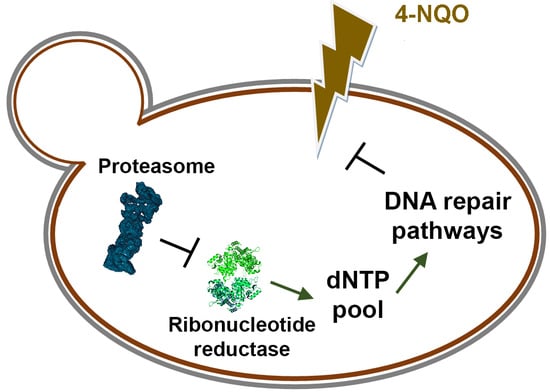
Yeast Ribonucleotide Reductase Is a Direct Target of the Proteasome and Provides Hyper Resistance to the Carcinogen 4-NQO
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Strains
2.2. Media, Culture, and Assay Conditions
2.3. LC-MS/MS Data Acquisition
2.4. LC-MS/MS Data Processing
2.5. Real-Time qPCR Analysis
2.6. Cloning of RNR Genes
2.7. Western Blot Analysis of RNR Subunits
2.8. Cycloheximide Chase
2.9. Creation of Yeast Mutants and Inhibitor Plate Assay
3. Results
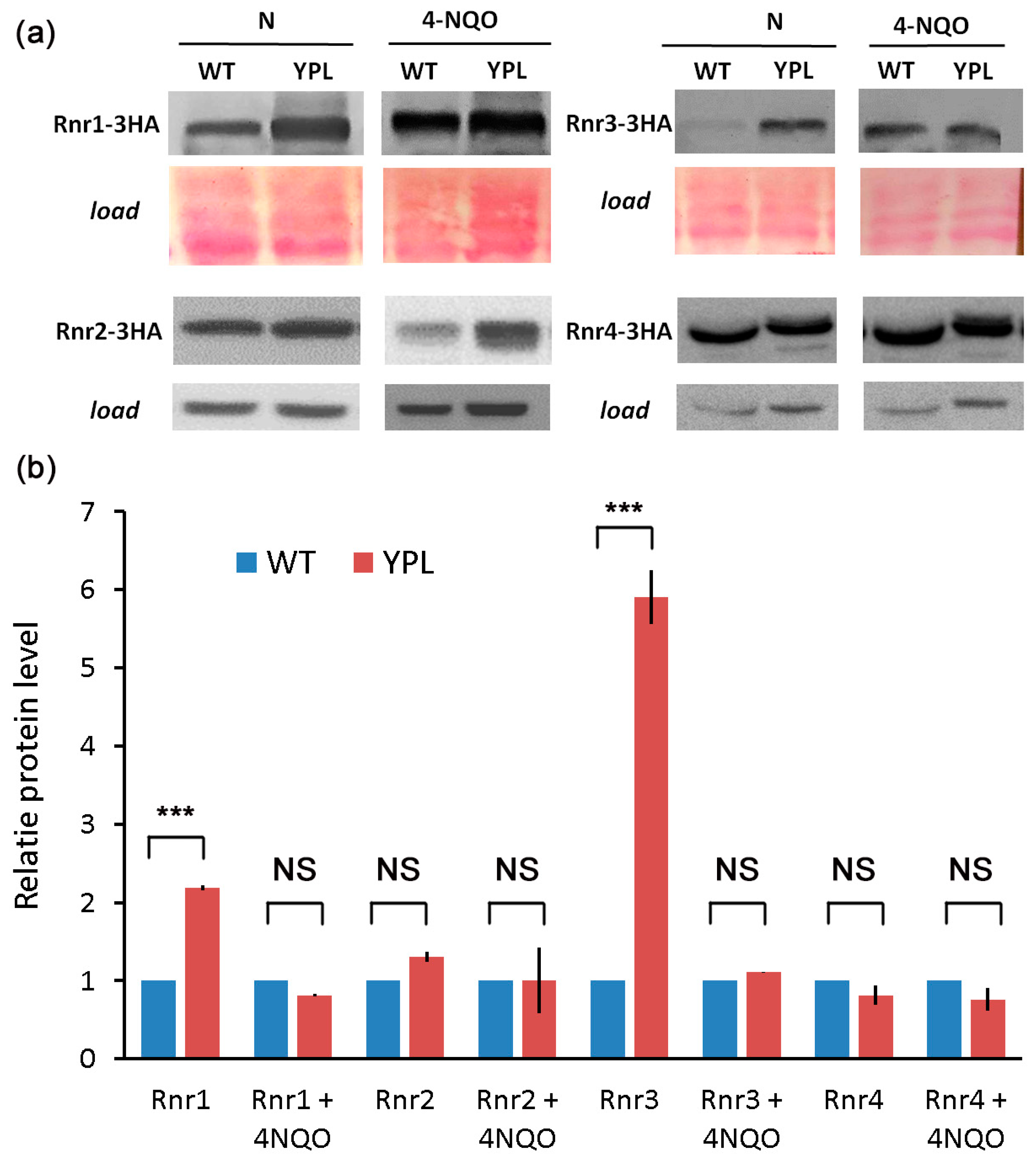
3.1. Proteomic Analysis Indicates Increased Levels of RNR Subunits in the YPL Strain Compared to the WT Strain
3.2. Deregulation of the Proteasome Leads to Enhanced Levels of Large RNR Subunits
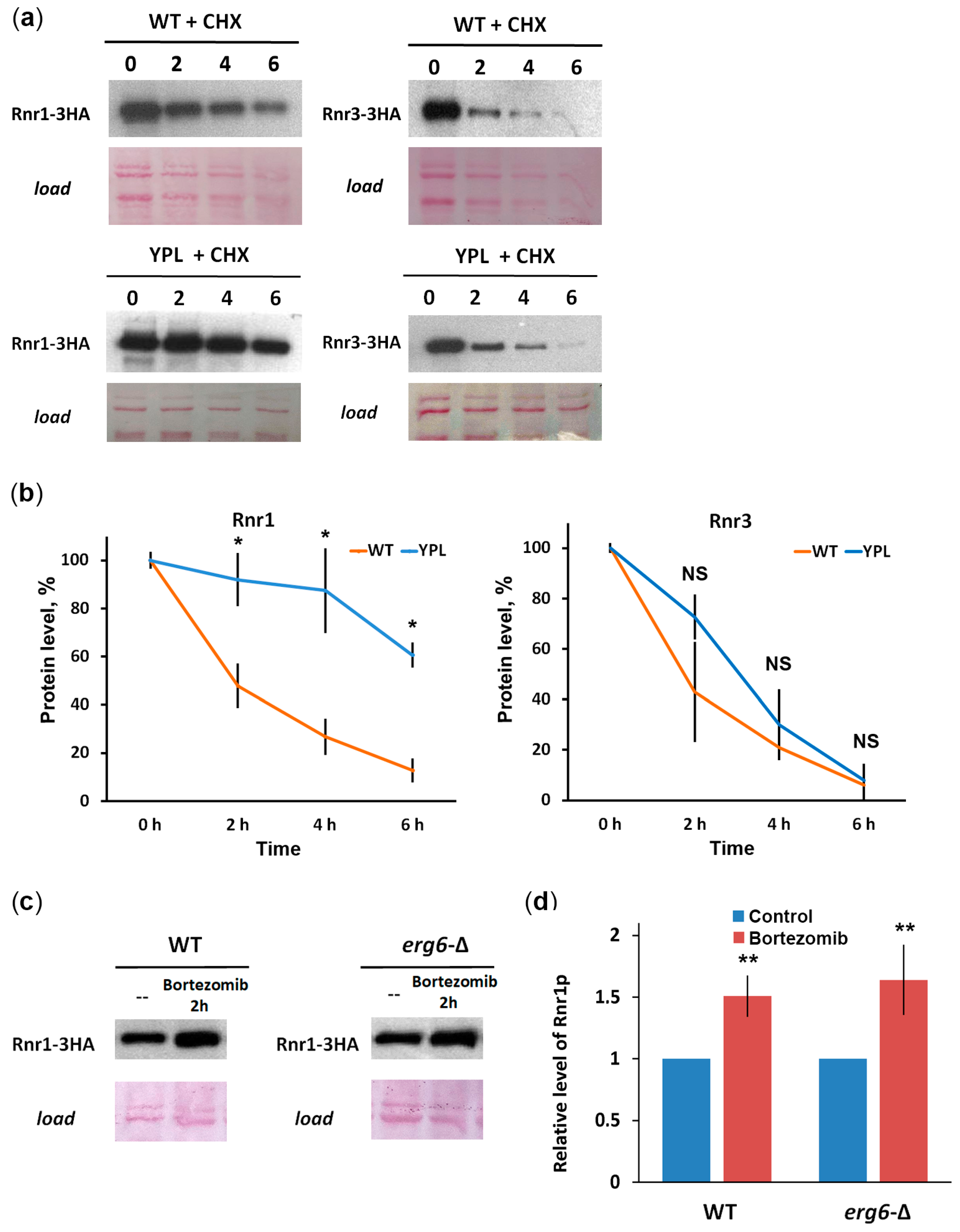
3.3. Rnr1 Is a Proteasome Substrate
3.4. HU Reverses the 4-NQO Hyper Resistance Phenotype in the YPL Strain
3.5. Lack of RNR Regulatory Proteins Sensitizes WT and YPL Strains to HU and 4-NQO
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yousefzadeh, M.; Henpita, C.; Vyas, R.; Soto-Palma, C.; Robbins, P.; Niedernhofer, L. DNA damage-how and why we age? eLife 2021, 10, e62852. [Google Scholar] [CrossRef] [PubMed]
- Konopka, A.; Atkin, J.D. The role of DNA damage in neural plasticity in physiology and neurodegeneration. Front. Cell. Neurosci. 2022, 16, 836885. [Google Scholar] [CrossRef] [PubMed]
- Alhmoud, J.F.; Woolley, J.F.; Al Moustafa, A.E.; Malki, M.I. DNA damage/repair management in cancers. Cancers 2020, 12, 1050. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, B.; Pothof, J.; Vijg, J.; Hoeijmakers, J.H.J. The central role of DNA damage in the ageing process. Nature 2021, 592, 695–703. [Google Scholar] [CrossRef]
- Kumari, S.; Sharma, S.; Advani, D.; Khosla, A.; Kumar, P.; Ambasta, R.K. Unboxing the molecular modalities of mutagens in cancer. Environ. Sci. Pollut. Res. Int. 2022, 29, 62111–62159. [Google Scholar] [CrossRef]
- Bouaoud, J.; De Souza, G.; Darido, C.; Tortereau, A.; Elkabets, M.; Bertolus, C.; Saintigny, P. The 4-NQO mouse model: An update on a well-established in vivo model of oral carcinogenesis. Methods Cell Biol. 2021, 163, 197–229. [Google Scholar] [CrossRef]
- Ge, S.; Zhang, J.; Du, Y.; Hu, B.; Zhou, Z.; Lou, J. Dynamic changes in the gene expression profile during rat oral carcinogenesis induced by 4-nitroquinoline 1-oxide. Mol. Med. Rep. 2016, 13, 2561–2569. [Google Scholar] [CrossRef]
- Choi, D.H.; Min, M.H.; Kim, M.J.; Lee, R.; Kwon, S.H.; Bae, S.H. Hrq1 facilitates nucleotide excision repair of DNA damage induced by 4-nitroquinoline-1-oxide and cisplatin in Saccharomyces cerevisiae. J. Microbiol. 2014, 52, 292–298. [Google Scholar] [CrossRef]
- Arima, Y.; Nishigori, C.; Takeuchi, T.; Oka, S.; Morimoto, K.; Utani, A.; Miyachi, Y. 4-Nitroquinoline 1-oxide forms 8-hydroxydeoxyguanosine in human fibroblasts through reactive oxygen species. Toxicol. Sci. 2006, 91, 382–392. [Google Scholar] [CrossRef]
- Yan, L.L.; Simms, C.L.; McLoughlin, F.; Vierstra, R.D.; Zaher, H.S. Oxidation and alkylation stresses activate ribosome-quality control. Nat. Commun. 2019, 10, 5611. [Google Scholar] [CrossRef]
- Karpov, D.S.; Spasskaya, D.S.; Tutyaeva, V.V.; Mironov, A.S.; Karpov, V.L. Proteasome inhibition enhances resistance to DNA damage via upregulation of Rpn4-dependent DNA repair genes. FEBS Lett. 2013, 587, 3108–3114. [Google Scholar] [CrossRef] [PubMed]
- Tallec, B.L.; Peyroche, A. Using DNA damage sensitivity phenotypes to characterize mutations affecting proteasome function. Methods Mol. Biol. 2012, 832, 363–371. [Google Scholar] [CrossRef]
- Spasskaya, D.S.; Nadolinskaia, N.I.; Tutyaeva, V.V.; Lysov, Y.P.; Karpov, V.L.; Karpov, D.S. Yeast Rpn4 links the proteasome and DNA repair via RAD52 regulation. Int. J. Mol. Sci. 2020, 21, 8097. [Google Scholar] [CrossRef]
- Karpov, D.S.; Spasskaya, D.S.; Nadolinskaia, N.I.; Tutyaeva, V.V.; Lysov, Y.P.; Karpov, V.L. Deregulation of the 19S proteasome complex increases yeast resistance to 4-NQO and oxidative stress via upregulation of Rpn4- and proteasome-dependent stress responsive genes. FEMS Yeast Res. 2019, 19, foz002. [Google Scholar] [CrossRef] [PubMed]
- Mannhaupt, G.; Schnall, R.; Karpov, V.; Vetter, I.; Feldmann, H. Rpn4p acts as a transcription factor by binding to PACE, a nonamer box found upstream of 26S proteasomal and other genes in yeast. FEBS Lett. 1999, 450, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Varshavsky, A. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: A negative feedback circuit. Proc. Natl. Acad. Sci. USA 2001, 98, 3056–3061. [Google Scholar] [CrossRef]
- Wang, X.; Xu, H.; Ju, D.; Xie, Y. Disruption of Rpn4-induced proteasome expression in Saccharomyces cerevisiae reduces cell viability under stressed conditions. Genetics 2008, 180, 1945–1953. [Google Scholar] [CrossRef]
- Greene, B.L.; Kang, G.; Cui, C.; Bennati, M.; Nocera, D.G.; Drennan, C.L.; Stubbe, J. Ribonucleotide reductases: Structure, chemistry, and metabolism suggest new therapeutic targets. Annu. Rev. Biochem. 2020, 89, 45–75. [Google Scholar] [CrossRef]
- Zhao, X.; Rothstein, R. The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc. Natl. Acad. Sci. USA 2002, 99, 3746–3751. [Google Scholar] [CrossRef]
- D’Angiolella, V.; Donato, V.; Forrester, F.M.; Jeong, Y.T.; Pellacani, C.; Kudo, Y.; Saraf, A.; Florens, L.; Washburn, M.P.; Pagano, M. Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell 2012, 149, 1023–1034. [Google Scholar] [CrossRef]
- Sanvisens, N.; Romero, A.M.; An, X.; Zhang, C.; de Llanos, R.; Martinez-Pastor, M.T.; Bano, M.C.; Huang, M.; Puig, S. Yeast Dun1 kinase regulates ribonucleotide reductase inhibitor Sml1 in response to iron deficiency. Mol. Cell. Biol. 2014, 34, 3259–3271. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Hou, W.; Wang, Z.Q.; Xu, X. Biogenesis of Iron-Sulfur Clusters and Their Role in DNA Metabolism. Front. Cell Dev. Biol. 2021, 9, 735678. [Google Scholar] [CrossRef]
- Andreson, B.L.; Gupta, A.; Georgieva, B.P.; Rothstein, R. The ribonucleotide reductase inhibitor, Sml1, is sequentially phosphorylated, ubiquitylated and degraded in response to DNA damage. Nucleic Acids Res. 2010, 38, 6490–6501. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.D.; Wang, J.; Stubbe, J.; Elledge, S.J. Dif1 is a DNA-damage-regulated facilitator of nuclear import for ribonucleotide reductase. Mol. Cell 2008, 32, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, X.; Chen, Z.; Bepler, G. Ubiquitination and degradation of ribonucleotide reductase M1 by the polycomb group proteins RNF2 and Bmi1 and cellular response to gemcitabine. PLoS ONE 2014, 9, e91186. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]
- Bubis, J.A.; Spasskaya, D.S.; Gorshkov, V.A.; Kjeldsen, F.; Kofanova, A.M.; Lekanov, D.S.; Gorshkov, M.V.; Karpov, V.L.; Tarasova, I.A.; Karpov, D.S. Rpn4 and proteasome-mediated yeast resistance to ethanol includes regulation of autophagy. Appl. Microbiol. Biotechnol. 2020, 104, 4027–4041. [Google Scholar] [CrossRef]
- Kessner, D.; Chambers, M.; Burke, R.; Agus, D.; Mallick, P. ProteoWizard: Open source software for rapid proteomics tools development. Bioinformatics 2008, 24, 2534–2536. [Google Scholar] [CrossRef]
- Levitsky, L.I.; Ivanov, M.V.; Lobas, A.A.; Bubis, J.A.; Tarasova, I.A.; Solovyeva, E.M.; Pridatchenko, M.L.; Gorshkov, M.V. IdentiPy: An extensible search engine for protein identification in shotgun proteomics. J. Proteome Res. 2018, 17, 2249–2255. [Google Scholar] [CrossRef]
- Elias, J.E.; Gygi, S.P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 2007, 4, 207–214. [Google Scholar] [CrossRef]
- Ivanov, M.V.; Levitsky, L.I.; Bubis, J.A.; Gorshkov, M.V. Scavager: A versatile postsearch validation algorithm for shotgun proteomics based on gradient boosting. Proteomics 2019, 19, e1800280. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Pirmoradian, M.; Zubarev, R.; Kall, L. Covariation of peptide abundances accurately reflects protein concentration differences. Mol. Cell. Proteom. 2017, 16, 936–948. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Kazakova, E.M.; Solovyeva, E.M.; Levitsky, L.I.; Bubis, J.A.; Emekeeva, D.D.; Antonets, A.A.; Nazarov, A.A.; Gorshkov, M.V.; Tarasova, I.A. Proteomics-based scoring of cellular response to stimuli for improved characterization of signaling pathway activity. Proteomics 2022, 23, e2200275. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Gietz, R.D.; Sugino, A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 1988, 74, 527–534. [Google Scholar] [CrossRef]
- Joska, T.M.; Mashruwala, A.; Boyd, J.M.; Belden, W.J. A universal cloning method based on yeast homologous recombination that is simple, efficient, and versatile. J. Microbiol. Methods 2014, 100, 46–51. [Google Scholar] [CrossRef]
- Gietz, R.D.; Schiestl, R.H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007, 2, 31–34. [Google Scholar] [CrossRef]
- Kushnirov, V.V. Rapid and reliable protein extraction from yeast. Yeast 2000, 16, 857–860. [Google Scholar] [CrossRef]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef]
- Mayan, M.D. Drug-induced permeabilization of S. cerevisiae. Curr. Protoc. Mol. Biol. 2010, 92, 13.2B.1–13.2B.4. [Google Scholar] [CrossRef] [PubMed]
- Bader, G.D.; Hogue, C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2. [Google Scholar] [CrossRef] [PubMed]
- Jorda, T.; Puig, S. Regulation of ergosterol biosynthesis in Saccharomyces cerevisiae. Genes 2020, 11, 795. [Google Scholar] [CrossRef]
- Zencir, S.; Dilg, D.; Rueda, M.P.; Shore, D.; Albert, B. Mechanisms coordinating ribosomal protein gene transcription in response to stress. Nucleic Acids Res. 2020, 48, 11408–11420. [Google Scholar] [CrossRef]
- Domkin, V.; Thelander, L.; Chabes, A. Yeast DNA damage-inducible Rnr3 has a very low catalytic activity strongly stimulated after the formation of a cross-talking Rnr1/Rnr3 complex. J. Biol. Chem. 2002, 277, 18574–18578. [Google Scholar] [CrossRef] [PubMed]
- Elledge, S.J.; Davis, R.W. Two genes differentially regulated in the cell cycle and by DNA-damaging agents encode alternative regulatory subunits of ribonucleotide reductase. Genes Dev. 1990, 4, 740–751. [Google Scholar] [CrossRef]
- Corcoles-Saez, I.; Ferat, J.L.; Costanzo, M.; Boone, C.M.; Cha, R.S. Functional link between mitochondria and Rnr3, the minor catalytic subunit of yeast ribonucleotide reductase. Microb. Cell 2019, 6, 286–294. [Google Scholar] [CrossRef] [PubMed]
- Maicher, A.; Kupiec, M. Rnr1’s role in telomere elongation cannot be replaced by Rnr3: A role beyond dNTPs? Curr. Genet. 2018, 64, 547–550. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.R.; Tsou, B.; Hu, S.; Ma, H.; Liu, X.; Yen, Y.; Ann, D.K. Autophagy induction causes a synthetic lethal sensitization to ribonucleotide reductase inhibition in breast cancer cells. Oncotarget 2016, 7, 1984–1999. [Google Scholar] [CrossRef] [PubMed]
- Musialek, M.W.; Rybaczek, D. Hydroxyurea-The Good, the Bad and the Ugly. Genes 2021, 12, 1096. [Google Scholar] [CrossRef]
- Chabes, A.; Domkin, V.; Thelander, L. Yeast Sml1, a protein inhibitor of ribonucleotide reductase. J. Biol. Chem. 1999, 274, 36679–36683. [Google Scholar] [CrossRef]
- Sluder, I.T.; Nitika; Knighton, L.E.; Truman, A.W. The Hsp70 co-chaperone Ydj1/HDJ2 regulates ribonucleotide reductase activity. PLoS Genet. 2018, 14, e1007462. [Google Scholar] [CrossRef]
- Elledge, S.J.; Davis, R.W. DNA damage induction of ribonucleotide reductase. Mol. Cell. Biol. 1989, 9, 4932–4940. [Google Scholar] [CrossRef]
- Tsaponina, O.; Barsoum, E.; Astrom, S.U.; Chabes, A. Ixr1 is required for the expression of the ribonucleotide reductase Rnr1 and maintenance of dNTP pools. PLoS Genet. 2011, 7, e1002061. [Google Scholar] [CrossRef]
- Endo-Ichikawa, Y.; Kohno, H.; Furukawa, T.; Ueda, T.; Ogawa, Y.; Tokunaga, R.; Taketani, S. Requirement of multiple DNA-protein interactions for inducible expression of RNR3 gene in Saccharomyces cerevisiae in response to DNA damage. Biochem. Biophys. Res. Commun. 1996, 222, 280–286. [Google Scholar] [CrossRef]
- Filatov, D.; Bjorklund, S.; Johansson, E.; Thelander, L. Induction of the mouse ribonucleotide reductase R1 and R2 genes in response to DNA damage by UV light. J. Biol. Chem. 1996, 271, 23698–23704. [Google Scholar] [CrossRef]
- Sabouri, N.; Viberg, J.; Goyal, D.K.; Johansson, E.; Chabes, A. Evidence for lesion bypass by yeast replicative DNA polymerases during DNA damage. Nucleic Acids Res. 2008, 36, 5660–5667. [Google Scholar] [CrossRef]
- Lee, Y.D.; Elledge, S.J. Control of ribonucleotide reductase localization through an anchoring mechanism involving Wtm1. Genes Dev. 2006, 20, 334–344. [Google Scholar] [CrossRef]
- Chabes, A.; Georgieva, B.; Domkin, V.; Zhao, X.; Rothstein, R.; Thelander, L. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell 2003, 112, 391–401. [Google Scholar] [CrossRef]
- Zhao, X.; Muller, E.G.; Rothstein, R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 1998, 2, 329–340. [Google Scholar] [CrossRef]
- Meurisse, J.; Bacquin, A.; Richet, N.; Charbonnier, J.B.; Ochsenbein, F.; Peyroche, A. Hug1 is an intrinsically disordered protein that inhibits ribonucleotide reductase activity by directly binding Rnr2 subunit. Nucleic Acids Res. 2014, 42, 13174–13185. [Google Scholar] [CrossRef]
- Aliabadi, F.; Sohrabi, B.; Mostafavi, E.; Pazoki-Toroudi, H.; Webster, T.J. Ubiquitin-proteasome system and the role of its inhibitors in cancer therapy. Open Biol. 2021, 11, 200390. [Google Scholar] [CrossRef]
- Ito, S. Proteasome Inhibitors for the Treatment of Multiple Myeloma. Cancers 2020, 12, 265. [Google Scholar] [CrossRef]
- Pu, J.J.; Savani, M.; Huang, N.; Epner, E.M. Mantle cell lymphoma management trends and novel agents: Where are we going? Ther. Adv. Hematol. 2022, 13, 20406207221080743. [Google Scholar] [CrossRef]
- Sin, C.F.; Man, P.M. The Role of proteasome inhibitors in treating acute lymphoblastic leukaemia. Front. Oncol. 2021, 11, 802832. [Google Scholar] [CrossRef]
- Van Stiphout, C.M.; Luu, A.K.; Viloria-Petit, A.M. Proteasome inhibitors and their potential applicability in osteosarcoma treatment. Cancers 2022, 14, 4544. [Google Scholar] [CrossRef]
- San Miguel, J.F.; Schlag, R.; Khuageva, N.K.; Dimopoulos, M.A.; Shpilberg, O.; Kropff, M.; Spicka, I.; Petrucci, M.T.; Palumbo, A.; Samoilova, O.S.; et al. Bortezomib plus melphalan and prednisone for initial treatment of multiple myeloma. N. Engl. J. Med. 2008, 359, 906–917. [Google Scholar] [CrossRef]
- Caponigro, F.; Lacombe, D.; Twelves, C.; Bauer, J.; Govaerts, A.S.; Marreaud, S.; Milano, A.; Anthoney, A. An EORTC phase I study of Bortezomib in combination with oxaliplatin, leucovorin and 5-fluorouracil in patients with advanced colorectal cancer. Eur. J. Cancer 2009, 45, 48–55. [Google Scholar] [CrossRef]
- Zhan, Y.; Jiang, L.; Jin, X.; Ying, S.; Wu, Z.; Wang, L.; Yu, W.; Tong, J.; Zhang, L.; Lou, Y.; et al. Inhibiting RRM2 to enhance the anticancer activity of chemotherapy. Biomed. Pharmacother. 2021, 133, 110996. [Google Scholar] [CrossRef]
- Wakisaka, N.; Yoshizaki, T.; Raab-Traub, N.; Pagano, J.S. Ribonucleotide reductase inhibitors enhance cidofovir-induced apoptosis in EBV-positive nasopharyngeal carcinoma xenografts. Int. J. Cancer 2005, 116, 640–645. [Google Scholar] [CrossRef]
- Xie, Y.; Wang, Y.; Xu, Z.; Lu, Y.; Song, D.; Gao, L.; Yu, D.; Li, B.; Chen, G.; Zhang, H.; et al. Preclinical validation and phase I trial of 4-hydroxysalicylanilide, targeting ribonucleotide reductase mediated dNTP synthesis in multiple myeloma. J. Biomed. Sci. 2022, 29, 32. [Google Scholar] [CrossRef]




| Name of the Strain | Genotype | Source |
|---|---|---|
| BY4742 | MAT α; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0 | Euroscarf, Oberursel, Germany |
| YPL | BY4742 derivative MAT α; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0, pre1–8 | [11] |
| WT△sml1 | BY4741 YML058W::LEU2 | This work |
| WT△ydj1 | BY4741 YNL064C::LEU2 | –//– |
| YPL△sml1 | BY4742 YML058W::LEU2 | –//– |
| YPL△ydj1 | YPL YNL064C::LEU2 | –//– |
| Protein Name | Log2FC | −Log10FDR 1 |
|---|---|---|
| RNR1 | 1.52 | 2.16 |
| RNR3 | 1.57 | 3.38 |
| MCM3 | 1.36 | 1.74 |
| MCM6 | 1.23 | 2.55 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spasskaya, D.S.; Kulagin, K.A.; Grineva, E.N.; Osipova, P.J.; Poddubko, S.V.; Bubis, J.A.; Kazakova, E.M.; Kusainova, T.T.; Gorshkov, V.A.; Kjeldsen, F.; et al. Yeast Ribonucleotide Reductase Is a Direct Target of the Proteasome and Provides Hyper Resistance to the Carcinogen 4-NQO. J. Fungi 2023, 9, 351. https://doi.org/10.3390/jof9030351
Spasskaya DS, Kulagin KA, Grineva EN, Osipova PJ, Poddubko SV, Bubis JA, Kazakova EM, Kusainova TT, Gorshkov VA, Kjeldsen F, et al. Yeast Ribonucleotide Reductase Is a Direct Target of the Proteasome and Provides Hyper Resistance to the Carcinogen 4-NQO. Journal of Fungi. 2023; 9(3):351. https://doi.org/10.3390/jof9030351
Chicago/Turabian StyleSpasskaya, Daria S., Kirill A. Kulagin, Evgenia N. Grineva, Pamila J. Osipova, Svetlana V. Poddubko, Julia A. Bubis, Elizaveta M. Kazakova, Tomiris T. Kusainova, Vladimir A. Gorshkov, Frank Kjeldsen, and et al. 2023. "Yeast Ribonucleotide Reductase Is a Direct Target of the Proteasome and Provides Hyper Resistance to the Carcinogen 4-NQO" Journal of Fungi 9, no. 3: 351. https://doi.org/10.3390/jof9030351
APA StyleSpasskaya, D. S., Kulagin, K. A., Grineva, E. N., Osipova, P. J., Poddubko, S. V., Bubis, J. A., Kazakova, E. M., Kusainova, T. T., Gorshkov, V. A., Kjeldsen, F., Karpov, V. L., Tarasova, I. A., & Karpov, D. S. (2023). Yeast Ribonucleotide Reductase Is a Direct Target of the Proteasome and Provides Hyper Resistance to the Carcinogen 4-NQO. Journal of Fungi, 9(3), 351. https://doi.org/10.3390/jof9030351