Evaluation of a Novel FKS1 R1354H Mutation Associated with Caspofungin Resistance in Candida auris Using the CRISPR-Cas9 System
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Sequence Analysis of the C. auris FKS Genes
2.3. Genetic Manipulation
2.4. In Vitro Growth Curve and Antifungal Susceptibility Testing
2.5. In Vivo Experiments
2.6. Statistical Analysis
3. Results
3.1. R1354H Mutation in FKS1 Causes Increased Echinocandin MICs in C. auris
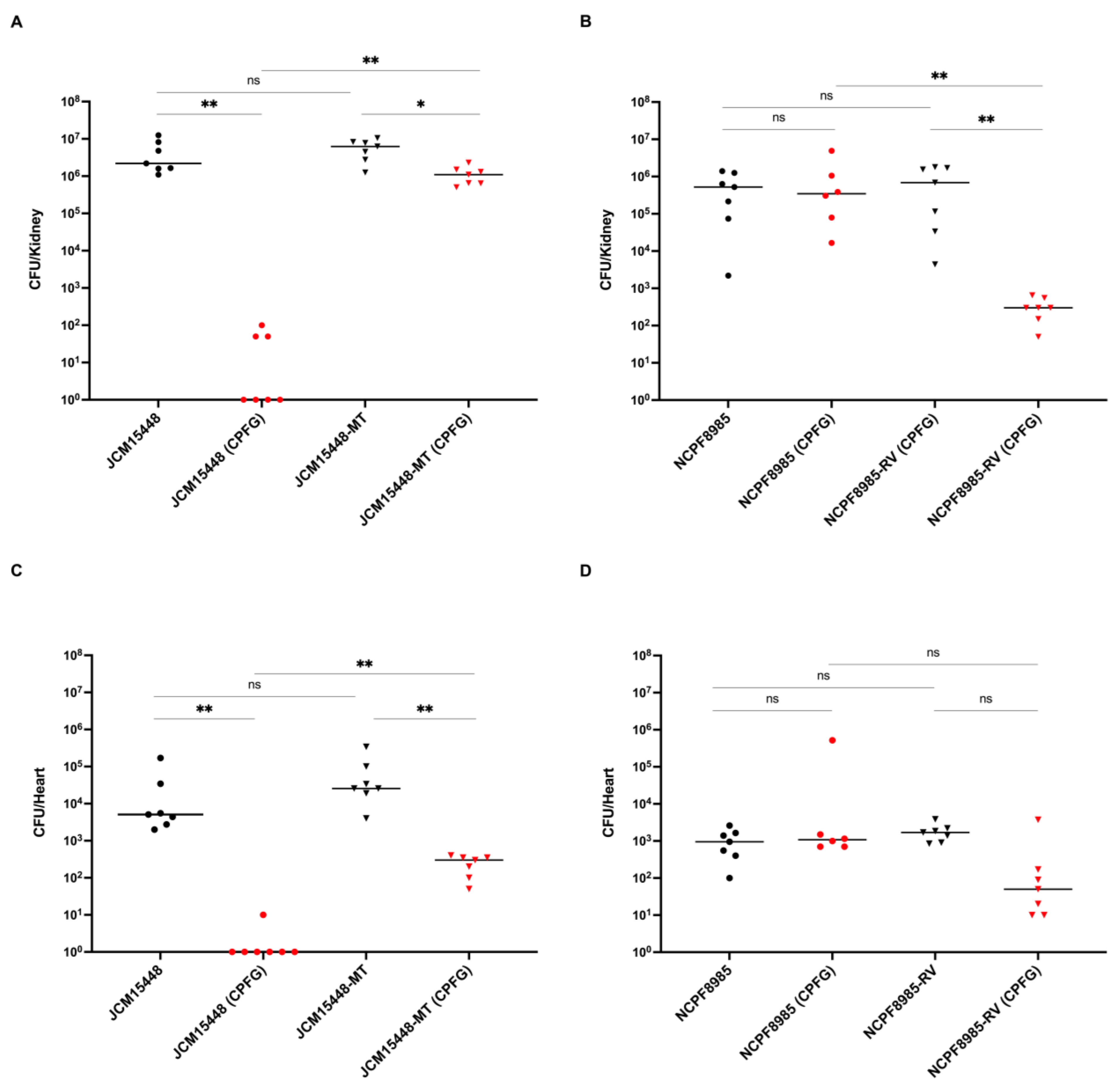
3.2. Efficacy of Caspofungin Treatment in Mice Infected with C. auris Strains Harboring the FKS1 R1354H Mutation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Satoh, K.; Makimura, K.; Hasumi, Y.; Nishiyama, Y.; Uchida, K.; Yamaguchi, H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol. Immunol. 2009, 53, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Sekyere, J.O. Candida auris: A systematic review and meta-analysis of current updates on an emerging multidrug-resistant pathogen. MicrobiologyOpen 2018, 7, e00578. [Google Scholar] [CrossRef] [PubMed]
- Eyre, D.W.; Sheppard, A.E.; Madder, H.; Moir, I.; Moroney, R.; Quan, T.P.; Griffiths, D.; George, S.; Butcher, L.; Morgan, M.; et al. A Candida auris outbreak and its control in an intensive care setting. N. Engl. J. Med. 2018, 379, 1322–1331. [Google Scholar] [CrossRef] [PubMed]
- Sherry, L.; Ramage, G.; Kean, R.; Borman, A.; Johnson, E.M.; Richardson, M.D.; Rautemaa-Richardson, R. Biofilm-forming capability of highly virulent, Multidrug-Resistant Candida auris. Emerg. Infect. Dis. 2017, 23, 328–331. [Google Scholar] [CrossRef] [PubMed]
- Borman, A.M.; Szekely, A.; Johnson, E.M. Isolates of the emerging pathogen Candida auris present in the UK have several geographic origins. Med. Mycol. 2017, 55, 563–567. [Google Scholar] [CrossRef] [PubMed]
- Chowdhary, A.; Sharma, C.; Meis, J.F. Candida auris: A rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLOS Pathog. 2017, 13, e1006290. [Google Scholar] [CrossRef]
- Chowdhary, A.; Tarai, B.; Singh, A.; Sharma, A. Multidrug-resistant Candida auris infections in critically ill coronavirus disease patients, India, April–July 2020. Emerg. Infect. Dis. 2020, 26, 2694–2696. [Google Scholar] [CrossRef]
- De Almeida, J.N., Jr.; Francisco, E.C.; Hagen, F.; Brandão, I.B.; Pereira, F.M.; Dias, P.H.P.; de Miranda Costa, M.M.; de Souza Jordão, R.T.; de Groot, T.; Colombo, A.L. Emergence of Candida auris in Brazil in a COVID-19 Intensive Care Unit. J. Fungi 2021, 7, 220. [Google Scholar] [CrossRef]
- Hanson, B.M.; Dinh, A.Q.; Tran, T.T.; Arenas, S.; Pronty, D.; Gershengorn, H.B.; Ferreira, T.; Arias, C.A.; Shukla, B.S. Candida auris invasive infections during a COVID-19 case surge. Antimicrob. Agents Chemother. 2021, 65, e0114621. [Google Scholar] [CrossRef]
- Janniger, E.J.; Kapila, R. Public health issues with Candida auris in COVID-19 patients. World Med. Health Policy. 2021, 13, 766–772. [Google Scholar] [CrossRef]
- CDC. Antibiotic Resistance Threats in the United States, 2019; United States Department of Health and Human Services, Centers for Disease Control and Prevention (US): Atlanta, GA, USA, 2019. [Google Scholar]
- Lyman, M.; Forsberg, K.; Sexton, D.J.; Chow, N.A.; Lockhart, S.R.; Jackson, B.R.; Chiller, T. Worsening Spread of Candida auris in the United States, 2019 to 2021. Ann. Intern. Med. 2023, 176, 489–495. [Google Scholar] [CrossRef] [PubMed]
- Chow, N.A.; Munoz, J.F.; Gade, L.; Berkow, E.L.; Li, X. Tracing the evolutionary history and global expansion of Candida auris using population genomic analyses. mBio 2020, 11, e03364-19. [Google Scholar] [CrossRef] [PubMed]
- Escandon, P.; Chow, N.A.; Caceres, D.H.; Gade, L.; Berkow, E.L. Molecular epidemiology of Candida auris in Colombia reveals a highly related, countrywide colonization with regional patterns in amphotericin B resistance. Clin. Infect. Dis. 2019, 68, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Kean, R.; Ramage, G. Combined antifungal resistance and biofilm tolerance: The global threat of Candida auris. mSphere 2019, 4, e00458-19. [Google Scholar] [CrossRef]
- Chakrabarti, A.; Sood, P. On the emergence, spread and resistance of Candida auris: Host, pathogen and environmental tipping points. J. Med. Microbiol. 2021, 70, 001318. [Google Scholar] [CrossRef]
- Lockhart, S.R.; Etienne, K.A.; Vallabhaneni, S.; Farooqi, J.; Chowdhary, A.; Govender, N.P.; Colombo, A.L.; Calvo, B.; Cuomo, C.A.; Desjardins, C.A.; et al. Simultaneous Emergence of Multidrug-Resistant Candida auris on 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses. Clin. Infect. Dis. 2017, 64, 134–140. [Google Scholar] [CrossRef]
- Kathuria, S.; Singh, P.K.; Sharma, C.; Prakash, A.; Masih, A.; Kumar, A.; Meis, J.F.; Chowdhary, A. Multidrug-Resistant Candida auris Misidentified as Candida haemulonii: Characterization by matrix-assisted laser desorption ionization-time of flight mass spectrometry and DNA Sequencing and Its antifungal Susceptibility Profile Variability by Vitek 2, CLSI Broth microdilution, and Etest Method. J. Clin. Microbiol. 2015, 53, 1823–1830. [Google Scholar] [CrossRef]
- Chen, J.; Tian, S.; Han, X.; Chu, Y.; Wang, Q.; Zhou, B.; Shang, H. Is the superbug fungus really so scary? A systematic review and meta-analysis of global epidemiology and mortality of Candida auris. BMC Infect. Dis. 2020, 20, 827. [Google Scholar] [CrossRef]
- Pandya, N.; Cag, Y.; Pandak, N.; Pekok, A.U.; Poojary, A.; Ayoade, F.; Fasciana, T.; Giammanco, A.; Caskurlu, H.; Rajani, D.P.; et al. International Multicentre Study of Candida auris Infections. J. Fungi. 2021, 7, 878. [Google Scholar] [CrossRef]
- Park, S.; Kelly, R.; Kahn, J.N.; Robles, J.; Hsu, M.J.; Register, E.; Li, W.; Vyas, V.; Fan, H.; Abruzzo, G.; et al. Specific substitutions in the echinocandin target Fks1p account for reduced susceptibility of rare laboratory and clinical Candida sp. isolates. Antimicrob. Agents Chemother. 2005, 49, 3264–3273. [Google Scholar] [CrossRef]
- Garcia-Effron, G.; Park, S.; Perlin, D.S. Correlating echinocandin MIC and kinetic inhibition of fks1 mutant glucan synthases for Candida albicans: Implications for interpretive breakpoints. Antimicrob. Agents Chemother. 2009, 53, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Perlin, D.S. Resistance to echinocandin-class antifungal drugs. Drug Resist. Updat. 2007, 10, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Kordalewska, M.; Lee, A.; Park, S.; Berrio, I.; Chowdhary, A.; Zhao, Y.; Perlin, D.S. Understanding echinocandin resistance in the emerging pathogen Candida auris. Antimicrob. Agents Chemother. 2018, 62, e00238-18. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Paul, R.A.; Rudramurthy, S.M.; Kashyap, N.; Bhattacharya, S.; Soman, R.; Shankarnarayan, S.A.; Chavan, D.; Singh, S.; Das, P.; et al. Impact of FKS1 genotype on echinocandin in vitro susceptibility in Candida auris and in vivo response in a murine model of infection. Antimicrob. Agents Chemother. 2022, 66, e0165221. [Google Scholar] [CrossRef] [PubMed]
- Grahl, N.; Demers, E.G.; Crocker, A.W.; Hogan, D.A. Use of RNA-protein complexes for genome editing in non-albicans Candida species. mSphere. 2017, 2, e00218-17. [Google Scholar] [CrossRef]
- Li, J.; Coste, A.T.; Liechti, M.; Bachmann, D.; Sanglard, D.; Lamoth, F. Novel ERG11 and TAC1b mutations associated with azole resistance in Candida auris. Antimicrob. Agents Chemother. 2023, 65, e02663-20. [Google Scholar] [CrossRef]
- Shahana, S.; Childers, D.S.; Ballou, E.R.; Bohovych, I.; Odds, F.C.; Gow, N.A.; Brown, A.J. New Clox Systems for rapid and efficient gene disruption in Candida albicans. PLoS ONE 2014, 9, e100390. [Google Scholar] [CrossRef]
- Chen, Y.L.; Yu, S.J.; Huang, H.Y.; Chang, Y.L.; Lehman, V.N.; Silao, F.G.S.; Bigol, U.G.; Bungay, A.A.C.; Averette, A.; Heitman, J. Calcineurin controls hyphal growth, virulence, and drug tolerance of Candida tropicalis. Eukaryot. Cell 2014, 13, 844–854. [Google Scholar] [CrossRef]
- Committee on the Care and Use of Laboratory Animals of the Institute of Laboratory Animal Resources. CoLS, National Research Council. Guide-for-the-Care-and-Use-of-Laboratory-Animals; National Academy: Washington, DC, USA, 2011. [Google Scholar]
- Forgács, L.; Borman, A.M.; Prépost, E.; Tóth, Z.; Kardos, G.; Kovács, R.; Szekely, A.; Nagy, F.; Kovacs, I.; Majoros, L. Comparison of in vivo pathogenicity of four Candida auris clades in a neutropenic bloodstream infection murine model. Emerg. Microbes Infect. 2020, 9, 1160–1169. [Google Scholar] [CrossRef]
- Andes, D. Use of an animal model of disseminated candidiasis in the evaluation of antifungal therapy. Methods Mol Med. 2005, 118, 111–128. [Google Scholar] [CrossRef]
- Sekizuka, T.; Iguchi, S.; Umeyama, T.; Inamine, Y.; Makimura, K.; Kuroda, M.; Miyazaki, Y.; Kikuchi, K. Clade II Candida auris possess genomic structural variations related to an ancestral strain. PLoS ONE 2019, 14, e0223433. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Hu, W.; Tamura, T.; Alshahni, M.M.; Satoh, K.; Yamanishi, C.; Naito, T.; Makimura, K. Investigation of the physiological, biochemical and antifungal susceptibility properties of Candida auris. Mycopathologia 2021, 186, 189–198. [Google Scholar] [CrossRef] [PubMed]

| Strain | Clade * | Genotype or Description | Reference or Source |
|---|---|---|---|
| JCM15448 | Ⅱ | Type strain, Parental strain (R1354) | Teikyo University [1] |
| NPCF8971 | Ⅰ | Parental strain (R1354) | National Collection of Pathogenic Fungi [5] |
| NPCF8985 | Ⅰ | Parental strain (H1354) | National Collection of Pathogenic Fungi [5] |
| JCM15448-MT | Ⅱ | FKS1c.4061G>A::NatR (R1354H) | This study |
| NPCF8971-MT | Ⅰ | FKS1c.4061G>A::NatR (R1354H) | This study |
| NPCF8985-RV | Ⅰ | FKS1c.4061A>G::NatR (H1354R) | This study |
| For FKS1 Sequencing | |
| Primer | Sequence (5′-3′) |
| CauFKS1-F | ATGTCTTACGATAACAATC |
| CauFKS1-R | TTAGAATGCCTTTGTAGTATAG |
| CauFKS1-F653 | GAAGTGGTTTTTCGCCTCG |
| CauFKS1-F1256 | AGAGATACATGAGATTGGGTG |
| CauFKS1-F1863 | TCTTTGGGTCACTGTGTTTG |
| CauFKS1-F2446 | CGTATCAGTTTCTTTGCTCAG |
| CauFKS1-F3048 | CGCCGAGTTTTTGTTGAGAG |
| CauFKS1-F3674 | GTATGACTGCCATGTTGAGAG |
| CauFKS1-F4241 | CAGACTTGACCGTTGGTGGTG |
| CauFKS1-F4878 | CTGTCTTGGAATGGCTTGTTG |
| CauFKS1-R178 | AGTTCTGGTAACCGTCCATG |
| CauFKS1-F67 | GAGTATTACCAACAGGGCG |
| CauFKS1-F5303 | TAGACAGATGGCACTCCACC |
| For genetic manipulation | |
| Primer | Sequence (5′-3′) |
| Cau1ST-F | CGCCGAGTTTTTGTTGAGAG |
| Cau1ST-R | CGGCGGGGACGAGGCAAGCTTACAGCTTGTAGAATTCTCGAAT |
| NatR-F | TATTATTCTATATTGGTTTGAATTAAGCTTGCCTCGTCCCCGCCGGGTCA |
| NatR-R | GCTTGTAGAATTCTCGAATAATGTAGACACTGGATGGCGGCGTTAGTATC |
| Cau3RD-F | GATACTAACGCCGCCATCCAGTGTCTACATTATTCGAGAATTCTACAAGCTG |
| Cau3RD-R | GGTAGCATTCAAGAACCAAGAC |
| Strain | MIC (μg/mL) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| AFG | MFG | CAS | 5FC | POS | VRC | ITC | FLC | AMB | |
| JCM15448 | 0.12 | 0.06 | 0.03 | <0.06 | 0.015 | 0.015 | 0.03 | 4 | 0.5 |
| JCM15448-MT | 0.5 | 0.25 | 0.12 | <0.06 | 0.015 | 0.015 | 0.03 | 2 | 0.5 |
| NCPF8971 | 0.12 | 0.12 | 0.12 | <0.06 | <0.008 | 0.25 | 0.06 | 128 | 1 |
| NCPF8971-MT | 1 | 0.5 | 2 | <0.06 | <0.008 | 0.25 | 0.06 | >256 | 1 |
| NCPF8985 | 2 | 2 | 8 | >64 | >8 | >8 | >16 | >256 | 1 |
| NCPF8985-RV | 0.5 | 0.5 | 2 | >64 | >8 | >8 | >16 | >256 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kiyohara, M.; Miyazaki, T.; Okamoto, M.; Hirayama, T.; Makimura, K.; Chibana, H.; Nakada, N.; Ito, Y.; Sumiyoshi, M.; Ashizawa, N.; et al. Evaluation of a Novel FKS1 R1354H Mutation Associated with Caspofungin Resistance in Candida auris Using the CRISPR-Cas9 System. J. Fungi 2023, 9, 529. https://doi.org/10.3390/jof9050529
Kiyohara M, Miyazaki T, Okamoto M, Hirayama T, Makimura K, Chibana H, Nakada N, Ito Y, Sumiyoshi M, Ashizawa N, et al. Evaluation of a Novel FKS1 R1354H Mutation Associated with Caspofungin Resistance in Candida auris Using the CRISPR-Cas9 System. Journal of Fungi. 2023; 9(5):529. https://doi.org/10.3390/jof9050529
Chicago/Turabian StyleKiyohara, Maiko, Taiga Miyazaki, Michiyo Okamoto, Tatsuro Hirayama, Koichi Makimura, Hiroji Chibana, Nana Nakada, Yuya Ito, Makoto Sumiyoshi, Nobuyuki Ashizawa, and et al. 2023. "Evaluation of a Novel FKS1 R1354H Mutation Associated with Caspofungin Resistance in Candida auris Using the CRISPR-Cas9 System" Journal of Fungi 9, no. 5: 529. https://doi.org/10.3390/jof9050529
APA StyleKiyohara, M., Miyazaki, T., Okamoto, M., Hirayama, T., Makimura, K., Chibana, H., Nakada, N., Ito, Y., Sumiyoshi, M., Ashizawa, N., Takeda, K., Iwanaga, N., Takazono, T., Izumikawa, K., Yanagihara, K., Kohno, S., & Mukae, H. (2023). Evaluation of a Novel FKS1 R1354H Mutation Associated with Caspofungin Resistance in Candida auris Using the CRISPR-Cas9 System. Journal of Fungi, 9(5), 529. https://doi.org/10.3390/jof9050529





