The Early Endocytosis Gene PAL1 Contributes to Stress Tolerance and Hyphal Formation in Candida albicans
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of the C. albicans Ortholog of PAL1
2.2. Deletion of C. albicans Pal1
2.3. Preparation of Genomic DNA and Plasmid Isolation
2.4. Strains, Media, and Cell Culture
2.5. Cell Growth Assay and Assays for Response to Environmental Stress and Filamentation
2.6. Fluorescence Microscopy of C. albicans Strains
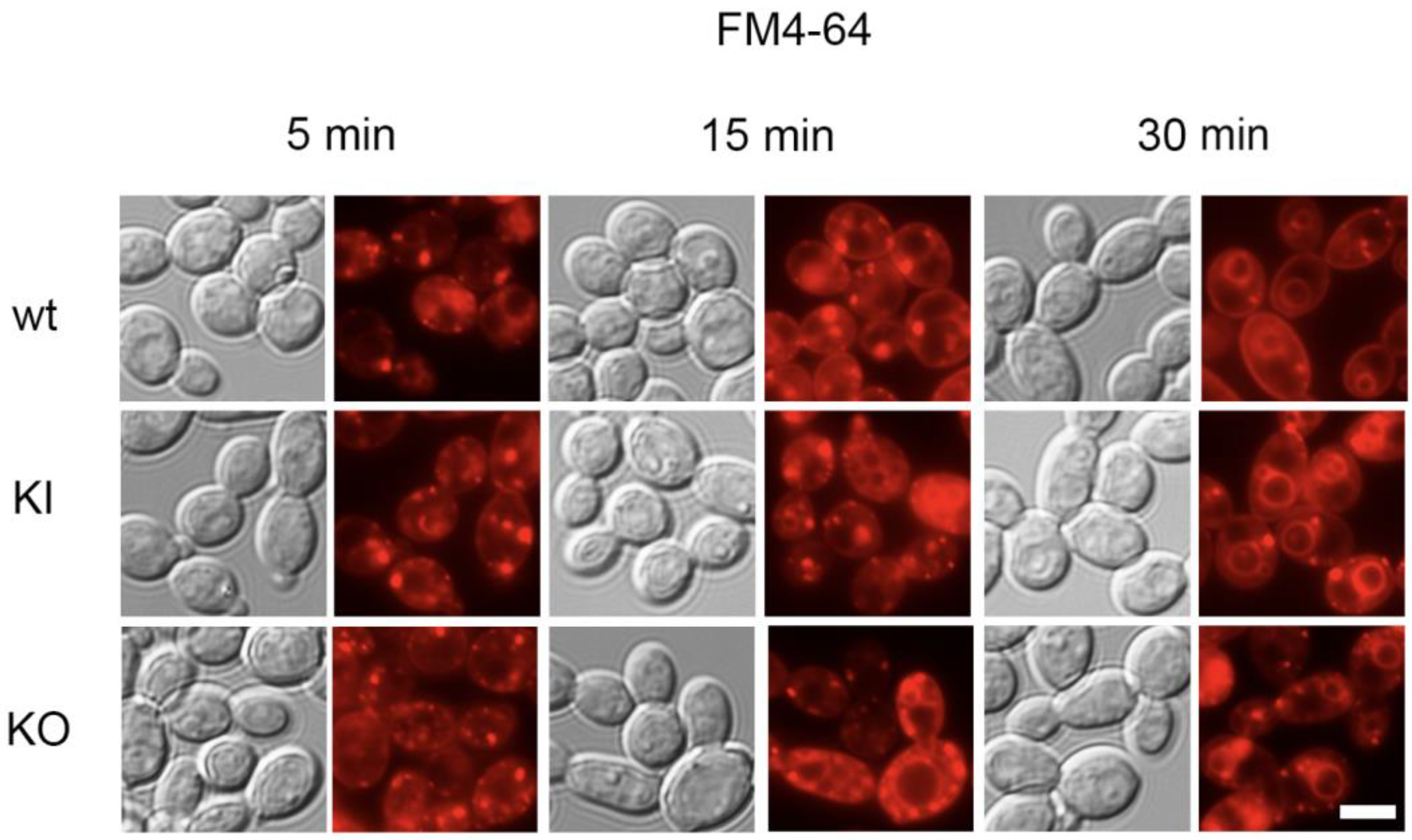
2.7. Analyses of Endocytosis with FM4-64
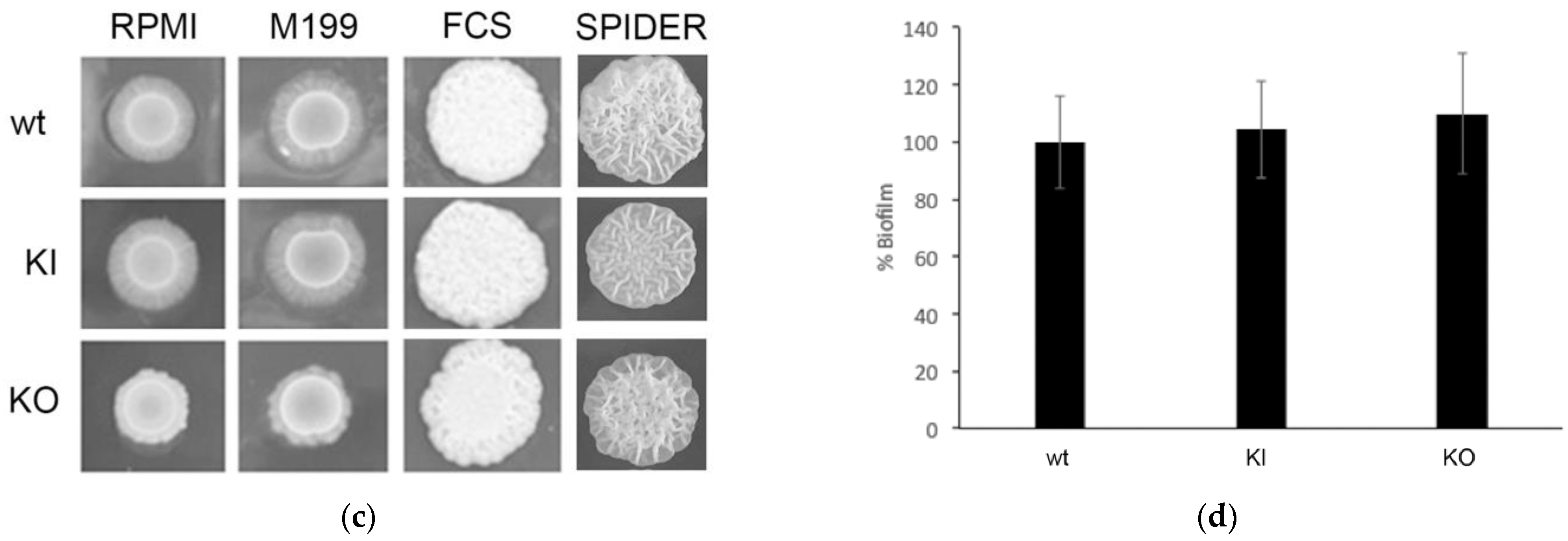
2.8. Analysis of Biofilm Formation in C. albicans Endocytosis Mutants
2.9. Immunofluorescence Microscopy of Co-Cultured VK-2 Cells with C. albicans Strains
2.10. Western Blotting and Detection of E-Cadherin
2.11. Live/Dead Viability Assay
3. Results
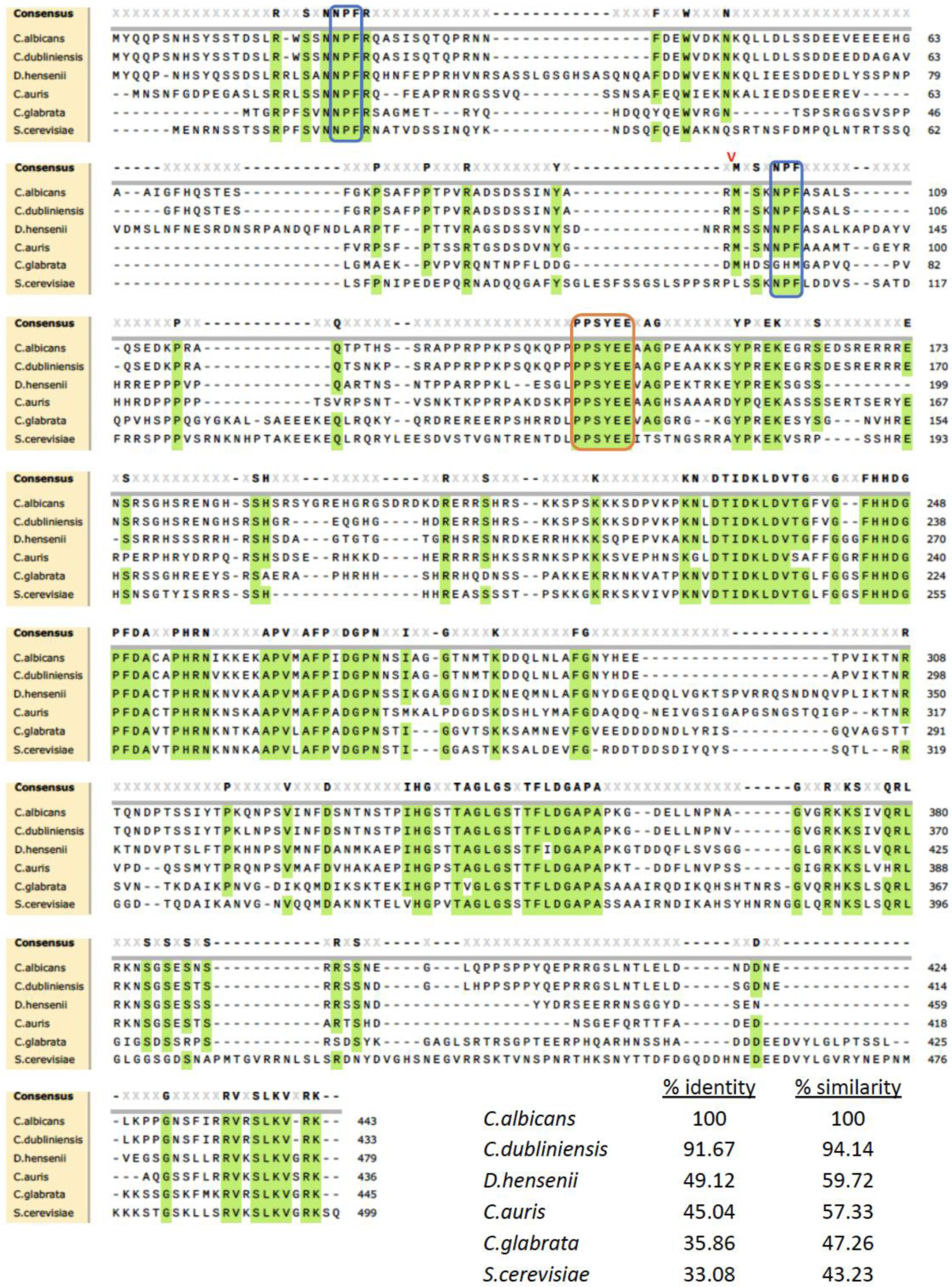
3.1. Identification and Comparison of S. cerevisiae PAL1 and C. albicans C3_01890C
3.2. Construction of C. albicans pal1Δ/Δ Mutant and Re-Integrant Strains
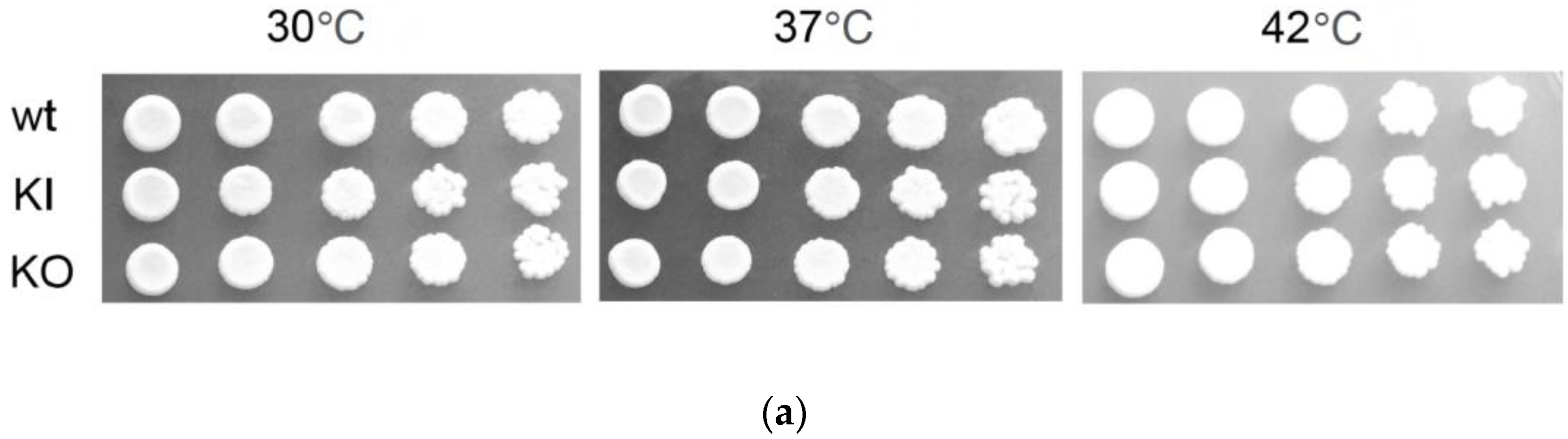
3.3. Contribution of C. albicans PAL1 to Growth and Viability
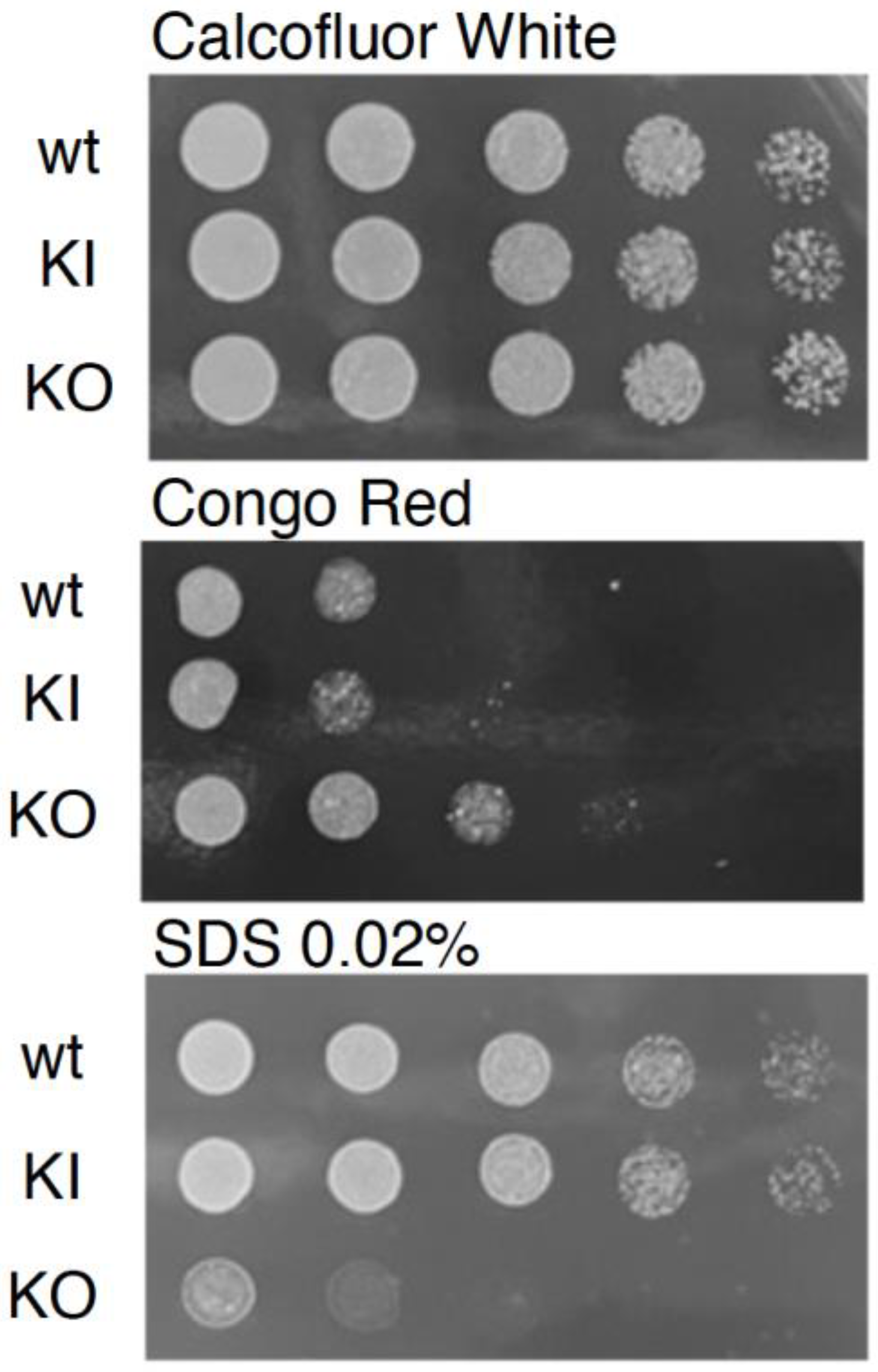
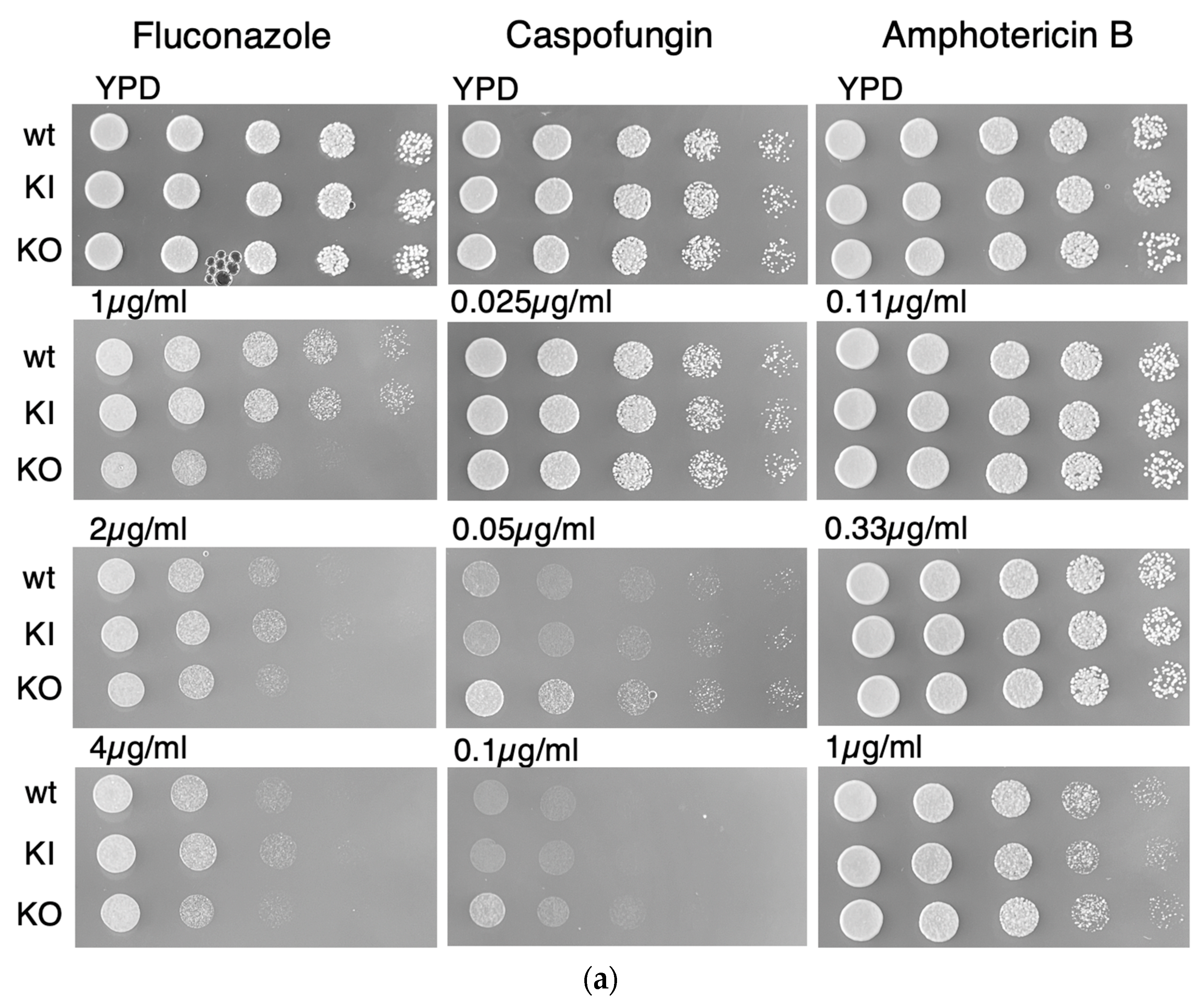
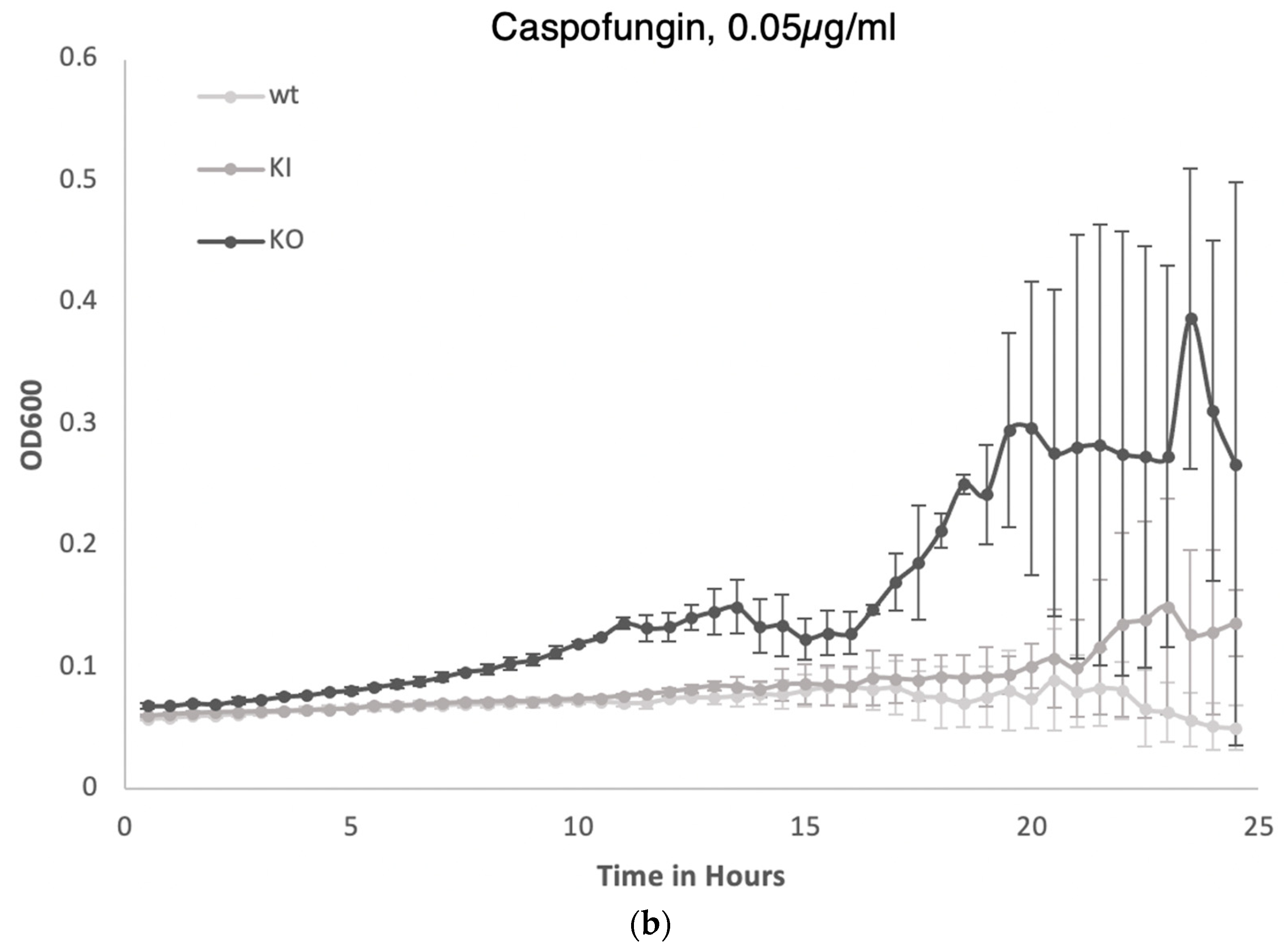
3.4. C. albicans PAL1 Affects Stress Tolerance
3.5. Membrane-Related Endocytosis Is Intact in the C. albicans pal1Δ/Δ Mutant
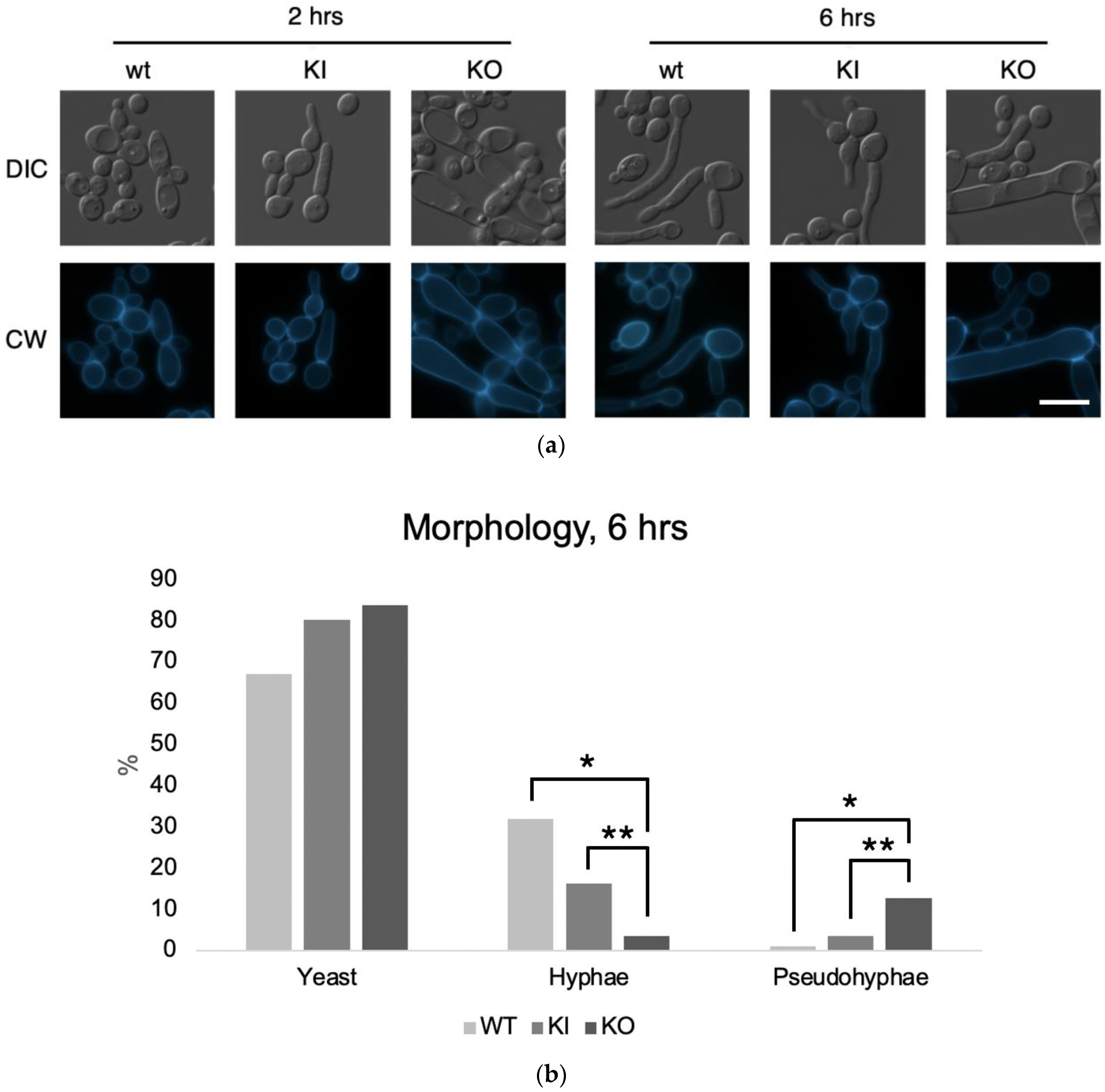
3.6. C. albicans PAL1 Is Required for Wild-Type Filamentation
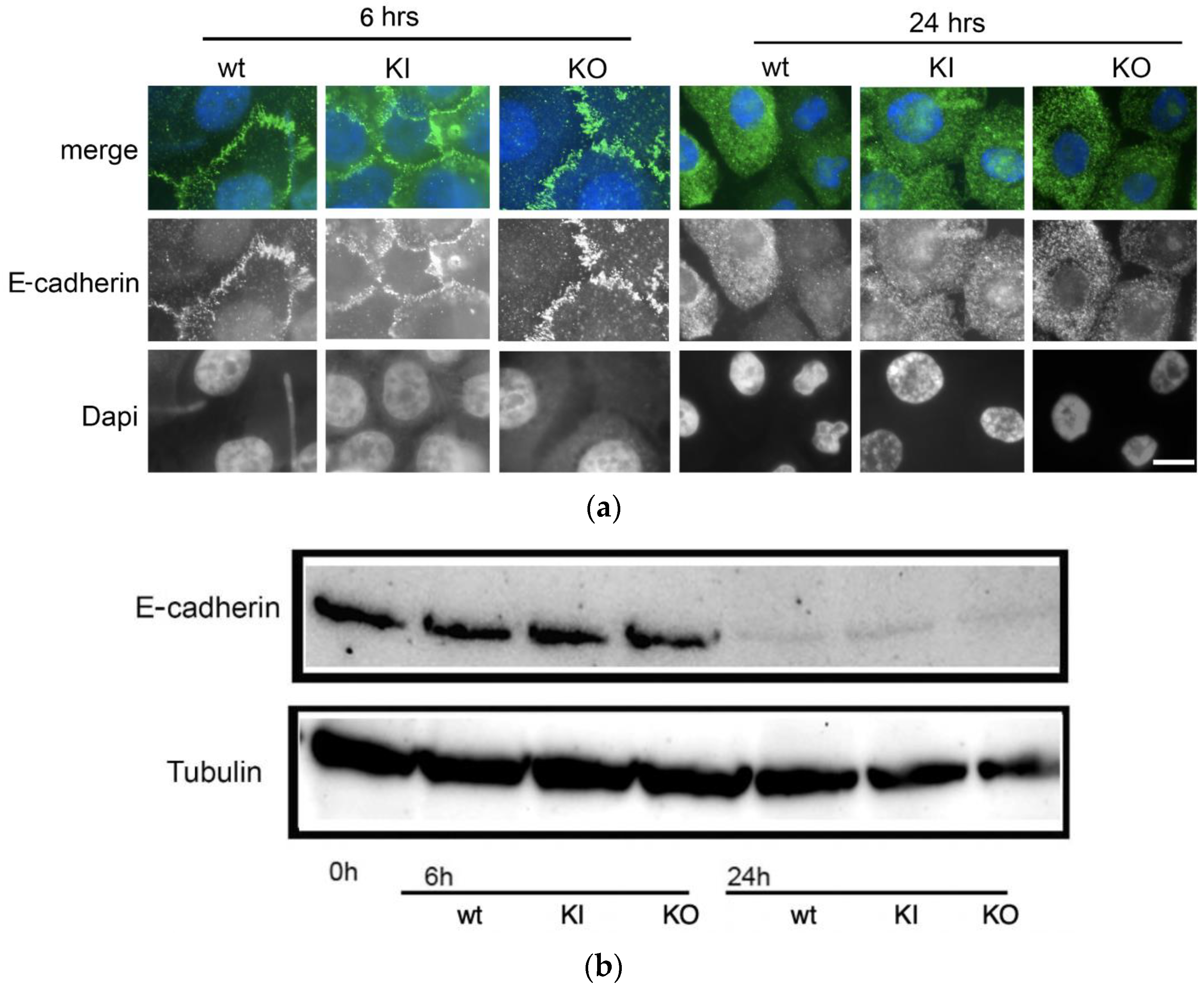
3.7. Dissolution of Cell–Cell Adhesions Is Unaffected in the C. albicans pal1Δ/Δ Mutant
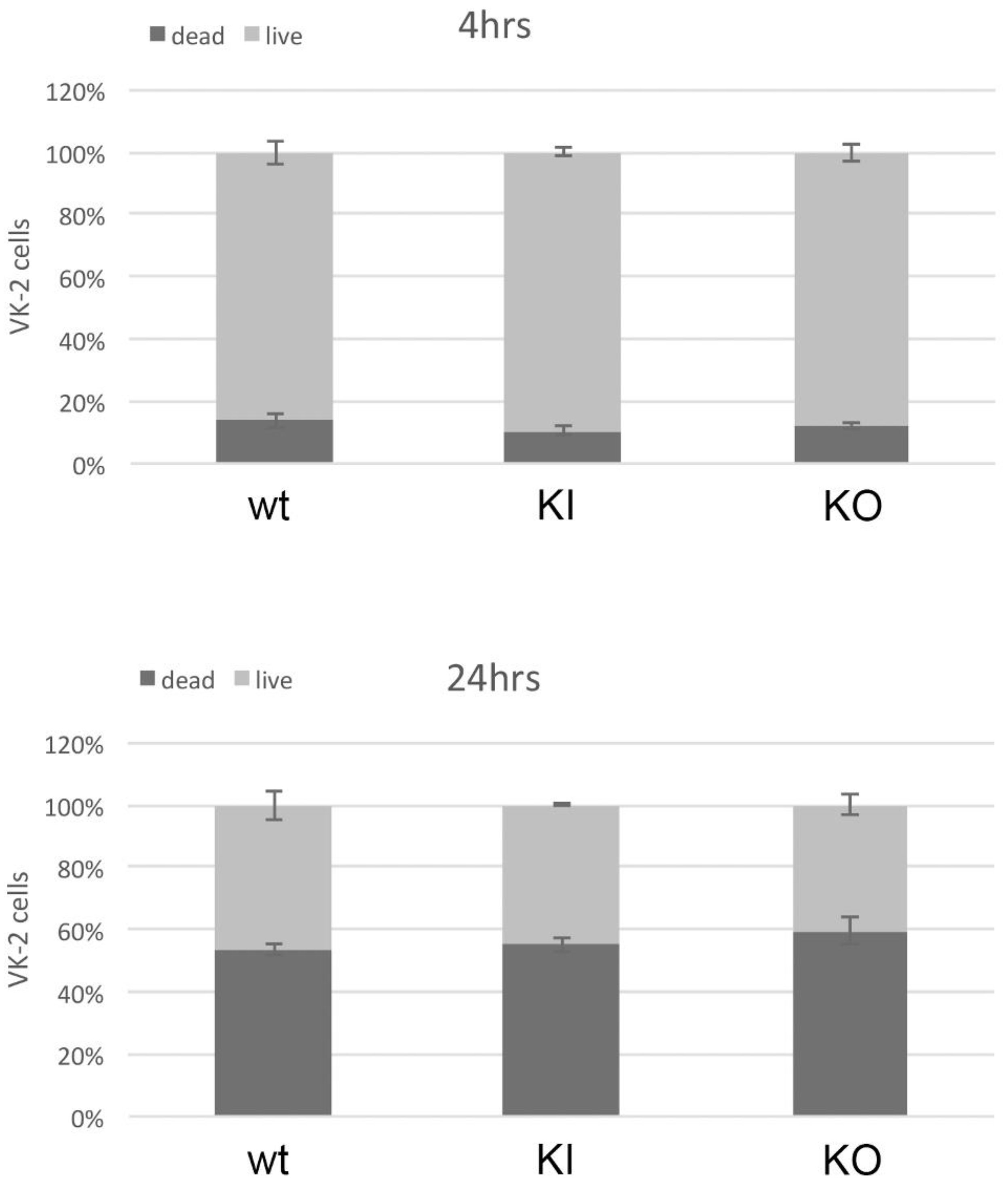
3.8. Ability to Kill Host Cells Does Not Diminish in the C. albicans pal1Δ/Δ Mutant
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wächtler, B.; Citiulo, F.; Jablonowski, N.; Förster, S.; Dalle, F.; Schaller, M.; Wilson, D.; Hube, B. Candida albicans-Epithelial Interactions: Dissecting the Roles of Active Penetration, Induced Endocytosis and Host Factors on the Infection Process. PLoS ONE 2012, 7, e36952. [Google Scholar] [CrossRef]
- Rollenhagen, C.; Mamtani, S.; Ma, D.; Dixit, R.; Eszterhas, S.; Lee, S.A. The Role of Secretory Pathways in Candida albicans Pathogenesis. J. Fungi 2020, 6, 26. [Google Scholar] [CrossRef]
- Goode, B.L.; Eskin, J.A.; Wendland, B. Actin and Endocytosis in Budding Yeast. Genetics 2015, 199, 315–358. [Google Scholar] [CrossRef] [PubMed]
- Tsui, C.; Kong, E.F.; Jabra-Rizk, M.A. Pathogenesis of Candida albicans Biofilm. Pathog. Dis. 2016, 74, ftw018. [Google Scholar] [CrossRef] [PubMed]
- Schaller, M.; Korting, H.C.; Schäfer, W.; Bastert, J.; Chen, W.; Hube, B. Secreted Aspartic Proteinase (Sap) Activity Contributes to Tissue Damage in a Model of Human Oral Candidosis. Mol. Microbiol. 1999, 34, 169–180. [Google Scholar] [CrossRef]
- Bernardo, S.M.; Khalique, Z.; Kot, J.; Jones, J.K.; Lee, S.A. Candida albicans VPS1 Contributes to Protease Secretion, Filamentation, and Biofilm Formation. Fungal Genet. Biol. 2008, 45, 861–877. [Google Scholar] [CrossRef]
- Lee, S.A.; Jones, J.; Hardison, S.; Kot, J.; Khalique, Z.; Bernardo, S.M.; Lazzell, A.; Monteagudo, C.; Lopez-Ribot, J. Candida albicans VPS4 Is Required for Secretion of Aspartyl Proteases and in Vivo Virulence. Mycopathologia 2009, 167, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Palanisamy, S.K.A.; Ramirez, M.A.; Lorenz, M.; Lee, S.A. Candida albicans PEP12 Is Required for Biofilm Integrity and In Vivo Virulence. Eukaryot. Cell 2010, 9, 266–277. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rollenhagen, C.; Agyeman, H.; Eszterhas, S.; Lee, S.A. Candida albicans END3 Mediates Endocytosis and Has Subsequent Roles in Cell Wall Integrity, Morphological Switching, and Tissue Invasion. Microbiol. Spectr. 2022, 10, e01880-21. [Google Scholar] [CrossRef]
- Rollenhagen, C.; Agyeman, H.; Eszterhas, S.; Lee, S.A. Candida albicans ENT2 Contributes to Efficient Endocytosis, Cell Wall Integrity, Filamentation, and Virulence. mSphere 2021, 6, e0070721. [Google Scholar] [CrossRef]
- Moreno-Ruiz, E.; Galán-Díez, M.; Zhu, W.; Fernández-Ruiz, E.; d’Enfert, C.; Filler, S.G.; Cossart, P.; Veiga, E. Candida albicans Internalization by Host Cells Is Mediated by a Clathrin-Dependent Mechanism. Cell Microbiol. 2009, 11, 1179–1189. [Google Scholar] [CrossRef]
- Carroll, S.Y.; Stimpson, H.E.M.; Weinberg, J.; Toret, C.P.; Sun, Y.; Drubin, D.G. Analysis of Yeast Endocytic Site Formation and Maturation through a Regulatory Transition Point. Mol. Biol. Cell 2012, 23, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Stimpson, H.E.M.; Toret, C.P.; Cheng, A.T.; Pauly, B.S.; Drubin, D.G. Early-Arriving Syp1p and Ede1p Function in Endocytic Site Placement and Formation in Budding Yeast. Mol. Biol. Cell 2009, 20, 4640–4651. [Google Scholar] [CrossRef] [PubMed]
- Kaksonen, M.; Toret, C.P.; Drubin, D.G. A Modular Design for the Clathrin- and Actin-Mediated Endocytosis Machinery. Cell 2005, 123, 305–320. [Google Scholar] [CrossRef]
- Wendland, B.; Steece, K.E.; Emr, S.D. Yeast Epsins Contain an Essential N-Terminal ENTH Domain, Bind Clathrin and Are Required for Endocytosis. EMBO J. 1999, 18, 4383–4393. [Google Scholar] [CrossRef] [PubMed]
- Wendland, B.; Emr, S.D. Pan1p, Yeast Eps15, Functions as a Multivalent Adaptor That Coordinates Protein–Protein Interactions Essential for Endocytosis. J. Cell Biol. 1998, 141, 71–84. [Google Scholar] [CrossRef]
- Sun, Y.; Leong, N.T.; Wong, T.; Drubin, D.G. A Pan1/End3/Sla1 Complex Links Arp2/3-Mediated Actin Assembly to Sites of Clathrin-Mediated Endocytosis. Mol. Biol. Cell 2015, 26, 3841–3856. [Google Scholar] [CrossRef]
- Feliciano, D.; Di Pietro, S.M. SLAC, a Complex between Sla1 and Las17, Regulates Actin Polymerization during Clathrin-Mediated Endocytosis. Mol. Biol. Cell 2012, 23, 4256–4272. [Google Scholar] [CrossRef]
- Bénédetti, H.; Raths, S.; Crausaz, F.; Riezman, H. The END3 Gene Encodes a Protein That Is Required for the Internalization Step of Endocytosis and for Actin Cytoskeleton Organization in Yeast. Mol. Biol. Cell 1994, 5, 1023–1037. [Google Scholar] [CrossRef]
- Tang, H.Y.; Cai, M. The EH-Domain-Containing Protein Pan1 Is Required for Normal Organization of the Actin Cytoskeleton in Saccharomyces cerevisiae. Mol. Cell Biol. 1996, 16, 4897–4914. [Google Scholar] [CrossRef]
- Sun, Y.; Leong, N.T.; Jiang, T.; Tangara, A.; Darzacq, X.; Drubin, D.G. Switch-like Arp2/3 Activation upon WASP and WIP Recruitment to an Apparent Threshold Level by Multivalent Linker Proteins in Vivo. eLife 2017, 6, e29140. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.; Hellwig, D.; Schaub, Y.; Bauer, J.; Walther, A.; Wendland, J. Functional Analysis of Candida albicans Genes Whose Saccharomyces cerevisiae Homologues Are Involved in Endocytosis. Yeast 2007, 24, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Walther, A.; Wendland, J. Polarized Hyphal Growth in Candida albicans Requires the Wiskott-Aldrich Syndrome Protein Homolog Wal1p. Eukaryot. Cell 2004, 3, 471–482. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ge, W.; Chew, T.G.; Wachtler, V.; Naqvi, S.N.; Balasubramanian, M.K. The Novel Fission Yeast Protein Pal1p Interacts with Hip1-Related Sla2p/End4p and Is Involved in Cellular Morphogenesis. Mol. Biol. Cell 2005, 16, 4124–4138. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.F.; Waterman, M.S. Identification of Common Molecular Subsequences. J. Mol. Biol. 1981, 147, 195–197. [Google Scholar] [CrossRef]
- Schultz, J.; Milpetz, F.; Bork, P.; Ponting, C.P. SMART, a Simple Modular Architecture Research Tool: Identification of Signaling Domains. Proc. Natl. Acad. Sci. USA 1998, 95, 5857–5864. [Google Scholar] [CrossRef]
- Nguyen, N.; Quail, M.M.F.; Hernday, A.D. An Efficient, Rapid, and Recyclable System for CRISPR-Mediated Genome Editing in Candida albicans. mSphere 2017, 2, e00149-17. [Google Scholar] [CrossRef]
- Liu, H.; Köhler, J.; Fink, G.R. Suppression of Hyphal Formation in Candida albicans by Mutation of a STE12 Homolog. Science 1994, 266, 1723–1726. [Google Scholar] [CrossRef]
- Pringle, J.R.; Adams, A.E.; Drubin, D.G.; Haarer, B.K. Immunofluorescence Methods for Yeast. Methods Enzymol. 1991, 194, 565–602. [Google Scholar] [CrossRef]
- Ramage, G.; López-Ribot, J.L. Techniques for Antifungal Susceptibility Testing of Candida albicans Biofilms. Methods Mol. Med. 2005, 118, 71–79. [Google Scholar] [CrossRef]
- Rollenhagen, C.; Wöllert, T.; Langford, G.M.; Sundstrom, P. Stimulation of Cell Motility and Expression of Late Markers of Differentiation in Human Oral Keratinocytes by Candida albicans. Cell Microbiol. 2009, 11, 946–966. [Google Scholar] [CrossRef]
- Kurien, B.T.; Scofield, R.H. Western Blotting. Methods 2006, 38, 283–293. [Google Scholar] [CrossRef]
- C. Albicans C3_01890C Summary. Available online: http://www.candidagenome.org/cgi-bin/locus.pl?locus=C3_01890C_A (accessed on 1 June 2020).
- de Beer, T.; Hoofnagle, A.N.; Enmon, J.L.; Bowers, R.C.; Yamabhai, M.; Kay, B.K.; Overduin, M. Molecular Mechanism of NPF Recognition by EH Domains. Nat. Struct. Biol. 2000, 7, 1018–1022. [Google Scholar] [CrossRef] [PubMed]
- Letscher-Bru, V.; Herbrecht, R. Caspofungin: The First Representative of a New Antifungal Class. J. Antimicrob. Chemother. 2003, 51, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, Q.; Ma, K.; Shi, P.; Liu, W.; Huang, Z. Fluconazole Inhibits Cellular Ergosterol Synthesis to Confer Synergism with Berberine against Yeast Cells. J. Glob. Antimicrob. Resist. 2018, 13, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Vasioukhin, V.; Bauer, C.; Yin, M.; Fuchs, E. Directed Actin Polymerization Is the Driving Force for Epithelial Cell-Cell Adhesion. Cell 2000, 100, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Gumbiner, B.M. Regulation of Cadherin-Mediated Adhesion in Morphogenesis. Nat. Rev. Mol. Cell Biol. 2005, 6, 622–634. [Google Scholar] [CrossRef] [PubMed]
- Ene, I.V.; Cheng, S.-C.; Netea, M.G.; Brown, A.J.P. Growth of Candida albicans Cells on the Physiologically Relevant Carbon Source Lactate Affects Their Recognition and Phagocytosis by Immune Cells. Infect. Immun. 2013, 81, 238–248. [Google Scholar] [CrossRef]
- Malavia, D.; Lehtovirta-Morley, L.E.; Alamir, O.; Weiß, E.; Gow, N.A.R.; Hube, B.; Wilson, D. Zinc Limitation Induces a Hyper-Adherent Goliath Phenotype in Candida albicans. Front. Microbiol. 2017, 8, 2238. [Google Scholar] [CrossRef]
- Moreno-Velásquez, S.D.; Seidel, C.; Juvvadi, P.R.; Steinbach, W.J.; Read, N.D. Caspofungin-Mediated Growth Inhibition and Paradoxical Growth in Aspergillus Fumigatus Involve Fungicidal Hyphal Tip Lysis Coupled with Regenerative Intrahyphal Growth and Dynamic Changes in β-1,3-Glucan Synthase Localization. Antimicrob. Agents Chemother. 2017, 61, e00710-17. [Google Scholar] [CrossRef]
- Ram, A.F.J.; Klis, F.M. Identification of Fungal Cell Wall Mutants Using Susceptibility Assays Based on Calcofluor White and Congo Red. Nat. Protoc. 2006, 1, 2253–2256. [Google Scholar] [CrossRef]
- Matsumori, N.; Tahara, K.; Yamamoto, H.; Morooka, A.; Doi, M.; Oishi, T.; Murata, M. Direct Interaction between Amphotericin B and Ergosterol in Lipid Bilayers As Revealed by 2H NMR Spectroscopy. J. Am. Chem. Soc. 2009, 131, 11855–11860. [Google Scholar] [CrossRef]
- Warenda, A.J.; Konopka, J.B. Septin Function in Candida albicans Morphogenesis. Mol. Biol. Cell 2002, 13, 2732–2746. [Google Scholar] [CrossRef] [PubMed]
- Warenda, A.J.; Kauffman, S.; Sherrill, T.P.; Becker, J.M.; Konopka, J.B. Candida albicans Septin Mutants Are Defective for Invasive Growth and Virulence. Infect. Immun. 2003, 71, 4045–4051. [Google Scholar] [CrossRef] [PubMed]
- Gale, C.; Gerami-Nejad, M.; McClellan, M.; Vandoninck, S.; Longtine, M.S.; Berman, J. Candida albicans Int1p Interacts with the Septin Ring in Yeast and Hyphal Cells. Mol. Biol. Cell 2001, 12, 3538–3549. [Google Scholar] [CrossRef]
- Gulati, M.; Nobile, C.J. Candida albicans Biofilms: Development, Regulation, and Molecular Mechanisms. Microbes Infect. 2016, 18, 310–321. [Google Scholar] [CrossRef]
- Moyes, D.L.; Richardson, J.P.; Naglik, J.R. Candida albicans-Epithelial Interactions and Pathogenicity Mechanisms: Scratching the Surface. Virulence 2015, 6, 338–346. [Google Scholar] [CrossRef]

| Name | Sequence | Reference |
|---|---|---|
| PAL1 inner F primer | caa acc ca acca cgc aat aat | This study |
| PAL1 inner R primer | gct cac ttc act aac tca ctc tc | This study |
| PAL1 outer 1F primer | gca acg ccc acg att tta tt | This study |
| PAL1 outer 1R primer | ccg tca tta tca tct tct ccc a | This study |
| PAL1 outer 2F primer | tag cat gat gca ctt cca caa c | This study |
| PAL1 outer 2R primer | cca cag ccg tca tta tca tct t | This study |
| PAL1 outer 3F primer | ctt cca caa cgg gaa aca agt a | This study |
| PAL1 outer 3R primer | at tcc cat tca gtc gtg gg | This study |
| PAL1 outer F/donor KI primer | ctt cac ata tcc ccc gct cc | This study |
| PAL1 outer R/donor KI primer | acg att tgt tgg tgc cct tac | This study |
| PAL1 KO donor U.95 | cac ttc ttc caa tgt acc aac aac cat caa atc att cat att cgt gaa aag gga taa gac aaa acg att taa gac aat tca aac acc aaa taa gg | This study |
| PALI KO donor L.95 | cct tat ttg gtg ttt gaa ttg tct taa atc gtt ttg tct tat ccc ttt tca cga ata tga atg att tga tgg ttg ttg gta cat tgg aag aag tg | This study |
| PAL1 guide RNA oligo | cgt aaa cta ttt tta att tgc att cat caa aat tat tgc ggt ttt aga gct aga aat agc | This study |
| AHO1098 | caa att aaa aat agt tta cgc aag | [27] |
| AHO1099 | gtt tta gag cta gaa atg caa gtt | [27] |
| Name | Description | Reference |
|---|---|---|
| pADH137 | C.alb LEUpOUT CAS9 expression plasmid | [27] |
| pADH118 | C.alb LEUpOUT “blank” gRNA plasmid | [27] |
| pCRPal1 | C.alb LEUpOUT “blank” gRNA plasmid with Pal1 guide RNA | This study |
| Name | Description/Genotype | Reference |
|---|---|---|
| AHY940 | SC5314 LEU2 heterozygous knockout a/α leu2Δ/LEU2 | [27] |
| CRMYPAL1KO | SC5314 pal1 homozygous knockout a/α pal1Δ/pal1Δ | This study |
| CRMYPAL1KI | SC5314 PAL1 homozygous knock-in a/α pal1Δ/pal1Δ PAL1/PAL1 | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, M.; Ma, D.; Eszterhas, S.; Rollenhagen, C.; Lee, S.A. The Early Endocytosis Gene PAL1 Contributes to Stress Tolerance and Hyphal Formation in Candida albicans. J. Fungi 2023, 9, 1097. https://doi.org/10.3390/jof9111097
Yu M, Ma D, Eszterhas S, Rollenhagen C, Lee SA. The Early Endocytosis Gene PAL1 Contributes to Stress Tolerance and Hyphal Formation in Candida albicans. Journal of Fungi. 2023; 9(11):1097. https://doi.org/10.3390/jof9111097
Chicago/Turabian StyleYu, Miranda, Dakota Ma, Susan Eszterhas, Christiane Rollenhagen, and Samuel A. Lee. 2023. "The Early Endocytosis Gene PAL1 Contributes to Stress Tolerance and Hyphal Formation in Candida albicans" Journal of Fungi 9, no. 11: 1097. https://doi.org/10.3390/jof9111097
APA StyleYu, M., Ma, D., Eszterhas, S., Rollenhagen, C., & Lee, S. A. (2023). The Early Endocytosis Gene PAL1 Contributes to Stress Tolerance and Hyphal Formation in Candida albicans. Journal of Fungi, 9(11), 1097. https://doi.org/10.3390/jof9111097




