora | YFCC 8401 | Lepidoptera | OL468556 | OL468576 | OL473524 | OL739570 | OL473535 | [25] |
| Cordyceps caloceroides | MCA 2249 | Araneae | MF416578 | MF416525 | MF416470 | MF416632 | [27] | |
| Cordyceps cateniannulata | CBS 152.83 | Coleoptera | AY526465 | MG665226 | JQ425687 | [30,32] | ||
| Cordyceps cateniobliqua | YFCC 3367 | Coleoptera | MN576765 | MN576821 | MN576991 | MN576881 | MN576935 | [26] |
| Cordyceps cateniobliqua | YFCC 5935 | Lepidoptera | MN576766 | MN576822 | MN576992 | MN576882 | MN576936 | [26] |
| Cordyceps cateniobliqua | CBS 153.83 | Lepidoptera | AY526466 | JQ425688 | MG665236 | [30,32] | ||
| Cordyceps cf. ochraceostromata | ARSEF 5691 | Lepidoptera | EF468964 | EF468819 | EF468759 | EF468867 | EF468921 | [12] |
| Cordyceps cf. pruinosa | spat 08-115 | Lepidoptera | MF416586 | MF416532 | MF416476 | MF416635 | MF416439 | [27] |
| Cordyceps cf. pruinosa | spat 09-021 | Lepidoptera | MF416587 | MF416533 | MF416477 | MF416636 | [27] | |
| Cordyceps cf. pruinosa | NHJ 10627 | Lepidoptera | EF468967 | EF468822 | EF468763 | EF468870 | [12] | |
| Cordyceps cf. pruinosa | NHJ 10684 | Lepidoptera | EF468968 | EF468823 | EF468761 | EF468871 | [12] | |
| Cordyceps cf. pruinosa | EFCC 5693 | Lepidoptera | EF468966 | EF468821 | EF468762 | EF468869 | [12] | |
| Cordyceps cf. pruinosa | EFCC 5197 | Lepidoptera | EF468965 | EF468820 | EF468760 | EF468868 | [12] | |
| Cordyceps cf. takaomontana | NHJ 12623 | Lepidoptera | EF468984 | EF468838 | EF468778 | EF468884 | EF468932 | [12] |
| Cordyceps chaetoclavata | YHH 15101 | Lepidoptera | MN576722 | MN576778 | MN576948 | MN576838 | MN576894 | [26] |
| Cordyceps chiangdaoensis | BCC 68469 | Coleoptera | MF140732 | KT261403 | [30,33] | |||
| Cordyceps chiangdaoensis | YFCC 857 | Coleoptera | MW181781 | MW173993 | MW168234 | MW168200 | MW168217 | [31] |
| Cordyceps cicadae | GACP 07071701 | Hemiptera | MK761207 | MK761212 | MK770631 | [34] | ||
| Cordyceps cicadae | RCEF HP090724-31 | Hemiptera | MF416605 | MF416552 | MF416496 | MF416653 | MF416447 | [27] |
| Cordyceps cocoonihabita | YFCC 3415 | Lepidoptera | MN576723 | MN576779 | MN576949 | MN576839 | MN576895 | [26] |
| Cordyceps cocoonihabita | YFCC 3416 | Lepidoptera | MN576724 | MN576780 | MN576950 | MN576840 | MN576896 | [26] |
| Cordyceps coleopterorum | CBS 110.73 | Coleoptera | JF415965 | JF415988 | JF416028 | JN049903 | JF416006 | [35] |
| Cordyceps exasperata | MCA 2288 | Lepidoptera | MF416592 | MF416538 | MF416482 | MF416639 | [27] | |
| Cordyceps farinosa | CBS 111113 | – | AY526474 | MF416554 | MF416499 | MF416656 | MF416450 | [27,32] |
| Cordyceps fumosorosea | YFCC 4561 | Lepidoptera | MN576761 | MN576817 | MN576987 | MN576877 | MN576931 | [26] |
| Cordyceps fumosorosea | CBS 244.31 | Butter | MF416609 | MF416557 | MF416503 | MF416660 | MF416454 | [27] |
| Cordyceps fumosorosea | CBS 375.70 | Food | MF416501 | MF416658 | MF416452 | [27] | ||
| Cordyceps fumosorosea | CBS 107.10 | – | MG665227 | HM161735 | MG665237 | [28] | ||
| Cordyceps grylli | MFLU 17-1023 | Orthoptera | MK863048 | MK863055 | MK860193 | Unpublished | ||
| Cordyceps grylli | MFLU 17-1024 | Orthoptera | MK863049 | MK863056 | MK860194 | Unpublished | ||
| Cordyceps inthanonensis | BCC 79828 | Lepidoptera | – | MT017854 | MT017816 | MT017833 | [29] | |
| Cordyceps inthanonensis | BCC 56302 | Lepidoptera | MT003040 | MT017853 | MT017814 | MT017831 | [29] | |
| Cordyceps inthanonensis | BCC 55812 | Lepidoptera | MT003041 | – | MT017815 | MT017832 | [29] | |
| Cordyceps jakajanicola | BCC 79816 | Hemiptera | MN275696 | MN338479 | MN338484 | MN338489 | [36] | |
| Cordyceps jakajanicola | BCC 79817 | Hemiptera | MN275697 | MN338480 | MN338485 | MN338490 | [36] | |
| Cordyceps javanica | TBRC 7259 | Lepidoptera | MF140711 | MF140831 | MF140780 | MF140804 | [30] | |
| Cordyceps javanica | CBS 134.22 | Coleoptera | MF416610 | MF416558 | MF416504 | MF416661 | MF416455 | [27] |
| Cordyceps kuiburiensis | BCC 90322 | Araneidae | MK968816 | MK988032 | MK988030 | [36] | ||
| Cordyceps kuiburiensis | BCC 90323 | Araneidae | MK968817 | MK988033 | MK988031 | [36] | ||
| Cordyceps kyusyuensis | EFCC 5886 | Lepidoptera | EF468960 | EF468813 | EF468754 | EF468863 | EF468917 | [12] |
| Cordyceps lepidopterorum | TBRC 7263 | Lepidoptera | MF140699 | MF140819 | MF140768 | MF140792 | [30] | |
| Cordyceps lepidopterorum | TBRC 7264 | Lepidoptera | MF140700 | MF140820 | MF140769 | MF140793 | [30] | |
| Cordyceps longiphialide | YFCC 8402 | rotted wood | OL468557 | OL468577 | OL473525 | OL739571 | OL473536 | [25] |
| Cordyceps longiphialide | YFCC 8403 | rotted wood | OL468558 | OL468578 | OL473526 | OL739572 | OL473537 | [25] |
| Cordyceps militaris | YFCC 6587 | Lepidoptera | MN576762 | MN576818 | MN576988 | MN576878 | MN576932 | [26] |
| Cordyceps militaris | YFCC 5840 | Lepidoptera | MN576763 | MN576819 | MN576989 | MN576879 | MN576933 | [26] |
| Cordyceps morakotii | BCC 55820 | Hymenoptera | MF140730 | KT261399 | [33] | |||
| Cordyceps morakotii | BCC 68398 | Hymenoptera | MF140731 | KT261398 | [33] | |||
| Cordyceps nabanheensis | YFCC 8409 | Lepidoptera | OL468564 | OL468584 | OL473532 | OL739578 | OL473543 | [25] |
| Cordyceps nabanheensis | YFCC 8410 | Lepidoptera | OL468565 | OL468585 | OL473533 | OL739579 | OL473544 | [25] |
| Cordyceps neopruinosa | BCC 91361 | Lepidoptera | MT003047 | MT017858 | MT017838 | [29] | ||
| Cordyceps neopruinosa | BCC 91362 | Lepidoptera | MT003048 | MT017859 | MT017818 | MT017839 | [29] | |
| Cordyceps nidus | HUA 186125 | Araneae | KC610778 | KC610752 | KC610722 | KC610711 | [37] | |
| Cordyceps nidus | HUA 186186 | Araneae | KY360301 | KC610753 | KC610723 | KY360297 | [37] | |
| Cordyceps ninchukispora | EGS 38.165 | Plant | EF468991 | EF468846 | EF468795 | EF468900 | [12] | |
| Cordyceps ninchukispora | EGS 38.166 | Plant | EF468992 | EF468847 | EF468794 | EF468901 | [12] | |
| Cordyceps ningxiaensis | HMJAU 25074 | Diptera | KF309671 | [38] | ||||
| Cordyceps ningxiaensis | HMJAU 25076 | Diptera | KF309673 | [38] | ||||
| Cordyceps nototenuipes | YFCC 8404 | Lepidoptera | OL468559 | OL468579 | OL473527 | OL739573 | OL473538 | [25] |
| Cordyceps nototenuipes | YFCC 8405 | Lepidoptera | OL468560 | OL468580 | OL473528 | OL739574 | OL473539 | [25] |
| Cordyceps oncoperae | ARSEF 4358 | Lepidoptera | AF339581 | AF339532 | EF468785 | EF468891 | EF468936 | [12,39] |
| Cordyceps polyarthra | MCA 996 | Lepidoptera | MF416597 | MF416543 | MF416487 | MF416644 | [27] | |
| Cordyceps polyarthra | MCA 1009 | Lepidoptera | MF416598 | MF416544 | MF416488 | MF416645 | [27] | |
| Cordyceps polystromata | YFCC 1610885 | Lepidoptera | OQ878491 | OQ878487 | OQ868508 | OQ868514 | OQ868511 | This study |
| Cordyceps polystromata | YFCC 1610886 | Lepidoptera | OQ878492 | OQ878488 | OQ868509 | OQ868515 | OQ868512 | This study |
| Cordyceps pruinosa | ARSEF 5413 | Lepidoptera | AY184979 | AY184968 | DQ522351 | DQ522397 | DQ522451 | [40] |
| Cordyceps qingchengensis | MFLU 17-1022 | Lepidoptera | MK761206 | MK761211 | MK770630 | [34] | ||
| Cordyceps rosea | spat 09-053 | Lepidoptera | MF416590 | MF416536 | MF416480 | MF416637 | MF416442 | [27] |
| Cordyceps roseostromata | ARSEF 4871 | Coleoptera | AF339573 | AF339523 | [39] | |||
| Cordyceps sapaensis | YFCC 5833 | Lepidoptera | MN576764 | MN576820 | MN576990 | MN576880 | MN576934 | [26] |
| Cordyceps sapaensis | YFCC 1610884 | Lepidoptera | OQ878490 | OQ878486 | OQ868507 | OQ868513 | OQ868510 | This study |
| Cordyceps shuifuensis | YFCC 5230 | Lepidoptera | MN576721 | MN576777 | MN576947 | MN576837 | MN576893 | [26] |
| Cordyceps simaoensis | YFCC 8406 | Lepidoptera | OL468561 | OL468581 | OL473529 | OL739575 | OL473540 | [25] |
| Cordyceps simaoensis | YFCC 8407 | Lepidoptera | OL468562 | OL468582 | OL473530 | OL739576 | OL473541 | [25] |
| Cordyceps simaoensis | YFCC 8408 | Lepidoptera | OL468563 | OL468583 | OL473531 | OL739577 | OL473542 | [25] |
| Cordyceps sp. | CBS 102184 | Arachnid | AF339613 | AF339564 | EF468803 | EF468907 | EF468948 | [12,39] |
| Cordyceps sp. | EFCC 2535 | Coleoptera | EF468980 | EF468835 | EF468772 | [12] | ||
| Cordyceps spegazzinii | ARSF 7850 | Diptera | DQ196435 | [41] | ||||
| Cordyceps subtenuipes | YFCC 6051 | Lepidoptera | MN576719 | MN576775 | MN576945 | MN576835 | MN576891 | [26] |
| Cordyceps subtenuipes | YFCC 6084 | Lepidoptera | MN576720 | MN576776 | MN576946 | MN576836 | MN576892 | [26] |
| Cordyceps succavus | MFLU 18-1890 | Lepidoptera | MK086058 | MK086062 | MK084616 | MK079353 | [42] | |
| Cordyceps tenuipes | ARSEF 5135 | Lepidoptera | MF416612 | JF415980 | JF416020 | JN049896 | JF416000 | [27,35] |
| Cordyceps tenuipes | YFCC 4266 | Lepidoptera | MN576774 | MN576830 | MN577000 | MN576890 | MN576944 | [26] |
| Cordyceps yinjiangensis | YJ06221 | Hymenoptera | MT577003 | MT577002 | [43] | |||
| Liangia sinensis | YFCC 3103 | Fungi | MN576726 | MN576782 | MN576952 | MN576842 | MN576898 | [26] |
| Liangia sinensis | YFCC 3104 | Fungi | MN576727 | MN576783 | MN576953 | MN576843 | MN576899 | [26] |

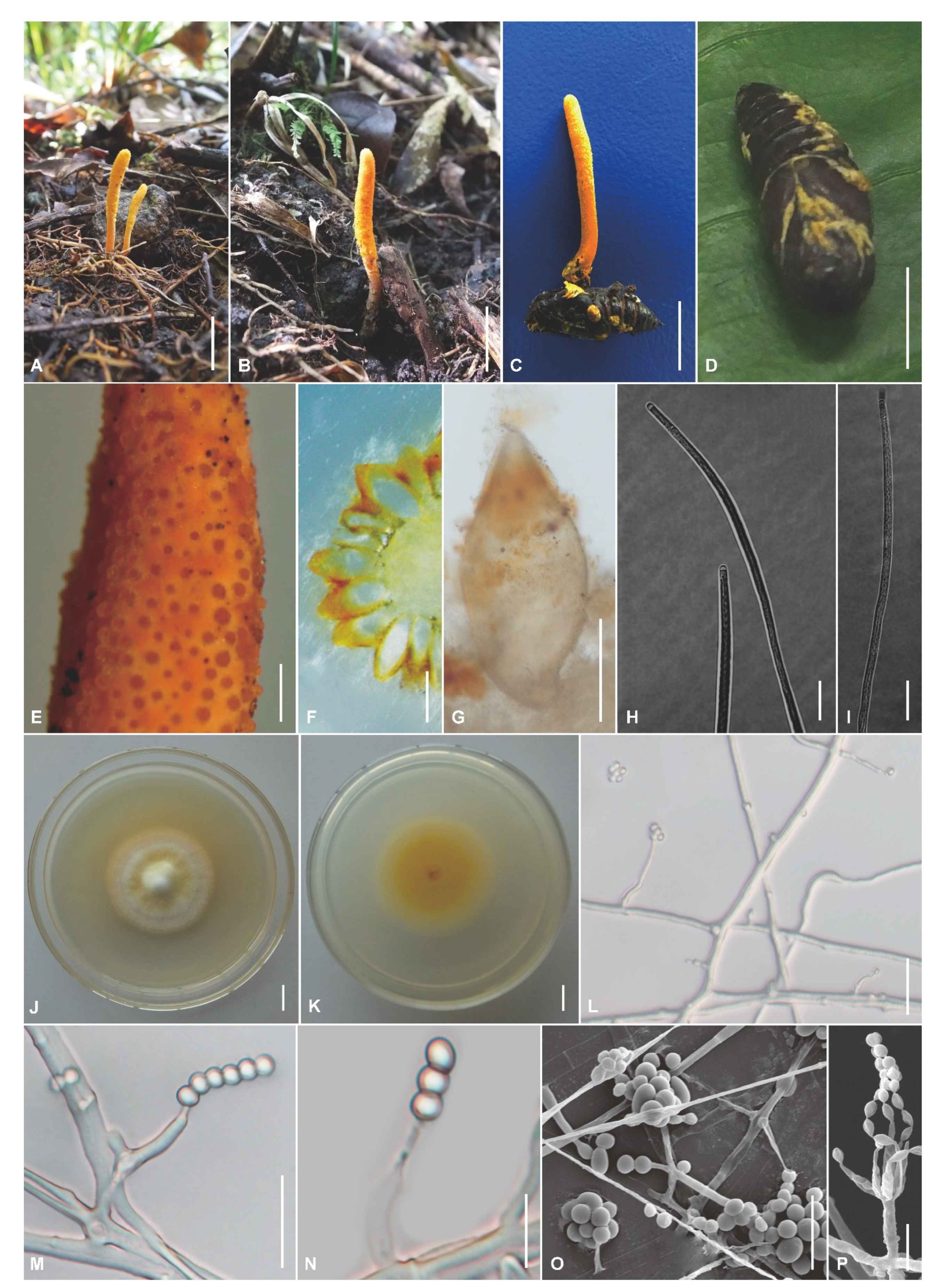
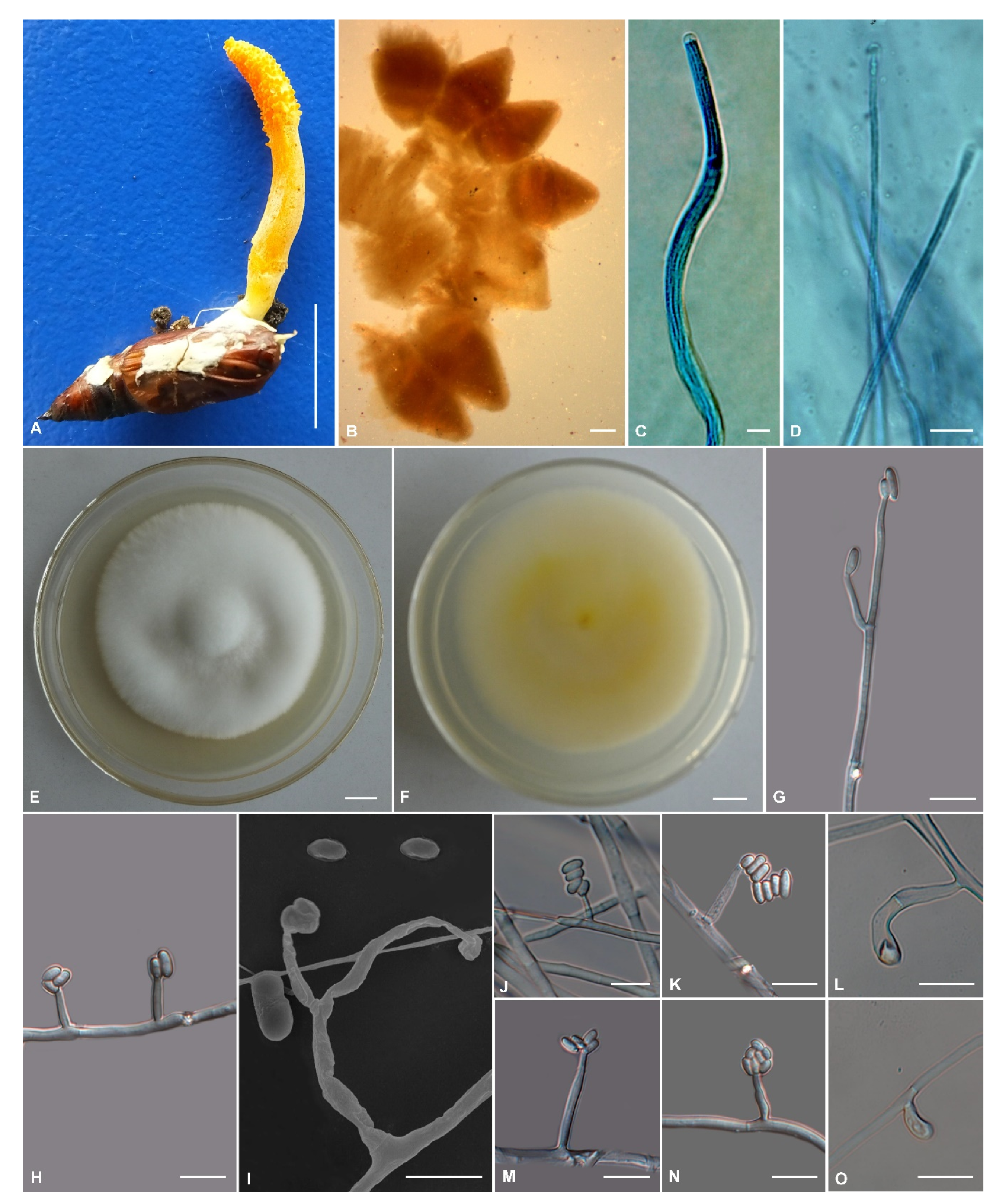
3.2. Morphological Features
3.3. Taxonomy

4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, Z.Z.; Luan, F.G.; Hywel-jones Nigel, L.; Zhang, S.L.; Chen, M.J.; Huang, B.; Sun, C.S.; Chen, Z.A.; Li, C.R.; Tan, Y.J.; et al. Biodiversity of cordycipitoid fungi associated with Isaria cicadae Miquel II: Teleomorph discovery and nomenclature of chanhua, an important medicinal fungus in China. Mycosystema 2021, 40, 95–107. [Google Scholar]
- Wang, Y.; Liu, Y.F.; Fan, Q.; Wang, Y.B.; Yu, H. Cordycipitoid Fungi Powders Promote Mycelial Growth and Bioactive-Metabolite Production in Liquid Cultures of the Stout Camphor Medicinal Mushroom Taiwanofungus camphoratus (Agaricomycetes). Int. J. Med. Mushrooms 2020, 22, 615–626. [Google Scholar] [CrossRef] [PubMed]
- Mascarin, G.M.; Kobori, N.N.; Quintela, E.D.; Delalibera, I.J. The virulence of entomopathogenic fungi against Bemisia tabaci biotype B (Hemiptera: Aleyrodidae) and their conidial production using solid substrate fermentation. Biol. Control 2013, 66, 209–218. [Google Scholar] [CrossRef]
- Rojas, V.M.A.; Iwanicki, N.S.A.; Alessandro, C.P.; Fatoretto, M.B.; Demétrio, C.G.B.; Delalibera, I.J. Characterization of Brazilian Cordyceps fumosorosea isolates: Conidial production, tolerance to ultraviolet-B radiation, and elevated temperature. J. Invertebr. Pathol. 2023, 197, 107888. [Google Scholar] [CrossRef]
- Cui, J.D. Biotechnological production and applications of Cordyceps militaris, a valued traditional Chinese medicine. Crit. Rev. Biotechnol. 2015, 35, 475–484. [Google Scholar] [CrossRef]
- Lou, H.W.; Lin, J.F.; Guo, L.Q.; Wang, X.W.; Tian, S.Q.; Liu, C.X.; Zhao, Y.; Zhao, R.Y. Advances in research on Cordyceps militaris degeneration. Appl. Microbiol. Biotechnol. 2019, 103, 7835–7841. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Luo, F.; Shang, Y.; Chen, P.; Lu, Y.; Wang, C. Fungal cordycepin biosynthesis is coupled with the production of the safeguard molecule pentostatin. Cell Chem. Biol. 2017, 24, 1479–1489. [Google Scholar] [CrossRef]
- Nurmamat, E.; Xiao, H.; Zhang, Y.; Jiao, Z. Effects of different temperatures on the chemical structure and antitumor activities of polysaccharides from Cordyceps militaris. Polymers 2018, 10, 430. [Google Scholar] [CrossRef]
- Kunhorm, P.; Chaicharoenaudomrung, N.; Noisa, P. Enrichment of cordycepin for cosmeceutical applications: Culture systems and strategies. Appl. Microbiol. Biotechnol. 2019, 103, 1681–1691. [Google Scholar] [CrossRef]
- Shrestha, B.; Tanaka, E.; Han, J.G.; Oh, J.S.; Han, S.K.; Lee, K.H.; Sung, G.H. A Brief Chronicle of the Genus Cordyceps Fr., the Oldest Valid Genus in Cordycipitaceae (Hypocreales, Ascomycota). Mycobiology 2014, 42, 93–99. [Google Scholar] [CrossRef]
- Fries, E.M. Observationes Mycologicae Praecipue ad Illustrandam Floram Suecicam; Pars secunda (Cancellans issue); G. Bonnieri: Copenhagen, Denmark, 1818. [Google Scholar]
- Sung, G.H.; Hywel-Jones, N.L.; Sung, J.M.; Luangsa-ard, J.J.; Shrestha, B.; Spatafora, J.W. Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud. Mycol. 2007, 57, 5–59. [Google Scholar] [CrossRef]
- Zou, W.Q.; Tang, D.X.; Xu, Z.H.; Huang, O.; Wang, Y.B.; Tran, N.-L.; Yu, H. Multigene phylogeny and morphology reveal Ophiocordyceps hydrangea sp. nov. and Ophiocordyceps bidoupensis sp. nov. (Ophiocordycipitaceae). MycoKeys 2022, 92, 109–130. [Google Scholar] [CrossRef]
- Lao, T.D.; Le, T.A.H.; Truong, N.B. Morphological and genetic characteristics of the novel entomopathogenic fungus Ophiocordyceps langbianensis (Ophiocordycipitaceae, Hypocreales) from Lang Biang Biosphere Reserve, Vietnam. Sci. Rep. 2021, 11, 1412. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.H.; Tran, N.L.; Wang, Y.; Zhang, G.D.; Dao, V.M.; Nguyen, T.T.; Wang, Y.B.; Yu, H. Phylogeny and morphology of Ophiocordyceps puluongensis sp. nov. (Ophiocordycipitaceae, Hypocreales), a new fungal pathogen on termites from Vietnam. J. Invertebr. Pathol. 2022, 192, 107771. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.B.; Yu, H.; Dai, Y.D.; Wu, C.K.; Zeng, W.B.; Yuan, F.; Liang, Z.Q. Polycephalomyces agaricus, a new hyperparasite of Ophiocordyceps sp. infecting melolonthid larvae in southwestern China. Mycol. Prog. 2015, 14, 70. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed]
- Rehner, S.A.; Samuels, G.J. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol. Res. 1994, 98, 625–634. [Google Scholar] [CrossRef]
- Bischoff, J.F.; Rehner, S.A.; Humber, R.A. Metarhizium frigidum sp. nov.: A cryptic species of M. anisopliae and a member of the M. flavoviride complex. Mycologia 2006, 98, 737–745. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP*: Phylogenetic Analysis USING parsimony (*and Other Methods), Version 4.0a166; Sinauer Associates: Sunderland, MA, USA, 2019. [Google Scholar]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar]
- Dong, Q.Y.; Wang, Y.; Wang, Z.Q.; Tang, D.X.; Zhao, Z.Y.; Wu, H.J.; Yu, H. Morphology and Phylogeny Reveal Five Novel Species in the Genus Cordyceps (Cordycipitaceae, Hypocreales) From Yunnan, China. Front. Microbiol. 2022, 13, 846909. [Google Scholar] [CrossRef]
- Wang, Y.B.; Wang, Y.; Fan, Q.; Duan, D.E.; Zhang, G.D.; Dai, R.Q.; Dai, Y.D.; Zeng, W.B.; Chen, Z.H.; Li, D.D.; et al. Multigene phylogeny of the family Cordycipitaceae (Hypocreales): New taxa and the new systematic position of the Chinese cordycipitoid fungus Paecilomyces hepiali. Fungal Divers. 2020, 103, 1–46. [Google Scholar] [CrossRef]
- Kepler, R.M.; Luangsa-Ard, J.J.; Hywel-Jones, N.L.; Quandt, C.A.; Sung, G.H.; Rehner, S.A.; Aime, M.C.; Henkel, T.W.; Sanjuan, T.; Zare, R.; et al. A phylogenetically-based nomenclature for Cordycipitaceae (Hypocreales). IMA Fungus 2017, 8, 335–353. [Google Scholar] [CrossRef] [PubMed]
- Luangsa-ard, J.J.; Hywel-Jones, N.L.; Manoch, L.; Samson, R.A. On the relationships of Paecilomyces sect. Isarioidea species. Mycol. Res. 2005, 109, 581–589. [Google Scholar] [CrossRef]
- Mongkolsamrit, S.; Noisripoom, W.; Tasanathai, K.; Khonsanit, A.; Thanakitpipattana, D.; Himaman, W.; Kobmoo, N.; Luangsa-ard, J.J. Molecular phylogeny and morphology reveal cryptic species in Blackwellomyces and Cordyceps (Cordycipitaceae) from Thailand. Mycol. Prog. 2020, 19, 957–983. [Google Scholar] [CrossRef]
- Mongkolsamrit, S.; Noisripoom, W.; Thanakitpipattana, D.; Wutikhun, T.; Spatafora, J.W.; Luangsa-ard, J. Disentangling cryptic species with isaria-like morphs in Cordycipitaceae. Mycologia 2018, 110, 230–257. [Google Scholar] [CrossRef]
- Wang, Y.; Fan, Q.; Wang, D.; Zou, W.Q.; Tang, D.X.; Hongthong, P.; Yu, H. Species Diversity and Virulence Potential of the Beauveria bassiana Complex and Beauveria scarabaeidicola Complex. Front. Microbiol. 2022, 13, 841604. [Google Scholar] [CrossRef]
- Luangsa-ard, J.J.; Hywel-Jones, N.L.; Samson, R.A. The polyphyletic nature of Paecilomyces sensu lato based on 18S-generated rDNA phylogeny. Mycologia 2004, 96, 773–780. [Google Scholar] [CrossRef]
- Tasanathai, K.; Thanakitpipattana, D.; Noisripoom, W.; Khonsanit, A.; Kumsao, J.; Luangsa-ard, J.J. Two new Cordyceps species from a community forest in Thailand. Mycol. Prog. 2016, 15, 28. [Google Scholar] [CrossRef]
- Zha, L.S.; Wen, T.C.; Huang, S.K.; Boonmee, S.; Eungwanichayapant, P.D. Taxonomy and biology of Cordyceps qingchengensis sp. nov. and its allies. Phytotaxa 2019, 416, 14–24. [Google Scholar] [CrossRef]
- Kepler, R.M.; Sung, G.H.; Ban, S.; Nakagiri, A.; Chen, M.J.; Huang, B.; Li, Z.; Spatafora, J.W. New teleomorph combinations in the entomopathogenic genus Metacordyceps. Mycologia 2012, 104, 182–197. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.W.; Wingfield, M.J.; Lombard, L.; Roets, F.; Swart, W.J.; Alvarado, P.; Carnegie, A.J.; Moreno, G.; Luangsa-Ard, J.; Thangavel, R.; et al. Fungal Planet description sheets: 951–1041. Persoonia 2019, 43, 223–425. [Google Scholar] [CrossRef]
- Chirivi, J.; Danies, G.; Sierra, R.; Schauer, N.; Trenkamp, S.; Restrepo, S.; Sanjuan, T. Metabolomic profile and nucleoside composition of Cordyceps nidus sp. nov.(Cordycipitaceae): A new source of active compounds. PLoS ONE 2017, 12, e0179428. [Google Scholar] [CrossRef]
- Yan, J.Q.; Bau, T. Cordyceps ningxiaensis sp. nov., a new species from dipteran pupae in Ningxia Hui Autonomous Region of China. Nova Hedwig. 2015, 100, 251–258. [Google Scholar] [CrossRef]
- Sung, G.H.; Spatafora, J.W.; Zare, R.; Gams, W. A revision of Verticillium sect. Prostrata. II. Phylogenetic analyses of SSU and LSU nuclear rDNA sequences from anamorphs and teleomorphs of the Clavicipitaceae. Nova Hedwig. 2001, 72, 311–328. [Google Scholar] [CrossRef]
- Spatafora, J.W.; Sung, G.-H.; Sung, G.H.; Hywel-Jones, N.; White, J.F. Phylogenetic evidence for an animal pathogen origin of ergot and the grass endophytes. Mol. Ecol. 2007, 16, 1701–1711. [Google Scholar] [CrossRef]
- Torres, M.S.; White, J.; Bischoff, J.F. Cordyceps spegazzinii sp. nov., a new species of the C. militaris group. Mycotaxon 2005, 94, 253–263. [Google Scholar]
- Hyde, K.D.; Tennakoon, D.S.; Jeewon, R.; Bhat, D.J.; Maharachchikumbura, S.S.; Rossi, W.; Leonardi, M.; Lee, H.B.; Mun, H.Y.; Houbraken, J.; et al. Fungal diversity notes 1036–1150: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2019, 96, 1–242. [Google Scholar] [CrossRef]
- Li, Y.P.; Chen, W.H.; Han, Y.F.; Liang, J.D.; Liang, Z.Q. Cordyceps yinjiangensis, a new ant-pathogenic fungus. Phytotaxa 2020, 453, 284–292. [Google Scholar] [CrossRef]
- Yang, Z.L.; Yu, H.; Chen, Z.H.; Wang, Y.B. Study on the biological and ecological habits of populations of Cordyceps militaris in middle of Yunnan. Edible Fungi China 2012, 30, 43–47. [Google Scholar]
- Kobayasi, Y.; Shimizu, D. Cordyceps species from Japan 5. Bull. Natn. Sci. Mus. Tokyo Ser. B 1982, 8, 111–123. [Google Scholar]
- Kobayasi, Y. Revision of the genus Cordyceps and its allies 1. Bull. Natn. Sci. Mus. Tokyo Ser. B 1981, 7, 1–13. [Google Scholar]
- Kobayasi, Y.; Shimizu, D. Cordyceps species from Japan 6. Bull. Natn. Sci. Mus. Tokyo Ser. B 1983, 9, 1–21. [Google Scholar]
- Wright, P.J. Cordyceps oncoperae sp. nov. (Ascomycota) Infecting Oncopera spp. (Lepidoptera: Hepialidae). J. Invertebr. Pathol. 1993, 61, 211–213. [Google Scholar] [CrossRef]
| Species | Stromata (mm) | Fertile Parts (mm) | Perithecia (μm) | Asci (μm) | Ascospores (μm) | Part-Spores (μm) | Phialides (μm) | Conidia (μm) | References |
|---|---|---|---|---|---|---|---|---|---|
| Cordyceps chaetoclavata | Solitary, 23 × 0.8 | Clavate, covered by a spinous surface, 5.6 × 0.7–1.1 | Superficial, 402–610 × 280–427 | Cylindrical, 274–385 × 3.7–4.8 | Filiform, multiseptate, 127–260 × 0.9–1.2 | Cylindrical, 3–12 μm long | [26] | ||
| Cordyceps inthanonensis | Multiple, 6–25 mm long | Cylindrical to clavate, 3–5 mm wide | Semi-immersed, ovoid, 600–720 × 220–420 | Cylindrical, 450–600 × 4–6 | Filiform, multiseptate, 400–550 μm long | Cylindrical, 3–4 × 1–1.5 | Solitary, cylindrical at the base tapering to the apex, (12)14–18.5(20) × 1.5–3 | Cylindrical, 4–7(9) × 1.5–2 | [29] |
| Cordyceps kyusyuensis | Multiple, 15–20 mm long | Cylindrical, 10–12 mm long | Semi-superficial, ovoid, 410–580 × 210–330 | 4 μm wide | 4–5 × 1 | Ovoid, 2 × 1.5 | [46] | ||
| Cordyceps militaris | Solitary or in groups of 2–3, 15–90 mm long | Clavate, 8–45 × 3.4–6.5 | Semi-immersed, ovoid, 230–556 × 113–319 µm | Cylindrical, 200.0–480.6 × 2.9–4.7 | Filiform, multiseptate | 1.8–4.2 × 0.7–1.6 | Solitary or verticillate, Verticillium-type: 2.8–29.5 × 0.8–3.4, Paecilomyces-type: 5.8–16.5 × 1.4–3.1 | Subglobose to ellipsoidal, 1.8–5.6 × 1.4–3.2 | This study |
| Cordyceps ningxiaensis | 1 to 2 in a group | Spherical to ovoid, 1.2–3 × 1.2–2.8 | Immersed, ellipsoid to ovoid, 288–400 × 103–240 | Cylindrical, 168–205 × (3.7–)4.1–5.5(–6.6) | Filiform, irregularly multiseptate | 3.6–7.8 × 1.0–1.4 | [38] | ||
| Cordyceps oncoperae | Solitary to multiple, up to 35 mm long | Clavate, usually with acute apices, 4–10 × 2–3 | Ovoid, 350–410 × 180–230(–380) | Cylindrical, (168–)200–224(–256) × (5–)6–6.5 | Filiform, multiseptate, 104–139 × 1.5–2 | [48] | |||
| Cordyceps polystromata | Multiple, 10–17 mm long | Cylindrical to clavate, covered by a spinous surface, 3–14 × 1.6–3.5 | Superficial, ovoid, 522–663 × 296–577 | Cylindrical, 54.2–172.8 × 4.1–6.5 | Filiform, multiseptate, 51.4–170.5 × 0.9–2.6 | 5.7–7.0 × 1.7–3.2 | Verticillate, 6.2–17.2 × 0.9–2.7 | Ovoid to ellipsoidal, 1.5–3.7 × 1.2–2.5 | This study |
| Cordyceps rosea | Solitary, 11 mm long | Clavate | Immersed, ovoid, 330–380 × 160–230 | 100 × 3–4 | Filiform, multiseptate, 120 × 1–1.5 | Navicular, 4–5 × 1 | [45] | ||
| Cordyceps roseostromata | Solitary to multiple | Subglobose to clavate, 1.2–5 × 1.5–2.2 | Superficial, pyriform, 280–300 × 140–160 | 3–3.5 μm wide | 4–5 × 1 | [47] | |||
| Cordyceps sapaensis | Solitary, 18–35 mm long | Banana-shaped, 8–15 × 3.3–4.8 | Superficial, crowded, ovoid, 587–743 × 341–396 | Cylindrical, 237.2–472.6 × 3.1–5.4 | Filiform, multiseptate, 230.2–457.8 × 1.6–2.1 | 2.0–4.8 × 1.3–2.2 | Solitary or verticillate, cylindrical, 5.1–28.5 µm long, 1.5–3.1 µm wide at the base, 1.3–2.1 µm wide at the apex | Ellipsoidal to cylindrical, 2.6–7.4 × 1.1–3.1 | This study |
| Cordyceps shuifuensis | Solitary, 25 mm long | Clavate, 4 × 1.5 | Pseudoimmersed, ovoid, 450–620 × 300–430 | Cylindrical, 275–510 × 3.5–5.2 | Filiform, multiseptate, 180–410 × 1.2–1.7 | Cylindrical, 2.8–6.5 μm long | Solitary or verticillate, cylindrical or subulate, 4.7–20 um long, 1.1–2.0 μm wide at the base, 0.4–2.1 μm wide at the apex | Macroconidia clavate to oblong-ovate, 5.1–11.8 × 1.3–2.4; microconidia globose to ellipsoidal, 1.8–3.0 × 1.6–2.5 | [26] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Dong, Q.-Y.; Luo, R.; Fan, Q.; Duan, D.-E.; Dao, V.-M.; Wang, Y.-B.; Yu, H. Molecular Phylogeny and Morphology Reveal Cryptic Species in the Cordyceps militaris Complex from Vietnam. J. Fungi 2023, 9, 676. https://doi.org/10.3390/jof9060676
Wang Y, Dong Q-Y, Luo R, Fan Q, Duan D-E, Dao V-M, Wang Y-B, Yu H. Molecular Phylogeny and Morphology Reveal Cryptic Species in the Cordyceps militaris Complex from Vietnam. Journal of Fungi. 2023; 9(6):676. https://doi.org/10.3390/jof9060676
Chicago/Turabian StyleWang, Yao, Quan-Ying Dong, Run Luo, Qi Fan, Dong-E Duan, Van-Minh Dao, Yuan-Bing Wang, and Hong Yu. 2023. "Molecular Phylogeny and Morphology Reveal Cryptic Species in the Cordyceps militaris Complex from Vietnam" Journal of Fungi 9, no. 6: 676. https://doi.org/10.3390/jof9060676
APA StyleWang, Y., Dong, Q.-Y., Luo, R., Fan, Q., Duan, D.-E., Dao, V.-M., Wang, Y.-B., & Yu, H. (2023). Molecular Phylogeny and Morphology Reveal Cryptic Species in the Cordyceps militaris Complex from Vietnam. Journal of Fungi, 9(6), 676. https://doi.org/10.3390/jof9060676






