Characterization of an α-Amylase from the Honeybee Chalk Brood Pathogen Ascosphaera apis
Abstract
1. Introduction
2. Materials and Methods
2.1. Microorganism and Preparation of Inoculum
2.2. Amylase Assay
2.3. Experimental Design and Statistical Analyses
2.3.1. Preliminary Experiments: The Effects of Carbon, Nitrogen, and Other Factors on Production of α-Amylase by A. apis
2.3.2. Plackett–Burman Design
2.3.3. The Steepest Ascent Experiment
2.3.4. Box–Behnken Design
2.3.5. Statistical Analysis
2.4. Purification of A. apis Amylase
2.5. Electrophoresis and Molecular Weight Determination
2.6. Characterization of the A. apis Amylase
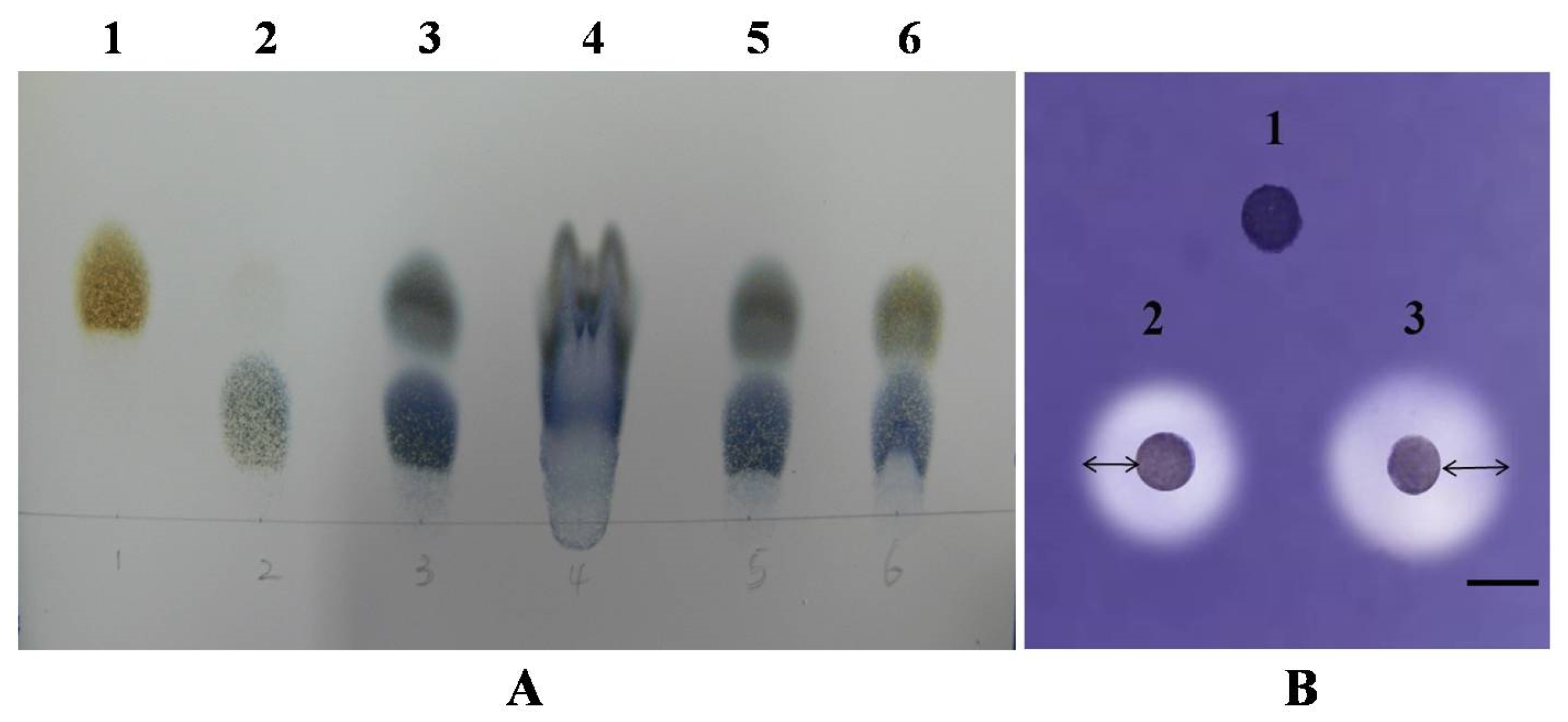
2.7. Determination of Amylase-Type and Amylase Plate Assay
2.8. Bioreactor Production of Amylase
3. Results
3.1. Preliminary Experiments: The Effects of Carbon, Nitrogen, and Other Factors on Production of α-Amylase by A. apis
3.2. Plackett–Burman Design
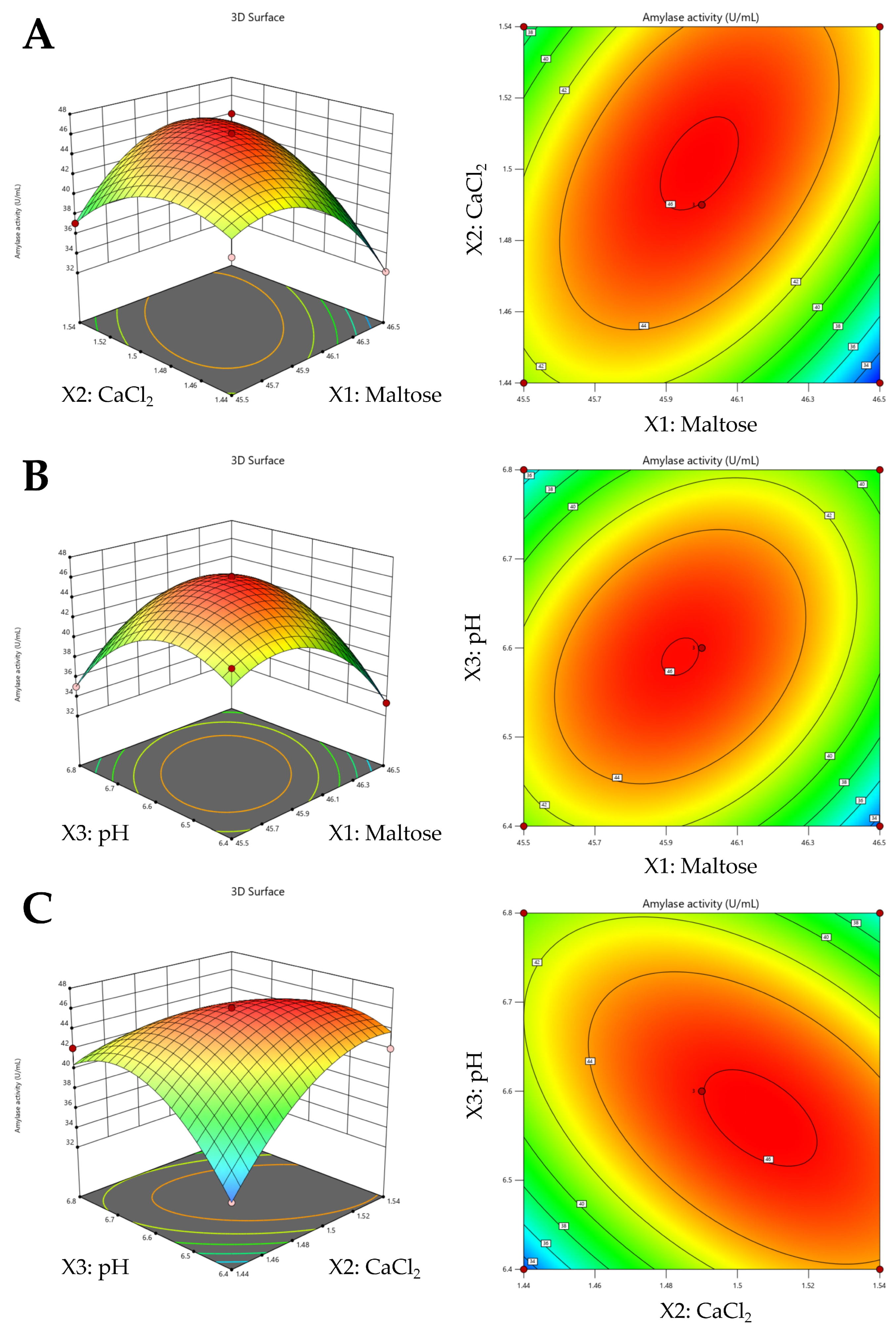
3.3. Optimization of Amylase Production by Response Surface Methodology
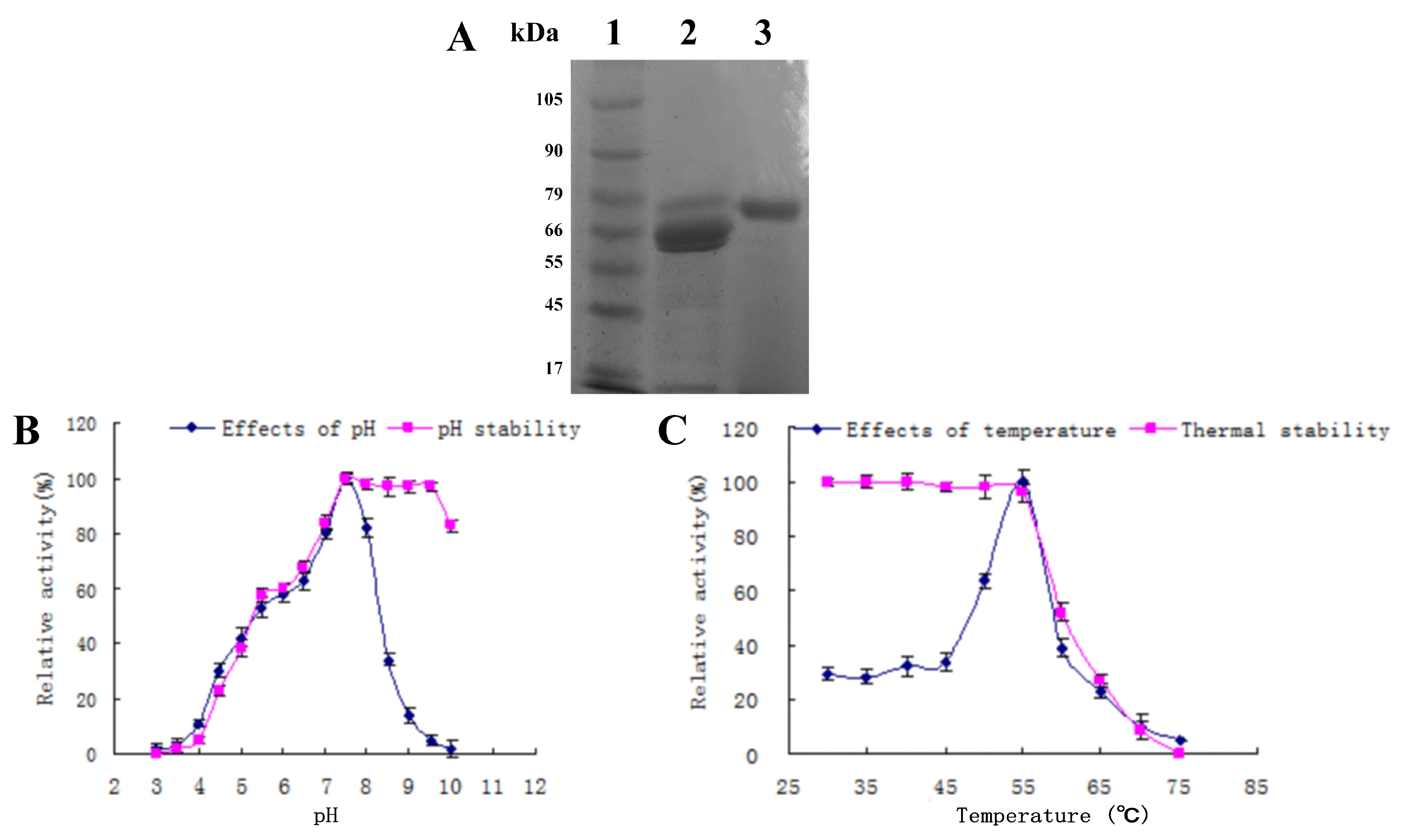
3.4. Enzyme Purification and Characterization
3.5. Characterization of the Purified Amylase
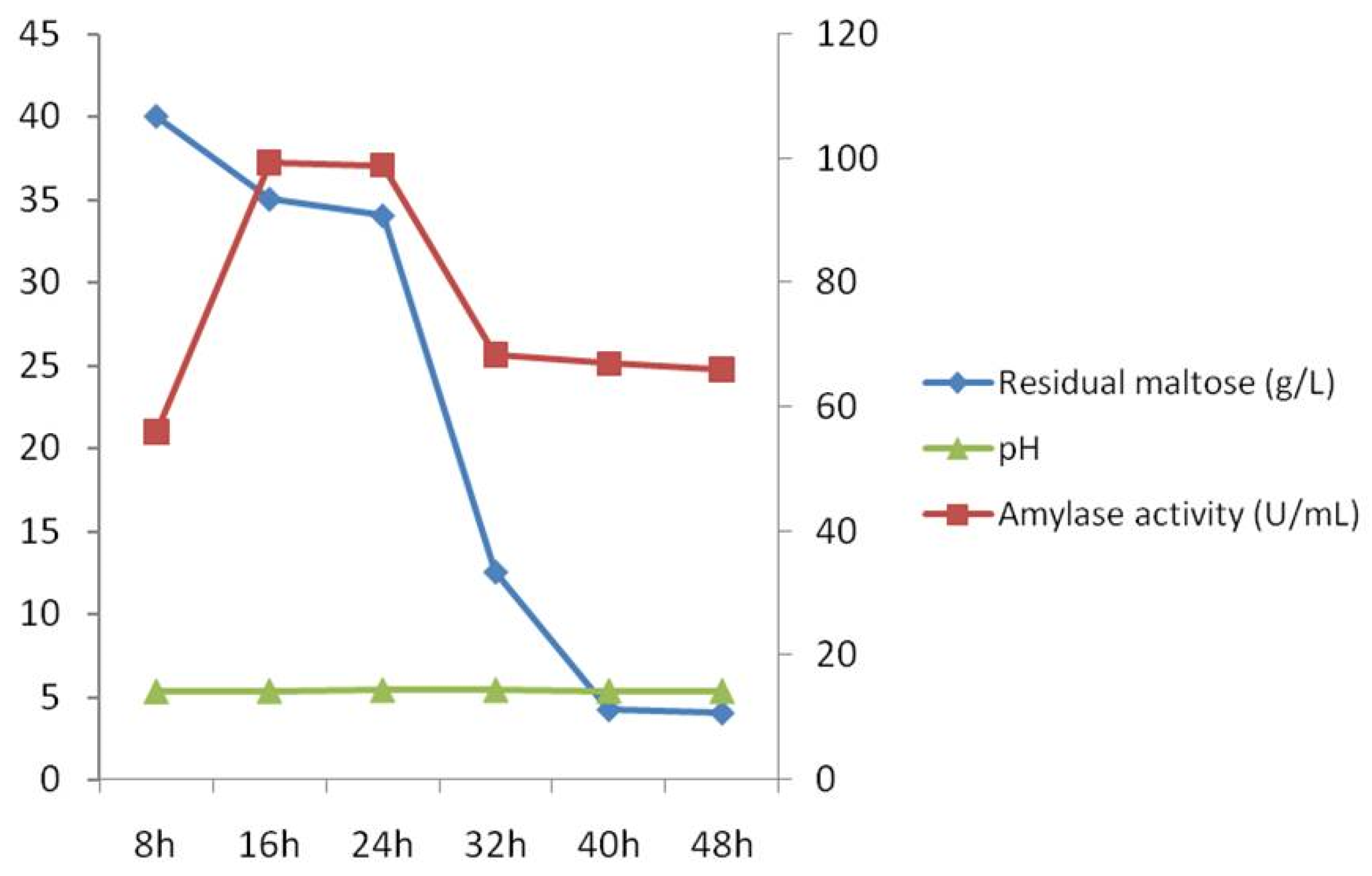
3.6. Bioreactor Production of the A. apis Amylase
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vojvodic, S.; Jensen, A.B.; James, R.R.; Boomsma, J.J.; Eilenberg, J. Temperature dependent virulence of obligate and facultative fungal pathogens of honeybee brood. Vet. Microbiol. 2011, 149, 200–205. [Google Scholar] [CrossRef]
- Krutmuang, P.; Rajula, J.; Pittarate, S.; Chatima, C.; Thungrabeab, M.; Mekchay, S.; Senthil-Nathan, S. The inhibitory action of plant extracts on the mycelial growth of Ascosphaera apis, the causative agent of chalkbrood disease in Honey bee. Toxicol. Rep. 2022, 9, 713–719. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Urquiza, A.; Keyhani, N.O. Action on the Surface: Entomopathogenic Fungi versus the Insect Cuticle. Insects 2013, 4, 357–374. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, Y.; Keyhani, N.O.; Zhu, S.; Wang, J.; Wang, J.; Jin, D.; Fan, Y. A perilipin affects lipid droplet homeostasis and aerial hyphal growth, but has only small effects on virulence in the insect pathogenic fungus Beauveria bassiana. J. Fungi 2022, 8, 634. [Google Scholar] [CrossRef] [PubMed]
- Aynalem, T.; Meng, L.; Getachew, A.; Wu, J.; Yu, H.; Tan, J.; Li, N.; Xu, S. StcU-2 gene mutation via CRISPR/Cas9 leads to misregulation of spore-cyst formation in Ascosphaera apis. Microorganisms 2022, 10, 2088. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Hou, M.; Qiu, Y.; Zhao, B.; Nie, H.; Su, S. Changes in antioxidant enzymes activity and metabolomic profiles in the guts of honey bee (Apis mellifera) larvae infected with Ascosphaera apis. Insects 2020, 11, 419. [Google Scholar] [CrossRef] [PubMed]
- Getachew, A.; Abejew, T.A.; Wu, J.; Xu, J.; Yu, H.; Tan, J.; Wu, P.; Tu, Y.; Kang, W.; Wang, Z.; et al. Transcriptome profiling reveals insertional mutagenesis suppressed the expression of candidate pathogenicity genes in honeybee fungal pathogen, Ascosphaera apis. Sci. Rep. 2020, 10, 7532. [Google Scholar] [CrossRef]
- Hrassnigg, N.; Brodschneider, R.; Fleischmann, P.H.; Crailsheim, K. Unlike nectar foragers, honeybee drones (Apis mellifera) are not able to utilize starch as fuel for flight. Apidologie 2005, 36, 547–557. [Google Scholar] [CrossRef][Green Version]
- Seow, E.K.; Tan, T.C.; Lee, L.K.; Easa, A.M. Effects of honey types and heating treatment on the textural, thermal, microstructural, and chemical properties of glutinous rice flour gels. J. Texture Stud. 2020, 51, 909–916. [Google Scholar] [CrossRef] [PubMed]
- Donkersley, P.; Rhodes, G.; Pickup, R.W.; Jones, K.C.; Wilson, K. Honeybee nutrition is linked to landscape composition. Ecol. Evol. 2014, 4, 4195–4206. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.A.; Nicolson, S.W.; Shafir, S. Nutritional physiology and ecology of honey bees. Annu. Rev. Entomol. 2018, 63, 327–344. [Google Scholar] [CrossRef] [PubMed]
- Masasa, M.; Kushmaro, A.; Chernova, H.; Shashar, N.; Guttman, L. Carbohydrate-active enzymes of a novel halotolerant Alkalihalobacillus species for hydrolysis of starch and other algal polysaccharides. Microbiol. Spectr. 2022, 10, e0107822. [Google Scholar] [CrossRef] [PubMed]
- Parashar, D.; Satyanarayana, T. Engineering a chimeric acid-stable α-amylase-glucoamylase (Amy-Glu) for one step starch saccharification. Int. J. Biol. Macromol. 2017, 99, 274–281. [Google Scholar] [CrossRef]
- Damaris, R.N.; Lin, Z.; Yang, P.; He, D. The rice alpha-amylase, conserved regulator of seed maturation and germination. Int. J. Mol. Sci. 2019, 20, 450. [Google Scholar] [CrossRef] [PubMed]
- Acet, Ö.; İnanan, T.; Acet, B.Ö.; Dikici, E.; Odabaşı, M. α-amylase immobilized composite cryogels: Some studies on kinetic and adsorption factors. Appl. Biochem. Biotechnol. 2021, 193, 2483–2496. [Google Scholar] [CrossRef]
- Farooq, M.A.; Ali, S.; Hassan, A.; Tahir, H.M.; Mumtaz, S.; Mumtaz, S. Biosynthesis and industrial applications of α-amylase: A review. Arch. Microbiol. 2021, 203, 1281–1292. [Google Scholar] [CrossRef] [PubMed]
- Kalia, S.; Bhattacharya, A.; Prajapati, S.K.; Malik, A. Utilization of starch effluent from a textile industry as a fungal growth supplement for enhanced α-amylase production for industrial application. Chemosphere 2021, 279, 130554. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.C.; Hu, H.F.; Ma, J.W.; Yan, Q.J.; Liu, H.J.; Jiang, Z.Q. A novel high maltose-forming α-amylase from Rhizomucor miehei and its application in the food industry. Food Chem. 2020, 305, 125447. [Google Scholar] [CrossRef] [PubMed]
- Nag, M.; Lahiri, D.; Garai, S.; Mukherjee, D.; Ray, R.R. Regulation of β-amylase synthesis: A brief overview. Mol. Biol. Rep. 2021, 48, 6503–6511. [Google Scholar] [CrossRef]
- Jung, Y.S.; Lee, B.H.; Yoo, S.H. Physical structure and absorption properties of tailor-made porous starch granules produced by selected amylolytic enzymes. PLoS ONE 2017, 12, e0181372. [Google Scholar] [CrossRef]
- David, L.C.; Lee, S.K.; Bruderer, E.; Abt, M.R.; Fischer-Stettler, M.; Tschopp, M.A.; Solhaug, E.M.; Sanchez, K.; Zeeman, S.C. Beta-AMYLASE9 is a plastidial nonenzymatic regulator of leaf starch degradation. Plant Physiol. 2022, 188, 191–207. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.Z.; Song, F.F.; Qiu, Y.F.; Li, X.X.; Guan, X. Optimization of the medium composition of a biphasic production system for mycelial growth and spore production of Aschersonia placenta using response surface methodology. J. Invertebr. Pathol. 2013, 112, 108–115. [Google Scholar] [CrossRef]
- Luo, L.; Wang, G.; Wang, Z.; Ma, J.; He, Y.; He, J.; Wang, L.; Liu, Y.; Xiao, H.; Xiao, Y.; et al. Optimization of Fenton process on removing antibiotic resistance genes from excess sludge by single-factor experiment and response surface methodology. Sci. Total Environ. 2021, 788, 147889. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.Y.; Song, X.Y.; Dong, D.; Keyhani, N.O.; Yao, L.D.; Zang, X.Y.; Dong, L.L.; Gu, Z.J.; Fu, D.L.; Liu, X.Z.; et al. Efficient production of Aschersonia placenta protoplasts for transformation using optimization algorithms. Can. J. Microbiol. 2016, 62, 579–587. [Google Scholar] [CrossRef] [PubMed]
- Rath, S.; Paul, M.; Behera, H.K.; Thatoi, H. Response surface methodology mediated optimization of lignin peroxidase from Bacillus mycoides isolated from Simlipal Biosphere Reserve, Odisha, India. J. Genet. Eng. Biotechnol. 2022, 20, 2. [Google Scholar] [CrossRef] [PubMed]
- Weremfo, A.; Abassah-Oppong, S.; Adulley, F.; Dabie, K.; Seidu-Larry, S. Response surface methodology as a tool to optimize the extraction of bioactive compounds from plant sources. J. Sci. Food Agric. 2023, 103, 26–36. [Google Scholar] [CrossRef]
- Qadi, M.; Jaradat, N.; Al-Lahham, S.; Ali, I.; Abualhasan, M.N.; Shraim, N.; Hussein, F.; Issa, L.; Mousa, A.; Zarour, A.; et al. Antibacterial, anticandidal, phytochemical, and biological evaluations of pellitory plant. Biomed. Res. Int. 2020, 2020, 6965306. [Google Scholar] [CrossRef] [PubMed]
- Kielkopf, C.L.; Bauer, W.; Urbatsch, I.L. Bradford assay for determining protein concentration. Cold Spring Harb. Protoc. 2020, 2020, 102269. [Google Scholar] [CrossRef]
- Pardhi, D.S.; Panchal, R.R.; Raval, V.H.; Rajput, K.N. Statistical optimization of medium components for biosurfactant production by Pseudomonas guguanensis D30. Prep. Biochem. Biotechnol. 2022, 52, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Zhou, Y.; Wang, Y.; Lin, M. Optimization of s-nitrosocaptopril monohydrate storage conditions based on response surface method. Molecules 2021, 26, 7533. [Google Scholar] [CrossRef] [PubMed]
- Narenderan, S.T.; Meyyanathan, S.N.; Karri, V.V.S.R. Experimental design in pesticide extraction methods: A review. Food Chem. 2019, 289, 384–395. [Google Scholar] [CrossRef]
- Matsumoto, H.; Haniu, H.; Komori, N. Determination of protein molecular weights on SDS-PAGE. Methods Mol. Biol. 2019, 1855, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Azhagesan, A.; Chandrasekaran, N.; Mukherjee, A. Multispectroscopy analysis of polystyrene nanoplastic interaction with diastase α-amylase. Ecotoxicol. Environ. Saf. 2022, 247, 114226. [Google Scholar] [CrossRef]
- Bukhteeva, I.; Hrunyk, N.I.; Yusypovych, Y.M.; Shalovylo, Y.I.; Kovaleva, V.; Nesmelova, I.V. Structure, dynamics, and function of PsDef2 defensin from Pinus sylvestris. Structure 2022, 30, 753–762. [Google Scholar] [CrossRef]
- Chen, W.N.; Tang, K.S.; Yeong, K.Y. Potential roles of α-amylase in Alzheimer’s disease: Biomarker and drug target. Curr. Neuropharmacol. 2022, 20, 1554–1563. [Google Scholar] [CrossRef]
- Ribeiro, M.M.; Rezende, M.I.; Baldo, C.; Sartori, D. Aspergillus welwitschiae: A potential amylases producer. Curr. Microbiol. 2022, 79, 307. [Google Scholar] [CrossRef]
- Balakrishnan, M.; Jeevarathinam, G.; Kumar, S.K.S.; Muniraj, I.; Uthandi, S. Optimization and scale-up of α-amylase production by Aspergillus oryzae using solid-state fermentation of edible oil cakes. BMC Biotechnol. 2021, 21, 33. [Google Scholar] [CrossRef] [PubMed]
- Elyasi Far, B.; Ahmadi, Y.; Yari Khosroshahi, A.; Dilmaghani, A. Microbial alpha-amylase production: Progress, challenges and perspectives. Adv. Pharm. Bull. 2020, 10, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Elsayed, E.A.; Ahmed Abdelwahed, N. Medium optimization by response surface methodology for improved cholesterol oxidase production by a newly isolated Streptomyces rochei NAM-19 strain. Biomed. Res. Int. 2020, 2020, 1870807. [Google Scholar] [CrossRef] [PubMed]
- Nam, Y.; Barnebey, A.; Kim, H.K.; Yannone, S.M.; Flint, S. Novel hyperthermoacidic archaeal enzymes for removal of thermophilic biofilms from stainless steel. J. Appl. Microbiol. 2023, 134, lxad106. [Google Scholar] [CrossRef]
- Babacan, S.; Rand, A.G. Characterization of honey amylase. J. Food Sci. 2007, 72, C050–C055. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Lee, C.; Lim, S.W.; Adhikari, A.; Andring, J.T.; McKenna, R.; Ghim, C.M.; Kim, C.U. Elucidating the role of metal ions in carbonic anhydrase catalysis. Nat. Commun. 2020, 11, 4557. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Grewal, J.; Sadaf, A.; Hemamalini, R.; Khare, S.K. Halophiles as a source of polyextremophilic α-aamylase for industrial applications. AIMS Microbiol. 2016, 2, 1–26. [Google Scholar] [CrossRef]
| Code | Medium Variable | Levels (g/L) | Statistics for Observed Data | ||
|---|---|---|---|---|---|
| Low (−1) | High (+1) | t | Pr > |t| | ||
| X1 | Maltose | 40 | 50 | 3.407537 | 0.0422 * |
| X2 | Yeast powder | 20 | 30 | −2.84837 | 0.0652 |
| X3 | Dummy | −1 | 1 | −1.13515 | 0.3388 |
| X4 | CaCl2 | 1.11 | 1.67 | 3.799317 | 0.0320* |
| X5 | ZnCl2 | 4.93 | 7.39 | −0.78043 | 0.4921 |
| X6 | Dummy | −1 | 1 | −1.1057 | 0.3496 |
| X7 | VE | 0.05 | 0.075 | −2.78186 | 0.0689 |
| X8 | pH | 7.0 | 7.5 | −4.78119 | 0.0174 * |
| Step Change Value | Maltose (gL−1) | CaCl2 (gL−1) | pH | Amylase Activity (U/mL) |
|---|---|---|---|---|
| X | 45.0 | 1.39 | 7.0 | 41.38 (1.62) |
| △ | 0.5 | 0.05 | −0.2 | 0 |
| X + 1△ | 45.5 | 1.44 | 6.8 | 44.18 (1.39) |
| X + 2△ | 46.0 | 1.49 | 6.6 | 46.68 (1.55) |
| X + 3△ | 46.5 | 1.54 | 6.4 | 44.61 (1.07) |
| X + 4△ | 47.0 | 1.59 | 6.2 | 43.65 (1.91) |
| X + 5△ | 47.5 | 1.64 | 6.0 | 41.38 (2.26) |
| Fraction | Total Activity (U) | Total Protein (mg) | Specific Activity (U/mg) | Purification (Fold) | Yield (%) |
|---|---|---|---|---|---|
| Crude enzyme | 2082.62 | 80.68 | 25.81 | 1 | 100 |
| 50% ammonium sulfate | 1008.77 | 9.97 | 101.18 | 3.92 | 24.96 |
| Ion exchange chromatography | 692.05 | 2.14 | 323.39 | 12.53 | 7.12 |
| Gel filtration chromatography | 597.55 | 1.61 | 371.15 | 14.38 | 2.51 |
| Metal Ion (10 mM) | Relative Activity (%) | Organic Solvent (10%) | Relative Activity (%) |
|---|---|---|---|
| None | 100 (0.14) | none | 100 (0.11) |
| Fe2+ | 143.25 (2.12) | DMSO | 99.14 (0.82) |
| K+ | 91.73 (1.96) | isopropanol | 96.98 (1.21) |
| Fe3+ | 81.91 (0.73) | dichloromethane | 95.31 (0.44) |
| Na+ | 77.46 (2.21) | acetone | 91.97 (1.15) |
| Mg2+ | 37.99 (0.65) | methanol | 91.49 (1.53) |
| Ca2+ | 37.99 (0.87) | n-butyl alcohol | 86.26 (0.77) |
| Zn2+ | 5.74 (0.38) | n-hexane | 83.67 (1.49) |
| Cu2+ | 0.00 | ethanol | 75.70 (0.68) |
| Mn2+ | 0.00 | chloroform | 65.63 (0.93) |
| Ba2+ | 0.00 | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Liu, S.; Yang, C.; Keyhani, N.O.; Pu, H.; Lin, L.; Li, X.; Jia, P.; Wu, D.; Pan, J.; et al. Characterization of an α-Amylase from the Honeybee Chalk Brood Pathogen Ascosphaera apis. J. Fungi 2023, 9, 1082. https://doi.org/10.3390/jof9111082
Li J, Liu S, Yang C, Keyhani NO, Pu H, Lin L, Li X, Jia P, Wu D, Pan J, et al. Characterization of an α-Amylase from the Honeybee Chalk Brood Pathogen Ascosphaera apis. Journal of Fungi. 2023; 9(11):1082. https://doi.org/10.3390/jof9111082
Chicago/Turabian StyleLi, Jincheng, Sen Liu, Chenjie Yang, Nemat O. Keyhani, Huili Pu, Longbin Lin, Xiaoxia Li, Peisong Jia, Dongmei Wu, Jieming Pan, and et al. 2023. "Characterization of an α-Amylase from the Honeybee Chalk Brood Pathogen Ascosphaera apis" Journal of Fungi 9, no. 11: 1082. https://doi.org/10.3390/jof9111082
APA StyleLi, J., Liu, S., Yang, C., Keyhani, N. O., Pu, H., Lin, L., Li, X., Jia, P., Wu, D., Pan, J., Stevenson, P. C., Fernández-Grandon, G. M., Zhang, L., Chen, Y., Guan, X., & Qiu, J. (2023). Characterization of an α-Amylase from the Honeybee Chalk Brood Pathogen Ascosphaera apis. Journal of Fungi, 9(11), 1082. https://doi.org/10.3390/jof9111082







