Phenotypic Array for Identification and Screening of Antifungals against Aspergillus Isolates from Respiratory Infections in KwaZulu Natal, South Africa
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Culture Maintenance
2.2. Microscopic and Culture Morphology Identification
2.3. DNA Identification
2.4. Phenotypic Identification
2.5. Minimum Inhibitory Concentration (MIC) of Voriconazole and Posaconazole
- Voriconazole: MIC ≤ 0.25; intermediate susceptibility > 0.25–2.0; resistant > 2.0
- Posaconazole: MIC ≤ 0.06; intermediate susceptibility > 0.06–0.25; resistant > 0.25
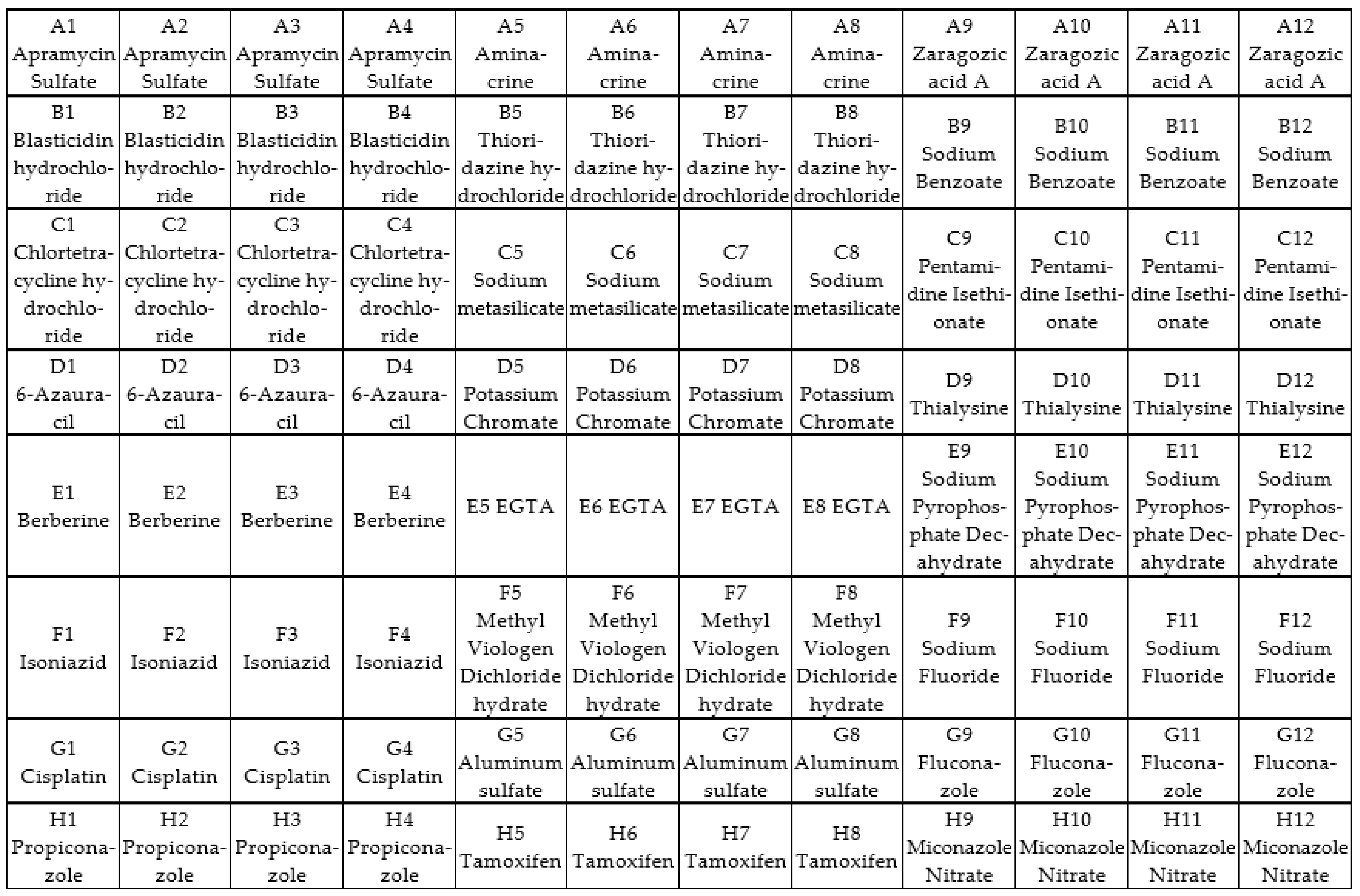
2.6. Screening of Antifungal Compounds Using Phenotypic Microarray
2.7. Identification of TR34/98 Mutations Associated with Azole Resistance
3. Results
3.1. Identification Using Traditional (Culture and Microscopy), Genetic and Phenotypic Methods
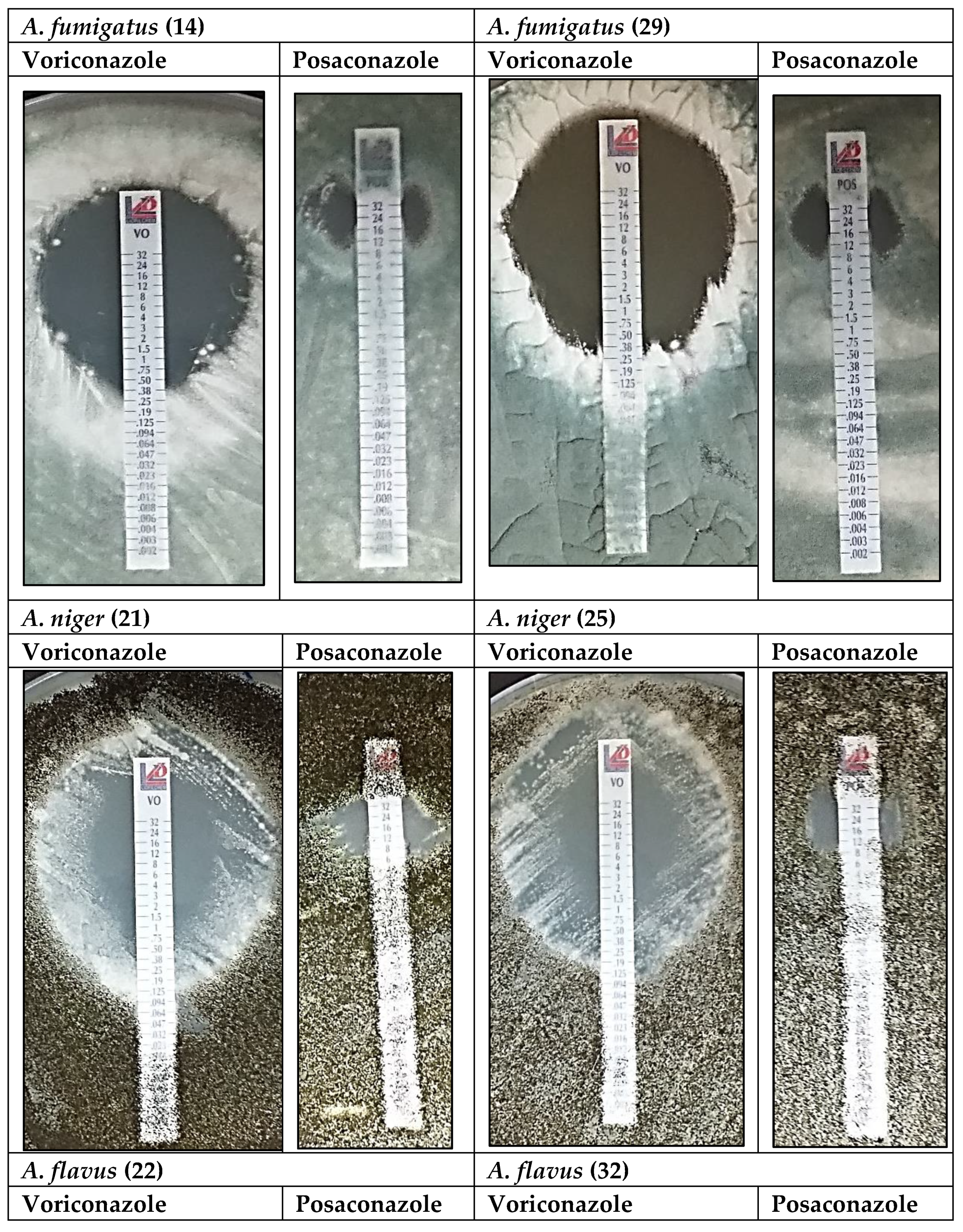
3.2. Antifungal Analysis: Voriconazole and Posaconazole
3.3. Phenotypic Microarray of Antifungal Compounds
3.4. Identification of TR34/98 Mutations Associated with Azole Resistance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Friedman, D.Z.; Schwartz, I.S. Emerging Fungal Infections: New Patients, New Patterns, and New Pathogens. J. Fungi 2019, 5, 67. [Google Scholar] [CrossRef] [PubMed]
- Bassetti, M.; Righi, E.; Ansaldi, F.; Merelli, M.; Scarparo, C.; Antonelli, M.; Garnacho-Montero, J.; Diaz-Martin, A.; Palacios-Garcia, I.; Luzzati, R.; et al. A multicenter multinational study of abdominal candidiasis: Epidemiology, outcomes and predictors of mortality. Intensive Care Med. 2015, 41, 1601–1610. [Google Scholar] [CrossRef] [PubMed]
- WHO. Fungal Priority Pathogens List to Guide Research, Development and Public Health Action; World Health Organization: Geneva, Switzerland, 2022; Available online: https://za.pinterest.com/pin/606789749769944470/ (accessed on 11 December 2022).
- Bongomin, F.; Gago, S.; Oladele, R.O.; Denning, D.W. Global and Multi-National Prevalence of Fungal Diseases—Estimate Precision. J. Fungi 2017, 3, 57. [Google Scholar] [CrossRef]
- Rybak, J.M.; Fortwendel, J.R.; Rogers, P.D. Emerging threat of triazole-resistant Aspergillus fumigatus. J. Antimicrob. Chemother. 2019, 74, 835–842. [Google Scholar] [CrossRef] [PubMed]
- Armstrong-James, D.; Meintjes, G.; Brown, G.D. A neglected epidemic: Fungal infections in HIV/AIDS. Trends Microbiol. 2014, 22, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Taccone, F.S.; Abeele, A.-M.V.D.; Bulpa, P.; Misset, B.; Meersseman, W.; Cardoso, T.; Paiva, J.-A.; Blasco-Navalpotro, M.; De Laere, E.; Dimopoulos, G.; et al. Epidemiology of invasive aspergillosis in critically ill patients: Clinical presentation, underlying conditions, and outcomes. Crit. Care 2015, 19, 7. [Google Scholar] [CrossRef]
- Schwartz, I.S.; Boyles, T.H.; Kenyon, C.R.; Hoving, J.C.; Brown, G.D.; Denning, D.W. The estimated burden of fungal disease in South Africa. S. Afr. Med. J. 2019, 109, 885. [Google Scholar] [CrossRef]
- Blackwell, M.; Hibbett, D.S.; Taylor, J.W.; Spatafora, J.W. Research coordination networks: A phylogeny for kingdom Fungi (Deep Hypha). Mycologia 2006, 98, 829–837. [Google Scholar] [CrossRef]
- Chojniak, J.; Jałowiecki, Ł.; Dorgeloh, E.; Hegedusova, B.; Ejhed, H.; Magnér, J.; Płaza, G. Application of the BIOLOG system for characterization of Serratia marcescens ss marcescens isolated from onsite wastewater technology (OSWT). Acta Biochim. Pol. 2015, 62, 799–805. [Google Scholar] [CrossRef]
- Pinzari, F.; Ceci, A.; Abu-Samra, N.; Canfora, L.; Maggi, O.; Persiani, A. Phenotype MicroArray™ system in the study of fungal functional diversity and catabolic versatility. Res. Microbiol. 2016, 167, 710–722. [Google Scholar] [CrossRef]
- Enoch, D.A.; Yang, H.; Aliyu, S.H.; Micallef, C. The Changing Epidemiology of Invasive Fungal Infections. Human Fungal Path. Ident. 2017, 1508, 17–65. [Google Scholar]
- Verweij, P.E.; Chowdhary, A.; Melchers, W.J.; Meis, J.F. Azole resistance in Aspergillus fumigatus: Can we retain the clinical use of mold-active antifungal azoles? Clin. Infect. Dis. 2016, 62, 362–368. [Google Scholar] [CrossRef]
- Lestrade, P.P.A.; Meis, J.F.; Melchers, W.J.G.; Verweij, P.E. Triazole resistance in Aspergillus fumigatus: Recent insights and challenges for patient management. Clin. Microbiol. Infect. 2019, 25, 799–806. [Google Scholar] [CrossRef] [PubMed]
- Zea, S.M.R.; Toyotome, T. Azole-resistant Aspergillus fumigatus as an emerging worldwide pathogen. Microbiol. Immunol. 2022, 66, 135–144. [Google Scholar]
- Arastehfar, A.; Carvalho, A.; Houbraken, J.; Lombardi, L.; Garcia-Rubio, R.; Jenks, J.; Rivero-Menendez, O.; Aljohani, R.; Jacobsen, I.; Berman, J. Aspergillus fumigatus and aspergillosis: From basics to clinics. Stud. Mycol. 2021, 100, 100115. [Google Scholar] [CrossRef]
- Perlin, D.S.; Shor, E.; Zhao, Y. Update on Antifungal Drug Resistance. Curr. Clin. Microbiol. Rep. 2015, 2, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Effron, G. Molecular Markers of Antifungal Resistance: Potential Uses in Routine Practice and Future Perspectives. J. Fungi 2021, 7, 197. [Google Scholar] [CrossRef]
- Mortensen, K.L.; Jensen, R.H.; Johansen, H.K.; Skov, M.; Pressler, T.; Howard, S.J.; Leatherbarrow, H.; Mellado, E.; Arendrup, M.C. Aspergillus Species and Other Molds in Respiratory Samples from Patients with Cystic Fibrosis: A Laboratory-Based Study with Focus on Aspergillus fumigatus Azole Resistance. J. Clin. Microbiol. 2011, 49, 2243–2251. [Google Scholar] [CrossRef]
- Garcia-Rubio, R.; Cuenca-Estrella, M.; Mellado, E. Triazole Resistance in Aspergillus Species: An Emerging Problem. Drugs 2017, 77, 599–613. [Google Scholar] [CrossRef]
- Alcazar-Fuoli, L.; Mellado, E.; Garcia-Effron, G.; Lopez, J.F.; Grimalt, J.O.; Cuenca-Estrella, J.M.; Rodriguez-Tudela, J.L. Ergosterol biosynthesis pathway in Aspergillus fumigatus. Steroids 2007, 73, 339–347. [Google Scholar] [CrossRef]
- Geddes-McAlister, J.; Shapiro, R.S. New pathogens, new tricks: Emerging, drug-resistant fungal pathogens and future prospects for antifungal therapeutics. Ann. N. Y. Acad. Sci. 2019, 1435, 57–78. [Google Scholar] [CrossRef] [PubMed]
- Sienkiewicz, B.M.; Łapiński, Ł.; Wiela-Hojeńska, A. Comparison of clinical pharmacology of voriconazole and posaconazole. Contemp. Oncol. 2016, 20, 365. [Google Scholar] [CrossRef] [PubMed]
- Melo, S.; Pungartnik, C.; Cascardo, J.; Brendel, M. Rapid and efficient protocol for DNA extraction and molecular identification of the basidiomycete Crinipellis perniciosa. Genet. Mol. Res. 2006, 5, 851–855. [Google Scholar] [PubMed]
- Hinrikson, H.P.; Hurst, S.F.; De Aguirre, L.; Morrison, C.J. Molecular methods for the identification of Aspergillus species. Med. Mycol. Supp/. 2005, 43, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Melchers, W.J.G.; Verweij, P.; Van den Hurk, P.; Van Belkum, A.; De Pauw, B.; Hoogkamp-Korstanje, J.A.A.; Meis, J.F.G. Primer-Mediated PCR for Detection of Aspergillus species. J. Clin. Microbiol. 1994, 32, 1710–1717. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Macedo, D.; Devoto, T.B.; Pola, S.; Finquelievich, J.L.; Cuestas, M.L.; Garcia-Effron, G. A Novel Combination of CYP51A Mutations Confers Pan-Azole Resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 2020, 64, e02501-19. [Google Scholar] [CrossRef]
- Beardsley, J.; Halliday, C.L.; Chen, S.C.; Sorrell, T.C. Responding to the emergence of antifungal drug resistance: Perspectives from the bench and the bedside. Future Microbiol. 2018, 13, 1175–1191. [Google Scholar] [CrossRef]
- Colombo, A.L.; Júnior, J.N.D.A.; A Slavin, M.; Chen, S.C.-A.; Sorrell, T.C. Candida and invasive mould diseases in non-neutropenic critically ill patients and patients with haematological cancer. Lancet Infect. Dis. 2017, 17, e344–e356. [Google Scholar] [CrossRef]
- Borman, A.M.; Linton, C.J.; Miles, S.-J.; Johnson, E.M. Molecular identification of pathogenic fungi. J. Antimicrob. Chemother. 2008, 61, i7–i12. [Google Scholar] [CrossRef]
- MacLeod, N.; Canty, R.J.; Polaszek, A. Morphology-Based Identification of Bemisia tabaci Cryptic Species Puparia via Embedded Group-Contrast Convolution Neural Network Analysis. Syst. Biol. 2021, 71, 1095–1109. [Google Scholar] [CrossRef] [PubMed]
- Leekha, S.; Terrell, C.L.; Edson, R.S. General Principles of Antimicrobial Therapy. Mayo Clin. Proc. 2011, 86, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.D.; Denning, D.W.; Levitz, S.M. Tackling human fungal infections. Science 2012, 336, 647. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.C.; Hawkins, N.J.; Sanglard, D.; Gurr, S.J. Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science 2018, 360, 739–742. [Google Scholar] [CrossRef] [PubMed]
- Perlin, D.S.; Rautemaa-Richardson, R.; Alastruey-Izquierdo, A. The global problem of antifungal resistance: Prevalence, mechanisms, and management. Lancet Infect. Dis. 2017, 17, e383–e392. [Google Scholar] [CrossRef] [PubMed]
- Jenks, J.D.; Hoenigl, M. Treatment of aspergillosis. J. Fungi 2018, 4, 98. [Google Scholar] [CrossRef]
- van de Veerdonk, F.L.; Kolwijck, E.; Lestrade, P.P.A.; Hodiamont, C.J.; Rijnders, B.J.A.; van Paassen, J.; Haas, P.-J.; dos Santos, C.O.; Kampinga, G.A.; Bergmans, D.C.J.J.; et al. Influenza-associated Aspergillosis in Critically Ill Patients. Am. J. Respir. Crit. Care Med. 2017, 196, 524–527. [Google Scholar] [CrossRef]
- Araujo, R.; Rodrigues, A.G. Variability of Germinative Potential among Pathogenic Species of Aspergillus. J. Clin. Microbiol. 2004, 42, 4335–4337. [Google Scholar] [CrossRef]
- Berkow, E.L.; Nunnally, N.S.; Bandea, A.; Kuykendall, R.; Beer, K.; Lockhart, S.R. Detection of TR34/L98H CYP51A mutation through passive surveillance for azole-resistant Aspergillus fumigatus in the United States from 2015 to 2017. Antimicrob. Agents Chem. 2018, 62, e02240-17. [Google Scholar] [CrossRef]
- Sharpe, A.R.; Lagrou, K.; Meis, J.F.; Chowdhary, A.; Lockhart, S.R.; E Verweij, P.; on behalf of the ISHAM/ECMM Aspergillus Resistance Surveillance working group. Triazole resistance surveillance in Aspergillus fumigatus. Med. Mycol. 2018, 56, S83–S92. [Google Scholar] [CrossRef]
- Kaur, R.; Mehra, B.; Dhakad, M.S.; Goyal, R.; Dewan, R. Pulmonary aspergillosis as opportunistic mycoses in a cohort of human immunodeficiency virus-infected patients: Report from a tertiary care hospital in North India. Int. J. Health Sci. 2017, 11, 45–50. [Google Scholar]
- Arendrup, M.C.; Verweij, P.E.; Mouton, J.W.; Lagrou, K.; Meletiadis, J. Multicentre validation of 4-well azole agar plates as a screening method for detection of clinically relevant azole-resistant Aspergillus fumigatus. J. Antimicrob. Chemother. 2017, 72, 3325–3333. [Google Scholar] [CrossRef] [PubMed]
- Herbrecht, R.; Denning, D.W.; Patterson, T.F.; Bennett, J.E.; Greene, R.E.; Oestmann, J.-W.; Kern, W.V.; Marr, K.A.; Ribaud, P.; Lortholary, O.; et al. Voriconazole versus Amphotericin B for Primary Therapy of Invasive Aspergillosis. N. Engl. J. Med. 2002, 347, 408–415. [Google Scholar] [CrossRef] [PubMed]
- Verweij, P.E.; Snelders, E.; Kema, G.H.; Mellado, E.; Melchers, W.J. Azole resistance in Aspergillus fumigatus: A side-effect of environmental fungicide use? Lancet Infect. Dis. 2009, 9, 789–795. [Google Scholar] [CrossRef]
- Snelders, E.; Camps, S.M.T.; Karawajczyk, A.; Schaftenaar, G.; Kema, G.H.; Van Der Lee, H.A.; Klaassen, C.H.; Melchers, W.J.G.; Verweij, P.E. Triazole Fungicides Can Induce Cross-Resistance to Medical Triazoles in Aspergillus fumigatus. PLoS ONE 2012, 7, e31801. [Google Scholar] [CrossRef]
- Seyedmousavi, S.; Mouton, J.W.; Melchers, W.J.; Brüggemann, R.J.; Verweij, P.E. The role of azoles in the management of azole-resistant aspergillosis: From the bench to the bedside. Drug Resist. Updat. 2014, 17, 37–50. [Google Scholar] [CrossRef]
- Garcia-Rubio, R.; Gonzalez-Jimenez, I.; Lucio, J.; Mellado, E. Aspergillus fumigatus Cross-Resistance between Clinical and Demethylase Inhibitor Azole Drugs. Appl. Environ. Microbiol. 2021, 87, e02539-20. [Google Scholar] [CrossRef]
- Becher, R.; Wirsel, S.G.R. Fungal cytochrome P450 sterol 14α-demethylase (CYP51) and azole resistance in plant and human pathogens. Appl. Microbiol. Biotechnol. 2012, 95, 825–840. [Google Scholar] [CrossRef]
- Meis, J.F.; Chowdhary, A.; Rhodes, J.L.; Fisher, M.C.; Verweij, P.E. Clinical implications of globally emerging azole resistance in Aspergillus fumigatus. Philos. Trans. R Soc. B Biol. Sci. 2016, 371, 20150460. [Google Scholar] [CrossRef]
- Jeanvoine, A.; Rocchi, S.; Reboux, G.; Crini, N.; Crini, G.; Millon, L. Azole-resistant Aspergillus fumigatus in sawmills of Eastern France. J. Appl. Microbiol. 2017, 123, 172–184. [Google Scholar] [CrossRef]
- Revie, N.M.; Iyer, K.R.; Robbins, N.; Cowen, L.E. Antifungal drug resistance: Evolution, mechanisms and impact. Curr. Opin. Microbiol. 2018, 45, 70–76. [Google Scholar] [CrossRef]
- Robbins, N.; Caplan, T.; Cowen, L.E. Molecular Evolution of Antifungal Drug Resistance. Annu. Rev. Microbiol. 2017, 71, 753–775. [Google Scholar] [CrossRef] [PubMed]
- Wambaugh, M.A.; Denham, S.T.; Ayala, M.; Brammer, B.; Stonhill, M.A.; Brown, J.C. Synergistic and antagonistic drug interactions in the treatment of systemic fungal infections. Elife 2020, 9, e54160. [Google Scholar] [CrossRef] [PubMed]
- Vuddanda, P.R.; Chakraborty, S.; Singh, S. Berberine: A potential phytochemical with multispectrum therapeutic activities. Expert Opin. Investig. Drugs 2010, 19, 1297–1307. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wei, J.; Xue, R.; Wu, J.-D.; Zhao, W.; Wang, Z.-Z.; Wang, S.-K.; Zhou, Z.-X.; Song, D.-Q.; Wang, Y.-M.; et al. Berberine lowers blood glucose in type 2 diabetes mellitus patients through increasing insulin receptor expression. Metabolism 2010, 59, 285–292. [Google Scholar] [CrossRef]
- Powers, K.T.; Stevenson-Jones, F.; Yadav, S.K.N.; Amthor, B.; Bufton, J.C.; Borucu, U.; Shen, D.; Becker, J.P.; Lavysh, D.; Hentze, M.W.; et al. Blasticidin S inhibits mammalian translation and enhances production of protein encoded by nonsense mRNA. Nucleic Acids Res. 2021, 49, 7665–7679. [Google Scholar] [CrossRef]
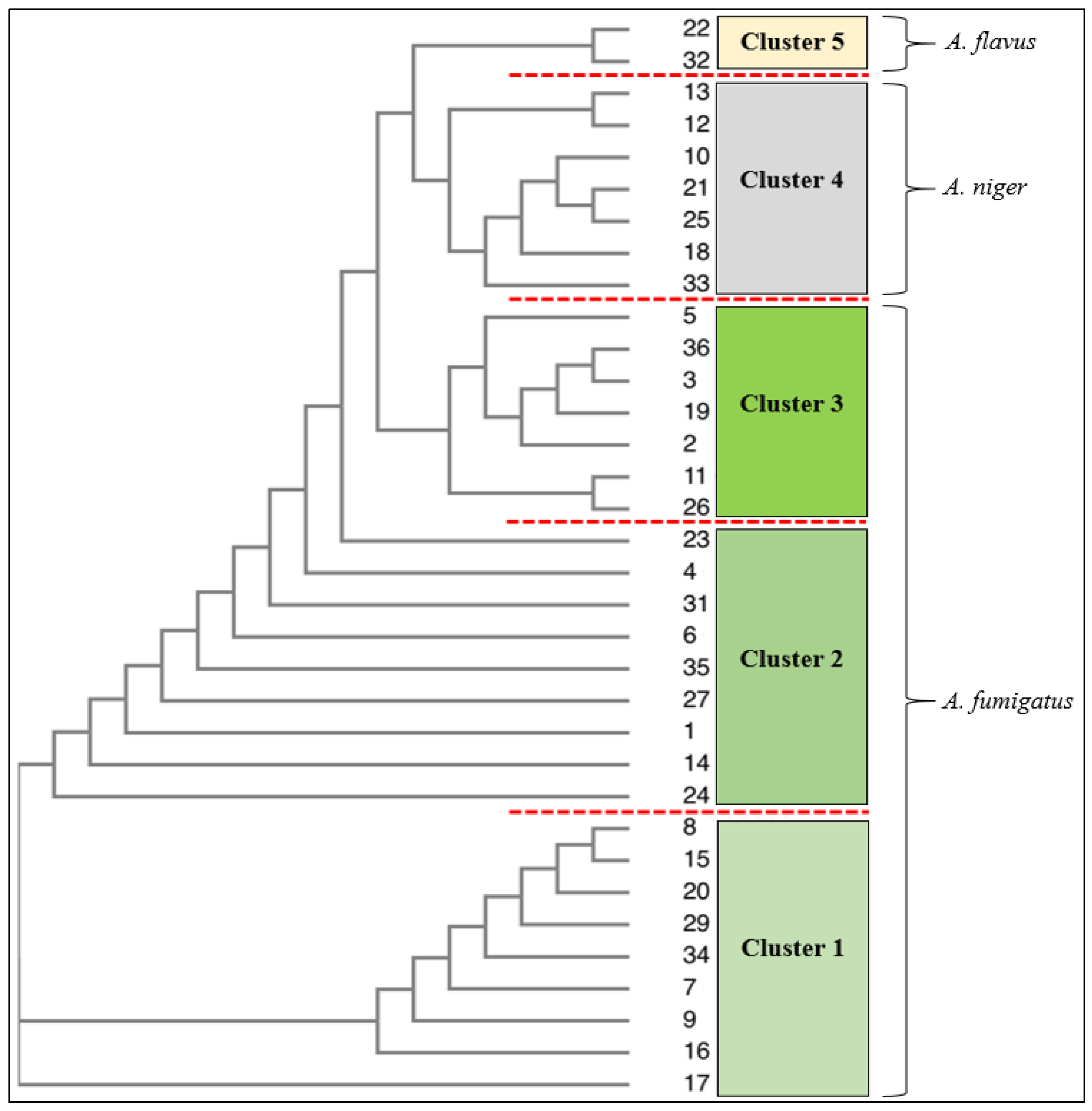


| Isolate | 18S rRNA/Microscope and Culture Techniques | Biolog Identification (48 h Incubation) | Similarity 1.0 = 100% |
|---|---|---|---|
| 1 | Aspergillus fumigatus | Neosartorya ficheri, Petromyces alliaceus, Aspergillus versicolor, Aspergillus clavatus, Aspergillus tamarii, Aspergillus parasiticus, | <0.1 |
| 2 | Aspergillus fumigatus | Neosartorya ficheri, Petromyces alliaceus, Aspergillus parasiticus, Aspergillus tamarii. | <0.1 |
| 3 | Aspergillus fumigatus | Neosartorya ficheri, Aspergillus tamarii, Aspergillus versicolor, Aspergillus puniceus, Petromyces alliaceus | <0.1 |
| 4 | Aspergillus fumigatus | Neosartorya ficheri, Petromyces alliaceus Aspergillus tamarii, Aspergillus versicolor, Aspergillus parasiticus | <0.1 |
| 5 | Aspergillus fumigatus | Neosartorya ficheri, Petromyces alliaceus Aspergillus tamarii, Aspergillus versicolor, Aspergillus parasiticus | <0.1 |
| 6 | Aspergillus fumigatus | Neosartorya ficheri, Petromyces alliaceus, Aspergillus puniceus, Aspergillus tamarii, Aspergillus parasiticus | <0.1 |
| 7 | Aspergillus fumigatus | Neosartorya ficheri, Petromyces alliaceus, Aspergillus tamarii, Aspergillus parasiticus, Aspergillus puniceus | <0.1 |
| 8 | Aspergillus fumigatus | Neosartorya ficheri, Petromyces alliaceus, Aspergillus tamarii, Aspergillus aureolatus, Aspergillus versicolor | <0.1 |
| 9 | Aspergillus fumigatus | Aspergillus parasiticus, Petromyces alliaceus, Neosartorya ficheri, Aspergillus tamarii, Chaetosartorya stomatoides. | <0.1 |
| 10 | Aspergillus niger | Aspergillus niger, Chaetosartorya stomatoides, Aspergillus ustus, Emericella variecola, Neosartorya ficheri | <0.1 A. niger = 0.006 |
| 11 | Aspergillus fumigatus | Petromyces alliaceus, Neosartorya ficheri, Aspergillus tamarii, Aspergillus parasiticus | <0.1 |
| 12 | Aspergillus niger/tubingensis | Aspergillus niger, Neosartorya ficheri,Aspergillus parasiticus, Aspergillus ustus, Chaetosartorya stomatoides, Aspergillus foetidus, Emericella variecola | <0.1 A. niger = 0.086 C. stromatoides = 0.138 |
| 13 | Aspergillus niger/tubingensis | Emericella striata, Emericella fruticulosa, Aspergillus wentii, Aspergillus phoenicis, Aspergillus brasiliensis, Emericella violacea, Aspergillus zonatus | <0.1 |
| 14 | Aspergillus fumigatus | Petromyces alliaceus, Neosartorya ficheri, Aspergillus tamarii, Aspergillus parasiticus, Aspergillus terricola, Aspergillus flavus | <0.1 A. terricola < 0.349 |
| 15 | Aspergillus fumigatus | Aspergillus terricola, Aspergillus flavus, Aspergillus pulverulentus, Chaetosartorya stomatoides, Aspergillus sydowii | <0.1 A. terricola = 0.565 |
| 16 | Aspergillus fumigatus | Aspergillus terricola, Aspergillus flavus, Aspergillus pulverulentus, Neosartorya ficheri, Aspergillus sydowii | <0.1 A. terricola = 0.371 |
| 17 | Aspergillus fumigatus | Aspergillus terricola, Aspergillus flavus, Neosartorya ficheri, Aspergillus sydowii | <0.1 A. terricola = 0.514 |
| 18 | Aspergillus niger | Aspergillus brasiliensis, Aspergillus kanagawaensis, Emericella striata, Aspergillus restrictus, Aspergillus zonatus, Emericella violacea | <0.1 |
| 19 | Aspergillus fumigatus | Aspergillus terricola, Aspergillus flavus, Neosartorya ficheri, Aspergillus versicolor, Chaetosartorya stomatoides | <0.1 A. terricola = 0.137 |
| 20 | Aspergillus fumigatus | Aspergillus puniceus, Chaetosartorya stomatoides, Aspergillus ustus, Aspergillus terricola, Aspergillus flavus, Neosartorya ficheri | <0.1 A. puniceus = 0.167 |
| 21 | Aspergillus niger/welwitschiae | Aspergillus foetidus, Emericella violacea, Aspergillus niger, Aspergillus brasiliensis, Emericella striata, Aspergillus ochraceus, Aspergillus carneus | <0.1 A. foetidus = 0.130 A. niger = 0.003 |
| 22 | Aspergillus flavus | Aspergillus terricola, Aspergillus flavus, Neosartorya ficheri, Aspergillus pulverulentus, | <0.1 A. terricola = 0.148 A. flavus = 0.002 |
| 23 | Aspergillus fumigatus | Aspergillus terricola, Aspergillus flavus, Chaetosartorya stomatoides, Aspergillus puniceus, Neosartorya ficheri, Aspergillus versicolor | <0.1 A. terricola =0.407 |
| 24 | Aspergillus fumigatus | Aspergillus terricola, Aspergillus flavus, Neosartorya ficheri, Aspergillus puniceus, Chaetosartorya stomatoides, | <0.1 N. ficheri = 0.186 |
| 25 | Aspergillus niger | Aspergillus foetidus, Aspergillus niger, Emericella violacea, Aspergillus wenti, Aaspergillus brevipes, Aspergillus carbonarius, Aspergillus brasiliensis, Chaetosartorya stomatoides | <0.1 A. foetidus = 0.116 A. brevipes = 0.145 A. niger = 0.015 |
| 26 | Aspergillus fumigatus | Aspergillus flavus, Neosartorya ficheri, Aspergillus terricola, Chaetosartorya stomatoides, Neosartorya ficheri | <0.1 A. flavus = 0.350 A. terricola = 0.205 |
| 27 | Aspergillus fumigatus | Aspergillus terricola, Aspergillus flavus, Aspergillus puniceus, Aspergillus versicolor, Neosartorya ficheri, Aspergillus pulverulentus | <0.1 A. terricola = 0.240 |
| 28 | Aspergillus fumigatus | Aspergillus terricola, Aspergillus flavus, Aspergillus versicolor, Aspergillus pulverulentus, Neosartorya ficheri | <0.1 A. terricola =0.332 |
| 29 | Aspergillus fumigatus | Aspergillus terricola, Aspergillus flavus, Aspergillus versicolor, Neosartorya ficheri, Aspergillus pulverulentus | <0.1 A. terricola = 0.238 |
| 30 | Aspergillus niger | Aspergillus niger, Emericella violacea, Aspergillus carbonarius, Aspergillus foetidus, Aspergillus brasiliensis, Emericella striata, Aspergillus zonatus, Aspergillus carbonarius | <0.1 A. niger = 0.008 |
| 31 | Aspergillus fumigatus | Aspergillus terricola, Aspergillus puniceus, Chaetosartorya stomatoides, Aspergillus flavus, Aspergillus ustus | <0.1 A. terricola =0.296 |
| 32 | Aspergillus flavus | Aspergillus terricola, Aspergillus flavus, Neosartorya ficheri, Aspergillus pulverulentus, Aspergillus puniceus, | <0.1 A. terricola = 0.347 A. flavus = 0.000 |
| 33 | Aspergillus niger | Aspergillus ustus, Aspergillus versicolor, Emericella violacea, Aspergillus sydowii, Aspergillus caesipitosus, Aspergillus foetidus, | <0.1 A. caespitosus = 0.187 |
| 34 | Aspergillus fumigatus | Aspergillus terricola, Aspergillus flavus, Aspergillus puniceus, Neosartorya ficheri | <0.1 A. flavus = 0.162 A. terricola =0.152 |
| 35 | Aspergillus fumigatus | Aspergillus terricola, Aspergillus flavus, Aspergillus pulverulentus, Chaetosartorya stomatoides | <0.1 A. terricola = 0.472 |
| 36 | Aspergillus fumigatus | Aspergillus puniceus, Neosartorya ficheri, Aspergillus ustus, Aspergillus flavus, Chaetosartorya stomatoides, Aspergillus fumigatus | <0.1 A. puniceus = 0.227 N. ficheri = 0.247 A. fumigatus = 0.000 |
| Voriconazole | Posaconazole | ||||
|---|---|---|---|---|---|
| Isolate No. | Species | Lowest Effective Concentration (µg/mL) | Concentration (µg/mL) for the Largest Inhibition Zone (mm) | Lowest Effective Concentration (µg/mL) | Concentration (µg/mL) for the Largest Inhibition Zone (mm) |
| 1 | A. fumigatus | 0.19 | 12.0–29 | 16.0 | 24.0–10 |
| 2 | A. fumigatus | 0.38 | 12.0–30 | 16.0 | 24.0–10 |
| 3 | A. fumigatus | 0.38 | 12.0–31 | 12.0 | 24.0–11 |
| 4 | A. fumigatus | 0.25 | 16.0–34 | 12.0 | 24.0–12 |
| 5 | A. fumigatus | 0.19 | 16.0–34 | 12.0 | 24.0–12 |
| 6 | A. fumigatus | 0.38 | 12.0–28 | 12.0 | 24-0–13 |
| 7 | A. fumigatus | 0.38 | 12.0–27 | 12.0 | 24.0–13 |
| 8 | A. fumigatus | 0.50 | 12.0–28 | 16.0 | 24.0–12 |
| 9 | A. fumigatus | 0.38 | 12.0–29 | 12.0 | 24.0–12 |
| 11 | A. fumigatus | 0.38 | 12.0–31 | 12.0 | 24.0–17 |
| 14 | A. fumigatus | 0.25 | 12.0–29 | 12.0 | 24.0–15 |
| 15 | A. fumigatus | 0.50 | 12.0–28 | 8.0 | 16.0–13 |
| 16 | A. fumigatus | 0.38 | 12.0–29 | 8.0 | 24.0–17 |
| 17 | A. fumigatus | 0.19 | 12.0–32 | 12.0 | 24.0–12 |
| 19 | A. fumigatus | 0.38 | 12.0–30 | 6.0 | 16.0–15 |
| 20 | A. fumigatus | 0.25 | 12.0–29 | 8.0 | 24.0–17 |
| 23 | A. fumigatus | 0.50 | 12.0–38 | 8.0 | 16.0–16 |
| 24 | A. fumigatus | 0.25 | 8..0–29 | 8.0 | 16.0–16 |
| 26 | A. fumigatus | 0.50 | 23.0–28 | 6.0 | 16.0–17 |
| 27 | A. fumigatus | 0.75 | 12.0–29 | 8.0 | 24.0–16 |
| 28 | A. fumigatus | 0.25 | 12.0–30 | 8.0 | 16.0–12 |
| 29 | A. fumigatus | 0.25 | 12.0–32 | 7.5 | 12.0–24 |
| 31 | A. fumigatus | 0.25 | 12.0–32 | 8.0 | 24.0–19 |
| 34 | A. fumigatus | 0.38 | 16.0–34 | 6.0 | 16.0–17 |
| 35 | A. fumigatus | 1.5 | 16.0–31 | 8.0 | 24.0–15 |
| 36 | A. fumigatus | 0.19 | 12.0–30 | 1.0 | 16.0–21 |
| 10 | Aspergillus niger | 0.19 | 12.0–39 | 8.0 | 16.0–13 |
| 12 | Aspergillus niger/tubingensis | 2.0 | 16.0–16 | 12.0 | 32.0–16 |
| 13 | Aspergillus niger/tubingensis | 1.5 | 12.0–21 | 12.0 | 16.0–13 |
| 18 | Aspergillus niger | 1.0 | 12.0–34 | 8.0 | 16.0–12 |
| 21 | Aspergillus niger/welwitschiae | 1.0 | 8.0–40 | 8.0 | 16.0–14 |
| 25 | Aspergillus niger | 1.0 | 8.0–39 | 8.0 | 24.0–14 |
| 30 | Aspergillus niger | 2.0 | 12.0–39 | 8.0 | 24.0–17 |
| 33 | Aspergillus niger | 1.5 | 8.0–43 | 12.0 | 34.0–13 |
| 22 | Aspergillus flavus | 0.5 | 8.0–39 | 6.0 | 24.0–21 |
| 32 | Aspergillus flavus | 1.5 | 12.0–31 | 6.0 | 24.0–18 |
| Isolate No. | Species | Resistant (R)/Susceptible (S)/ Intermediate Susceptibility (IS) | cyp51A Mutations | |
|---|---|---|---|---|
| VRC | POS | |||
| 1 | A. fumigatus | S | R | WT |
| 2 | A. fumigatus | IS | R | L98H |
| 3 | A. fumigatus | IS | R | L98H |
| 4 | A. fumigatus | S | R | L98H |
| 5 | A. fumigatus | S | R | WT |
| 6 | A. fumigatus | IS | R | L98H |
| 7 | A. fumigatus | IS | R | L98H |
| 8 | A. fumigatus | IS | R | WT |
| 9 | A. fumigatus | IS | R | L98H |
| 10 | A. niger | S | R | ND |
| 11 | A. fumigatus | IS | R | WT |
| 12 | A. niger | R | R | ND |
| 13 | A. niger | IS | R | ND |
| 14 | A. fumigatus | S | R | WT |
| 15 | A. fumigatus | IS | R | L98H |
| 16 | A. fumigatus | IS | R | WT |
| 17 | A. fumigatus | S | R | WT |
| 18 | A. niger | IS | R | ND |
| 19 | A. fumigatus | IS | R | WT |
| 20 | A. fumigatus | S | R | L98H |
| 21 | A. niger | IS | R | ND |
| 22 | A. flavus | IS | R | ND |
| 23 | A. fumigatus | IS | R | WT |
| 24 | A. fumigatus | S | R | L98H |
| 25 | A. niger | IS | R | ND |
| 26 | A. fumigatus | IS | R | WT |
| 27 | A. fumigatus | IS | R | L98H |
| 28 | A. fumigatus | S | R | WT |
| 29 | A. fumigatus | S | R | WT |
| 30 | A. niger | R | R | ND |
| 31 | A. fumigatus | S | R | WT |
| 32 | A. flavus | IS | R | ND |
| 33 | A. fumigatus | IS | R | WT |
| 34 | A. fumigatus | IS | R | L98H |
| 35 | A. fumigatus | IS | R | WT |
| 36 | A. fumigatus | S | R | WT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naicker, S.; Mohanlall, V.; Ngubane, S.; Mellem, J.; Mchunu, N.P. Phenotypic Array for Identification and Screening of Antifungals against Aspergillus Isolates from Respiratory Infections in KwaZulu Natal, South Africa. J. Fungi 2023, 9, 616. https://doi.org/10.3390/jof9060616
Naicker S, Mohanlall V, Ngubane S, Mellem J, Mchunu NP. Phenotypic Array for Identification and Screening of Antifungals against Aspergillus Isolates from Respiratory Infections in KwaZulu Natal, South Africa. Journal of Fungi. 2023; 9(6):616. https://doi.org/10.3390/jof9060616
Chicago/Turabian StyleNaicker, Sarla, Viresh Mohanlall, Sandile Ngubane, John Mellem, and Nokuthula Peace Mchunu. 2023. "Phenotypic Array for Identification and Screening of Antifungals against Aspergillus Isolates from Respiratory Infections in KwaZulu Natal, South Africa" Journal of Fungi 9, no. 6: 616. https://doi.org/10.3390/jof9060616
APA StyleNaicker, S., Mohanlall, V., Ngubane, S., Mellem, J., & Mchunu, N. P. (2023). Phenotypic Array for Identification and Screening of Antifungals against Aspergillus Isolates from Respiratory Infections in KwaZulu Natal, South Africa. Journal of Fungi, 9(6), 616. https://doi.org/10.3390/jof9060616





