Bioclimatic Origin Shapes Phylogenetic Structure of Tirmania (Pezizaceae): New Species and New Record from North Africa
Abstract
:1. Introduction
2. Materials and Methods
2.1. Field-Collected Fungal Specimens
2.2. Morphological Characterization
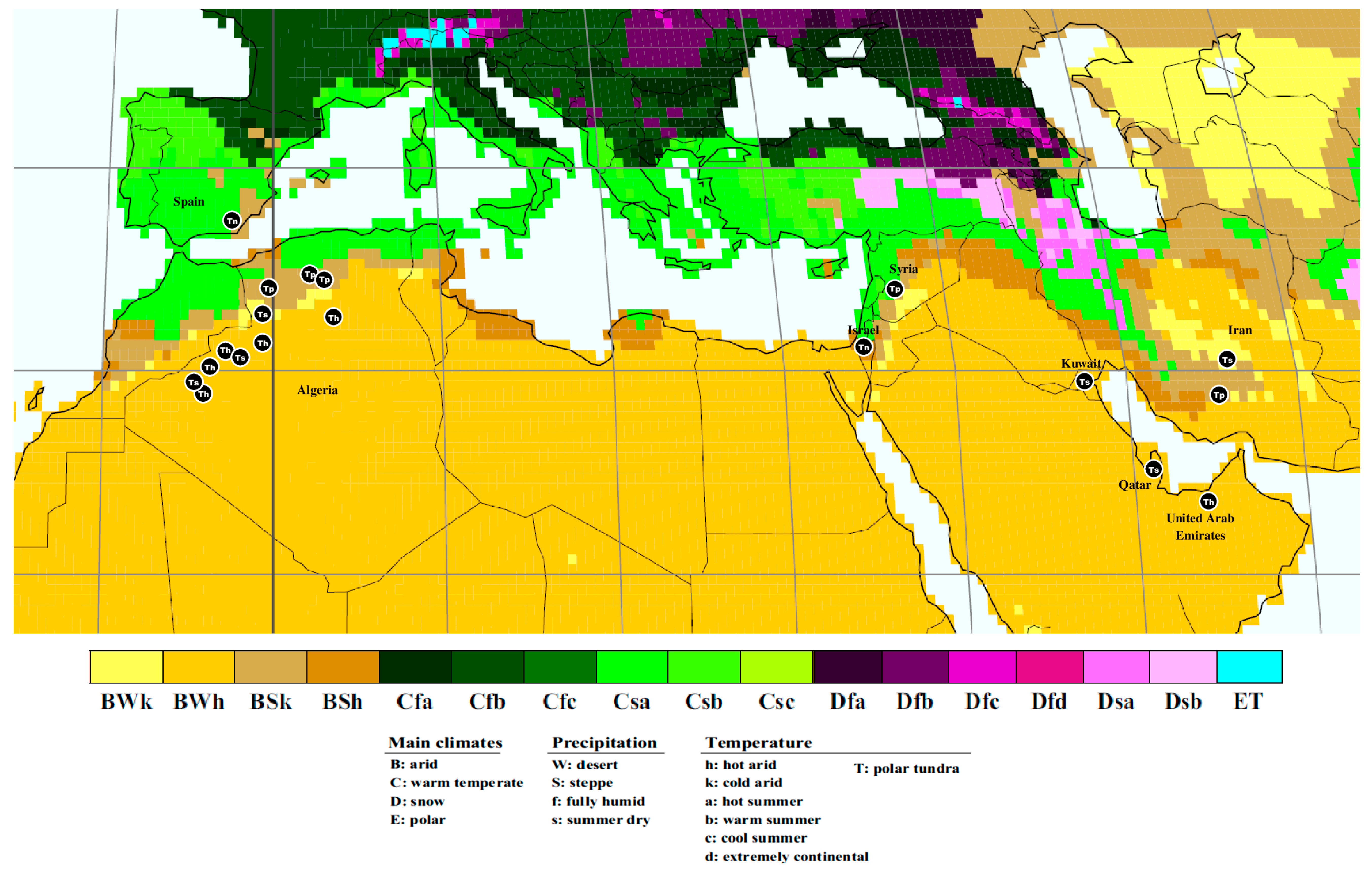
| Species | Voucher Specimen | Country of Origin | Locality | Host Plant | Bioclimatic Zone (Köppen–Geiger Climate Classification) | GenBank Accession IDs | |
|---|---|---|---|---|---|---|---|
| ITS | 28S LSU | ||||||
| Tirmania pinoyi lineage | |||||||
| Tirmania pinoyi | AH 49194 | Algeria | Naama, Mecheria, El Biodh | Helianthemum sp. | BSK | OM845235 | OM845236 |
| Tirmania pinoyi | TM2 | Algeria | Naama, Mecheria, El Biodh | Helianthemum sp. | BSK | OM845238 | OM854800 |
| Tirmania pinoyi | TM3 | Algeria | Naama, Mecheria, El Biodh | Helianthemum sp. | BSK | OM845239 | OM845241 |
| Tirmania pinoyi | AH 49192 | Algeria | Naama, Mecheria, Bougarne | Helianthemum sp. | BSK | OM845245 | N/A |
| Tirmania pinoyi | TM5 | Algeria | Naama, Mecheria, Bougarne | Helianthemum sp. | BSK | OM845247 | OM978172 |
| Tirmania pinoyi | TM6 | Algeria | Naama, Mecheria, Bougarne | Helianthemum sp. | BSK | OM978174 | OM978175 |
| Tirmania pinoyi | TM14 | Algeria | Tiaret, Ksar Chellala, Zmalet El-Amir Abdelkader, Bouchouat | Helianthemum hirtum, Helianthemum salicifolium | BSK | OM985023 | OM978180 |
| Tirmania pinoyi | TM15 | Algeria | Tiaret, Ksar Chellala, Zmalet El-Amir Abdelkader, Bouchouat | Helianthemum hirtum, Helianthemum salicifolium | BSK | OM978182 | OM978183 |
| Tirmania pinoyi | TM16 | Algeria | Tiaret, Ksar Chellala, Zmalet El-Amir Abdelkader, Bouchouat | Helianthemum hirtum, Helianthemum salicifolium | BSK | OM978184 | N/A |
| Tirmania pinoyi | TM17 | Algeria | Tiaret, Ksar Chellala, Zmalet El-Amir Abdelkader, Bouchouat | Helianthemum hirtum, Helianthemum salicifolium | BSK | OM978185 | N/A |
| Tirmania pinoyi | TM18 | Algeria | Tiaret, Ksar Chellala, Zmalet El-Amir Abdelkader, Bouchouat | Helianthemum hirtum Helianthemum salicifolium | BSK | OM985024 | OM978187 |
| Tirmania pinoyi | AH 49197 | Algeria | Tiaret, Ksar Chellala, Zmalet El-Amir Abdelkader, Bouchouat | Helianthemum hirtum Helianthemum salicifolium | BSK | OM854801 | OM845246 |
| Tirmania pinoyi | TM20 | Algeria | Tiaret, Hamadia, Rechaiga, Benhamed | Helianthemum hirtum | BSK | OM978186 | N/A |
| Tirmania pinoyi | TM21 | Algeria | Tiaret, Hamadia, Rechaiga, Benhamed | Helianthemum hirtum | BSK | OM985024 | N/A |
| Tirmania pinoyi | TM22 | Algeria | Tiaret, Hamadia, Rechaiga, Benhamed | Helianthemum hirtum | BSK | OM978189 | OM978191 |
| Tirmania pinoyi | TM24 | Algeria | Tiaret, Hamadia, Rechaiga, Benhamed | Helianthemum hirtum | BSK | OM978190 | OM978192 |
| Tirmania pinoyi | TM25 | Algeria | Tiaret, Hamadia, Rechaiga, Benhamed | Helianthemum hirtum | BSK | OM985026 | OM978193 |
| Tirmania pinoyi | TM26 | Algeria | Tiaret, Hamadia, Rechaiga, Benhamed | Helianthemum hirtum | BSK | OM985027 | OM985030 |
| Tirmania pinoyi | TM30 | Algeria | Tiaret, Hamadia, Rechaiga, Benhamed | Helianthemum hirtum | BSK | OM978207 | N/A |
| Tirmania pinoyi | Bd1 | Iran | Hormozgan, Bishederaz | Helianthemum salicifolium | BSK | GQ888697 * | N/A |
| Tirmania pinoyi | j601 | Syria | Damasco | N/F | BSK | MG920185 * | N/A |
| Tirmania pinoyi | STBD1 | Iran | Hormozgan, Bishederaz | Helianthemum salicifolium | BSK | HM352549 * | N/A |
| Tirmania honrubiae lineage | |||||||
| Tirmania honrubiae | TM34 | Algeria | Béchar, Béni Ounif, Oued Namous | Helianthemum lippii | BWH | OM978206 | OM978218 |
| Tirmania honrubiae | TM35 | Algeria | Béchar, Béni Ounif, Oued Namous | Helianthemum lippii | BWH | OM978219 | OP871363 |
| Tirmania honrubiae | AH 49191 | Algeria | Béchar, Béni Ounif, Oued Namous | Helianthemum lippii | BWH | OM985039 | OM985040 |
| Tirmania honrubiae | TM37 | Algeria | Béchar, Abadla, Hammaguir | Helianthemum lippii | BWH | OM985028 | N/A |
| Tirmania honrubiae | AH 49316 | Algeria | Béchar, Abadla, Hammaguir | Helianthemum lippii | BWH | OM985029 | N/A |
| Tirmania honrubiae | TM39 | Algeria | Béchar, Abadla, Hammaguir | Helianthemum lippii | BWH | OM985031 | OM978232 |
| Tirmania honrubiae | AH 49195 | Algeria | Béchar, Abadla, Hammaguir | Helianthemum lippii | BWH | OM985036 | OM985035 |
| Tirmania honrubiae | AH 49317 | Algeria | Béchar, Abadla, Hammaguir | Helianthemum lippii | BWH | OM985032 | N/A |
| Tirmania honrubiae | TM42 | Algeria | Béchar, Abadla, Oglat Braber | Helianthemum lippii | BWH | OM985034 | OM978234 |
| Tirmania honrubiae | AH 49319 | Algeria | Béchar, Abadla, Oglat Braber | Helianthemum lippii | BWH | OM985037 | OM978235 |
| Tirmania honrubiae | AH 49193 | Algeria | Béchar, Abadla, Oglat Braber | Helianthemum lippii | BWH | OM985038 | N/A |
| Tirmania honrubiae | TM45 | Algeria | Béchar, Abadla, Oglat Braber | Helianthemum lippii | BWH | OM985033 | N/A |
| Tirmania honrubiae | AH 49318 | Algeria | Béchar, Abadla, Oglat Braber | Helianthemum lippii | BWH | OM978236 | N/A |
| Tirmania honrubiae | TM47 | Algeria | Béni Abbès, Boulaadam | Helianthemum lippii | BWH | OP871121 | OP871282 |
| Tirmania honrubiae | TM48 | Algeria | Béni Abbès, Boulaadam | Helianthemum lippii | BWH | OM978238 | N/A |
| Tirmania honrubiae | TM49 | Algeria | Laghouat, Hassi R’Mel | Helianthemum sp. | BWH | OP871283 | OP871360 |
| Tirmania honrubiae | TM50 | Algeria | Laghouat, Hassi R’Mel | Helianthemum sp. | BWH | OP871359 | OP871362 |
| Tirmania honrubiae | j286 | United Arab Emirates | Abu Dhabi, Ghantoot | Helianthemum lippii | BWH | MG949283 * | N/A |
| Tirmania honrubiae | j294 | United Arab Emirates | Abu Dhabi, Ghantoot | Helianthemum lippii | BWH | MG949284 * | N/A |
| Tirmania honrubiae | j297 | United Arab Emirates | Abu Dhabi, Ghantoot | Helianthemum lippii | BWH | MG949282 * | N/A |
| Tirmania honrubiae | j299 | United Arab Emirates | Abu Dhabi, Ghantoot | Helianthemum lippii | BWH | MG949287 * | N/A |
| Tirmania honrubiae | j304 | United Arab Emirates | Abu Dhabi, Ghantoot | Helianthemum lippii | BWH | MG949285 * | N/A |
| Tirmania honrubiae | j344 | United Arab Emirates | Abu Dhabi, Seih Sadira | Helianthemum lippii | BWH | MG949288 * | N/A |
| Tirmania honrubiae | j359 | United Arab Emirates | Abu Dhabi, Ghantoot | Helianthemum lippii | BWH | MG949289 * | N/A |
| Tirmania honrubiae | j361 | United Arab Emirates | Abu Dhabi, Ghantoot | Helianthemum lippii | BWH | MG949286 * | N/A |
| Tirmania nivea lineage | |||||||
| Tirmania nivea | MA Fungi 37352 | Spain | Almeria, Tabernas Desert | Probably Helianthemum almeriense | BSK | OM978320 | OM978321 |
| Tirmania nivea | tab04 | Spain | Almeria, Tabernas Desert | N/F | BSK | AF276666 * | N/A |
| Tirmania nivea | 17084 | Israel | Ze’elim | N/F | BSH | JF908770 * | N/A |
| Tirmania sahariensis sp. nov. lineage | |||||||
| Tirmania sahariensis | TM9 | Algeria | Naama, Ain Sefra | Helianthemum lippii | BWK | OM985805 | OM985806 |
| Tirmania sahariensis | TM10 | Algeria | Naama, Ain Sefra | Helianthemum lippii | BWK | OM985804 | OM985791 |
| Tirmania sahariensis | AH 49198 | Algeria | Naama, Ain Sefra | Helianthemum lippii | BWK | OM985893 | OM985892 |
| Tirmania sahariensis | AH 49196 | Algeria | Naama, Ain Sefra | Helianthemum lippii | BWK | OM985807 | N/A |
| Tirmania sahariensis | TM13 | Algeria | Naama, Ain Sefra | Helianthemum lippii | BWK | OM985808 | N/A |
| Tirmania sahariensis | TM31 | Algeria | Béni Abbès, Tabelbala, Oued Daoura | Helianthemum lippii | BWH | OM985790 | N/A |
| Tirmania sahariensis | TM32 | Algeria | Béni Abbès, Tabelbala, Oued Daoura | Helianthemum lippii | BWH | OM985792 | N/A |
| Tirmania sahariensis | TM33 | Algeria | Béchar, Abadla, Hammaguir, Erg Serhen | Helianthemum lippii | BWH | OM985793 | N/A |
| Tirmania nivea | FaqZ1 | Qatar | N/F | N/F | BWH | KJ947347 * | N/A |
| Tirmania nivea | FaqZ2 | Qatar | N/F | N/F | BWH | KJ947357 * | N/A |
| Tirmania nivea | niv05 | Kuwait | N/F | Helianthemum salicifolium | BWH | AF276668 * | N/A |
| Tirmania nivea | Si2 | Iran | Kerman, Baghaat, Sirjan | Helianthemum salicifolium | BWK | FJ197820 * | N/A |
2.3. Genomic DNA Extraction, PCR Amplification, and Sequencing of Ribosomal DNA
2.4. Molecular Phylogeny Construction
3. Results
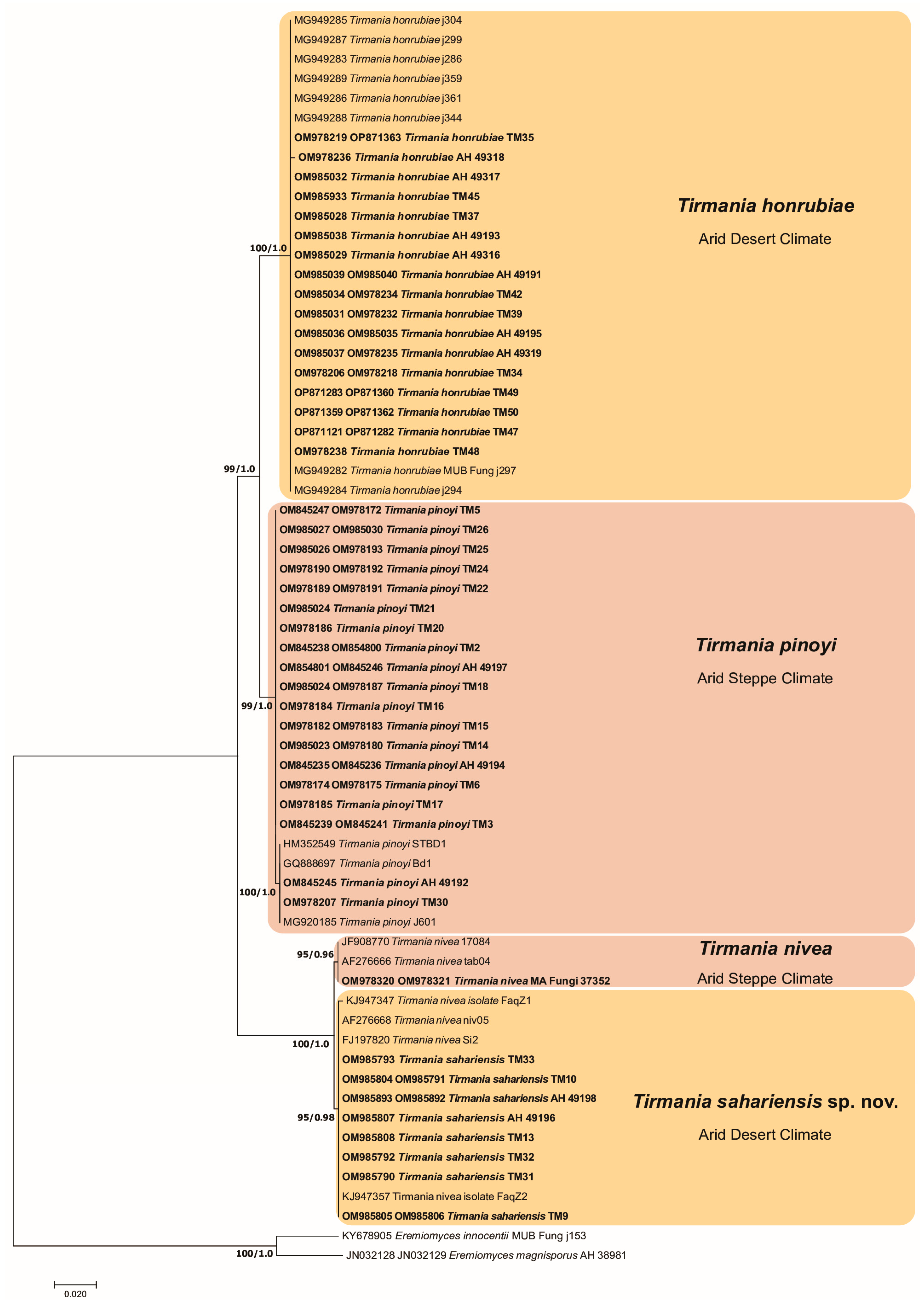
3.1. Phylogenetic Analysis
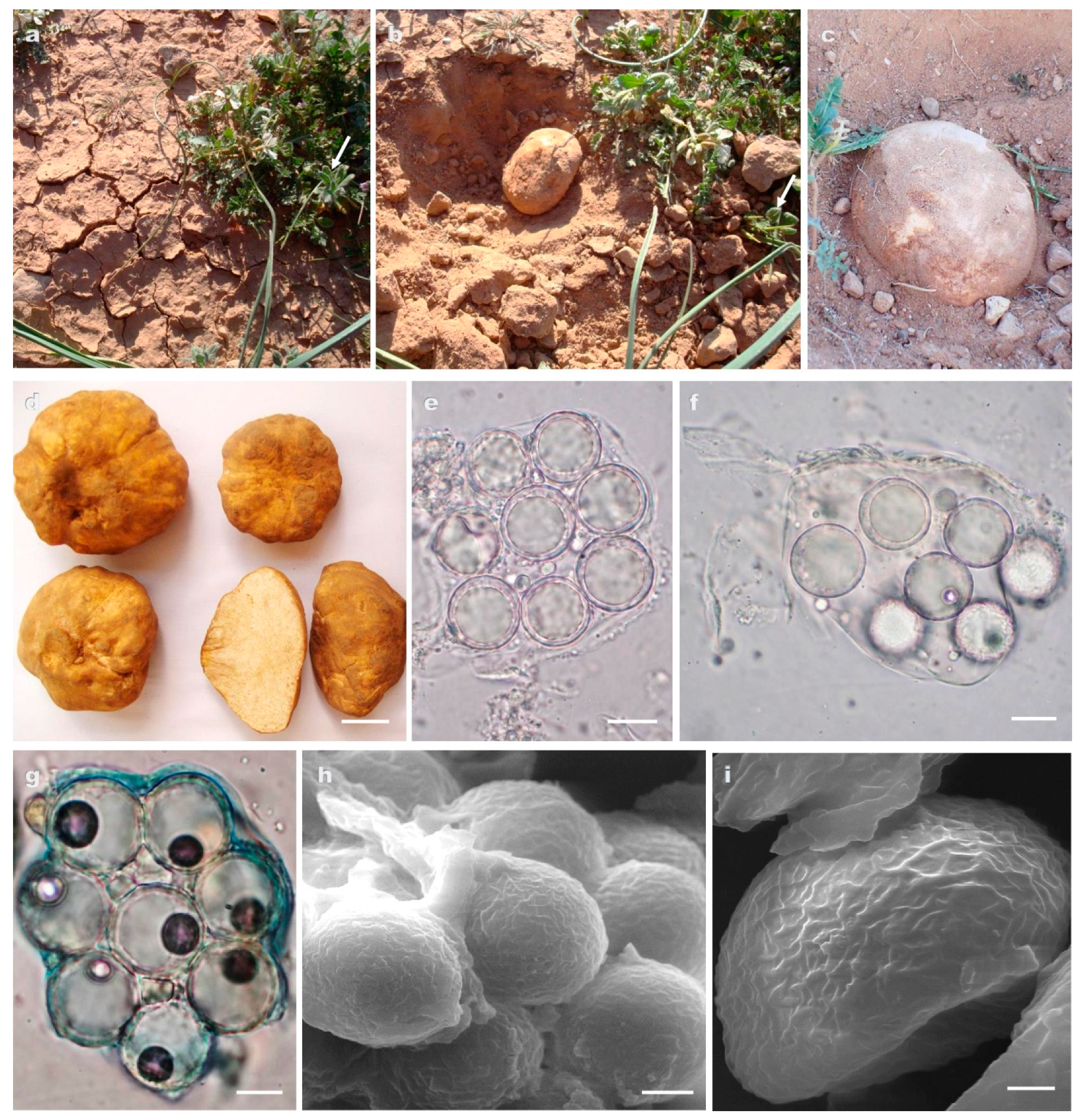
3.2. Taxonomy
- Peridium orangish yellow to pale brown–orange, spores globose roughened with ridges and low rounded warts……..………………………………… T. honrubiae (clade 1)
- Peridium light brown or yellowish brown, spores globose roughened with a not well-defined reticulum………………………………………………………T. pinoyi (clade 2)
- Peridium off white or yellow to cinnamon, spores smooth exclusively ellipsoid, asci (4–6)–8 spored …………………………………………………………………T. nivea (clade 3)
- Peridium off white or white yellowish, spores smooth ellipsoid or globose, asci 6–(8–10) spored..……………………………………………………………T. sahariensis (clade 4)
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alsheikh, A.M.; Trappe, J.M. Desert truffles: The genus Tirmania. Trans. Br. Mycol. Soc. 1983, 81, 83–90. [Google Scholar] [CrossRef]
- Alsheikh, A.M. Taxonomy and Mycorrhizal Ecology of the Desert Truffles in the Genus Terfezia. Ph.D. Thesis, Oregon State University, Corvallis, OR, USA, 1994; 239p. [Google Scholar]
- Díez, J.; Manjón, J.L.; Martin, F. Molecular phylogeny of the mycorrhizal desert truffles (Terfezia and Tirmania), host specificity and edaphic tolerance. Mycologia 2002, 94, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Zitouni-Haouar, F.E.-H.; Fortas, Z.; Chevalier, G. Morphological characterization of mycorrhizae formed between three Terfezia species (desert truffles) and several Cistaceae and Aleppo pine. Mycorrhiza 2014, 24, 397–403. [Google Scholar] [CrossRef]
- Zitouni-Haouar, F.E.-H.; Carlavilla, J.R.; Moreno, G.; Manjon, J.L.; Fortas, Z. Genetic diversity of the genus Terfezia (Pezizaceae, Pezizales): New species and new record from North Africa. Phytotaxa 2018, 334, 183–194. [Google Scholar] [CrossRef]
- Morte, A.; Gutiérrez, A.; Navarro-Ródenas, A. Advances in desert truffle mycorrhization and cultivation. In Mushrooms, Humans and Nature in a Changing World; Springer: Cham, Switzerland, 2020; pp. 205–219. [Google Scholar]
- Morte, A.; Kagan-Zur, V.; Navarro-Ródenas, A.; Sitrit, Y. Cultivation of desert truffles—A crop suitable for arid and semi-arid zones. Agronomy 2021, 11, 1462. [Google Scholar] [CrossRef]
- Chatin, A. Contribution à l′histoire naturelle de la truffe. Bull. Société Bot. Fr. 1891, 38, 54–64. [Google Scholar] [CrossRef]
- Chatin, A. La Truffe; J.B. Baillière et Fils: Paris, France, 1892. [Google Scholar]
- Patouillard, N. Énumération des champignons observés en Tunisie. In Exploration Scientifique de la Tunisie; Imprimerie Nationale: Paris, France, 1892. [Google Scholar]
- Trappe, J.M. A synopsis of the Carbomycetaceae and Terfeziaceae (Tuberales). Trans. Br. Mycol. Soc. 1971, 57, 85–92. [Google Scholar] [CrossRef]
- Trappe, J.M. The orders, families and genera of hypogeous ascomycotina (truffles and their relatives). Mycotaxon 1979, 9, 297–340. [Google Scholar]
- Hansen, K.; Læssøe, T.; Pfister, D.H. Phylogenetics of the Pezizaceae, with an emphasis on Peziza. Mycologia 2001, 93, 958–990. [Google Scholar] [CrossRef]
- Hansen, K.; Lobuglio, K.F.; Pfister, D.H. Evolutionary relationships of the cup-fungus genus Peziza and Pezizaceae inferred from multiple nuclear genes: RPB2, β-tubulin, and LSU rDNA. Mol. Phylogenet. Evol. 2005, 36, 1–23. [Google Scholar] [CrossRef]
- Ferdman, Y.; Aviram, S.; Roth-Bejerano, N.; Trappe, J.M.; Kagan-Zur, V. Phylogenetic studies of Terfezia pfeilii and Choiromyces echinulatus (Pezizales) support new genera for southern African truffles: Kalaharituber and Eremiomyces. Mycol. Res. 2005, 109, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Tedersoo, L.; Hansen, K.; Perry, B.A.; Kjøller, R. Molecular and morphological diversity of pezizalean ectomycorrhiza. New Phytol. 2006, 170, 581–596. [Google Scholar] [CrossRef] [PubMed]
- Læssøe, T.; Hansen, K. Truffle trouble: What happened to the Tuberales? Mycol. Res. 2007, 111, 1075–1099. [Google Scholar] [CrossRef]
- Kovács, G.M.; Trappe, J.M. Nomenclatural History and Genealogies of Desert Truffles. In Soil Biology; Kagan-Zur, V., Roth-Bejerano, N., Sitrit, Y., Morte, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; Volume 38, pp. 21–37. [Google Scholar] [CrossRef]
- Maire, R. Notes mycologiques. Ann. Mycol. 1906, 4, 329–335. [Google Scholar]
- Malençon, G. Champignons hypogés du Nord de, l’Afrique–I. Ascomycetes. Persoonia 1973, 7, 261–279. [Google Scholar]
- Moreno-Arroyo, B.; Gomez, J.; Calonge, F.D. Aportaciones a la micoflora hipogea Ibérica. Boletín Soc. Micol. Madr. 1997, 22, 91–95. [Google Scholar]
- Moreno, G.; Díez, J.; Manjon, J.L. Picoa lefebvrei and Tirmania nivea, two rare hypogeous fungi from Spain. Mycol. Res. 2000, 104, 378–381. [Google Scholar] [CrossRef]
- Kovács, G.M.; Balazs, T.K.; Calonge, F.D.; Martín, M.P. The diversity of Terfezia desert truffles: New species and a highly variable species complex with intrasporocarpic nrDNA ITS heterogeneity. Mycologia 2011, 103, 841–853. [Google Scholar] [CrossRef]
- Bordallo, J.J.; Rodríguez, A.; Honrubia, M.; Morte, A. Terfezia canariensis sp. nov. una nueva especie de trufa encontrada en las Islas Canarias. Cantarela 2012, 56, 1–8. [Google Scholar]
- Bordallo, J.J.; Rodríguez, A.; Muñoz-Mohedano, J.M.; Suz, L.M.; Honrubia, M.; Morte, A. Five new Terfezia species from the Iberian Peninsula. Mycotaxon 2013, 124, 189–208. [Google Scholar] [CrossRef]
- Bordallo, J.J.; Rodríguez, A.; Kaounas, V.; Camello, F.; Honrubia, M.; Morte, A. Two new Terfezia species from Southern Europe. Phytotaxa 2015, 230, 239–249. [Google Scholar] [CrossRef]
- Bordallo, J.J.; Rodriguez, A.; Santos-Silva, C.; Louro, R.; Muñoz-Mohedano, J.M.; Morte, A. Terfezia lusitanica, a new mycorrhizal species associated to Tuberaria guttata (Cistaceae). Phytotaxa 2018, 357, 141–147. [Google Scholar] [CrossRef]
- Bordallo, J.J.; Rodriguez, A.; Morte, A. Terfezia morenoi. Fungal Planet description sheets. Persoonia 2018, 40, 324–325. [Google Scholar] [CrossRef]
- Moreno, G.; Manjón, J.L.; Alvarado, P. A new Terfezia from Spain. Boletín Soc. Micol. Madr. 2019, 43, 55–60. [Google Scholar]
- Rodríguez, A.; Navarro-Ródenas, A.; Morte, A.; Cabero, J.; Luque, D. Terfezia dunensis. Fungal Planet description sheets. Persoonia 2019, 43, 410–411. [Google Scholar] [CrossRef]
- Louro, R.; Nobre, T.; Santos-Silva, C. Terfezia solaris-libera sp. nov., a new mycorrhizal species within the spiny-spored lineages. J. Mycol. Mycol. Sci. 2020, 3, 1–13. [Google Scholar] [CrossRef]
- Morte, A.; Bordallo, J.J.; Rodriguez, A. Tirmania honrubiae. Fungal Planet description sheets. Persoonia 2018, 40, 328–329. [Google Scholar] [CrossRef]
- Kottek, M.; Grieser, J.; Beck, C.; Rudolf, B.; Rubel, F. World map of the Köppen-Geiger climate classification updated. Meteorol. Z. 2006, 15, 259–263. [Google Scholar] [CrossRef]
- Langeron, K. Précis de Mycologie; Ed. Masson: Paris, France, 1952; pp. 367–397. [Google Scholar]
- Moreno, G.; Altés, A.; Ochoa, C.; Wright, J.E. Contribution to the study of the family Tulostomataceae in Baja California, Mexico. Mycologia 1995, 87, 96–120. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for Basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- Bidartondo, M.I.; Burghardt, B.; Gebauer, G.; Bruns, T.D.; Read, D.J. Changing partners in the dark: Isotopic and molecular evidence of ectomycorrhizal liaisons between forest orchids and trees. Proc. R. Soc. Lond. B 2004, 271, 1799–1806. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.D.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J., White, T.J., Eds.; Academic Press: London, UK, 1990; 482p. [Google Scholar]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed]
- Cubeta, M.A.; Echandi, E.; Abernethy, T.; Vilgalys, R. Characterization of anastomosis groups of binucleate Rhizoctonia species using restriction analysis of an amplified ribosomal RNA gene. Phytopathology 1991, 81, 1395–1400. [Google Scholar] [CrossRef]
- Zitouni-Haouar, F.E.-H.; Alvarado, P.; Sbissi, I.; Boudabous, A.; Fortas, Z.; Moreno, G.; Manjón, J.L.; Gtari, M. Contrasted genetic diversity, relevance of climate and host plants, and comments on the taxonomic problems of the genus Picoa (Pyronemataceae, Pezizales). PLoS ONE 2015, 10, e0138513. [Google Scholar] [CrossRef] [PubMed]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef] [PubMed]
- Mullis, K.B.; Faloona, F.A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987, 155, 335–350. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.H.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Jamali, S.; Banihashemi, Z. Hosts and Distribution of Desert Truffles in Iran, based on morphological and molecular criteria. J. Agric. Sci. Technol. 2012, 14, 1379–1396. [Google Scholar]
- Osmundson, T.W.; Robert, V.A.; Schoch, C.L.; Baker, L.J.; Smith, A.; Robich, G.; Mizzan, L.; Garbelotto, M.M. Filling gaps in biodiversity knowledge for macrofungi: Contributions and assessment of an herbarium collection DNA barcode sequencing project. PLoS ONE 2013, 8, e62419. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; Volume 14, pp. 1–8. [Google Scholar]
- Posada, D. jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Wiens, J.J.; Graham, C.H. Niche conservatism: Integrating evolution, ecology, and conservation biology. Annu. Rev. Ecol. Evol. Syst. 2005, 36, 519–539. [Google Scholar] [CrossRef]
- Chethana, K.T.; Manawasinghe, I.S.; Hurdeal, V.G.; Bhunjun, C.S.; Appadoo, M.A.; Gentekaki, E.; Raspé, O.; Promputtha, I.; Hyde, K.D. What are fungal species and how to delineate them? Fungal Divers. 2021, 109, 1–25. [Google Scholar] [CrossRef]
- Giraud, T.; Refrégier, G.; Le Gac, M.; de Vienne, D.M.; Hood, M.E. Speciation in fungi. Fungal Genet. Biol. 2008, 45, 791–802. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zitouni-Haouar, F.E.-H.; Bidartondo, M.I.; Moreno, G.; Carlavilla, J.R.; Manjón, J.L.; Neggaz, S.; Zitouni-Nourine, S.H. Bioclimatic Origin Shapes Phylogenetic Structure of Tirmania (Pezizaceae): New Species and New Record from North Africa. J. Fungi 2023, 9, 532. https://doi.org/10.3390/jof9050532
Zitouni-Haouar FE-H, Bidartondo MI, Moreno G, Carlavilla JR, Manjón JL, Neggaz S, Zitouni-Nourine SH. Bioclimatic Origin Shapes Phylogenetic Structure of Tirmania (Pezizaceae): New Species and New Record from North Africa. Journal of Fungi. 2023; 9(5):532. https://doi.org/10.3390/jof9050532
Chicago/Turabian StyleZitouni-Haouar, Fatima El-Houaria, Martin I. Bidartondo, Gabriel Moreno, Juan Ramón Carlavilla, José Luis Manjón, Samir Neggaz, and Saida Hanane Zitouni-Nourine. 2023. "Bioclimatic Origin Shapes Phylogenetic Structure of Tirmania (Pezizaceae): New Species and New Record from North Africa" Journal of Fungi 9, no. 5: 532. https://doi.org/10.3390/jof9050532
APA StyleZitouni-Haouar, F. E.-H., Bidartondo, M. I., Moreno, G., Carlavilla, J. R., Manjón, J. L., Neggaz, S., & Zitouni-Nourine, S. H. (2023). Bioclimatic Origin Shapes Phylogenetic Structure of Tirmania (Pezizaceae): New Species and New Record from North Africa. Journal of Fungi, 9(5), 532. https://doi.org/10.3390/jof9050532






