Regulatory Mechanism of Peroxisome Number Reduction Caused by FgPex4 and FgPex22-like Deletion in Fusarium graminearum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strains and Growth Assays
2.2. Strain Construction
2.3. Statistics of the Number of Peroxisomes
2.4. Quantitative Real-Time PCR
2.5. Autophagy and Pexophagy Detection
2.6. Polyubiquitination Detection of Pex5
2.7. Detection of the Recovery of Peroxisome Function
2.8. Determination of Matrix Protein Introduction
3. Results
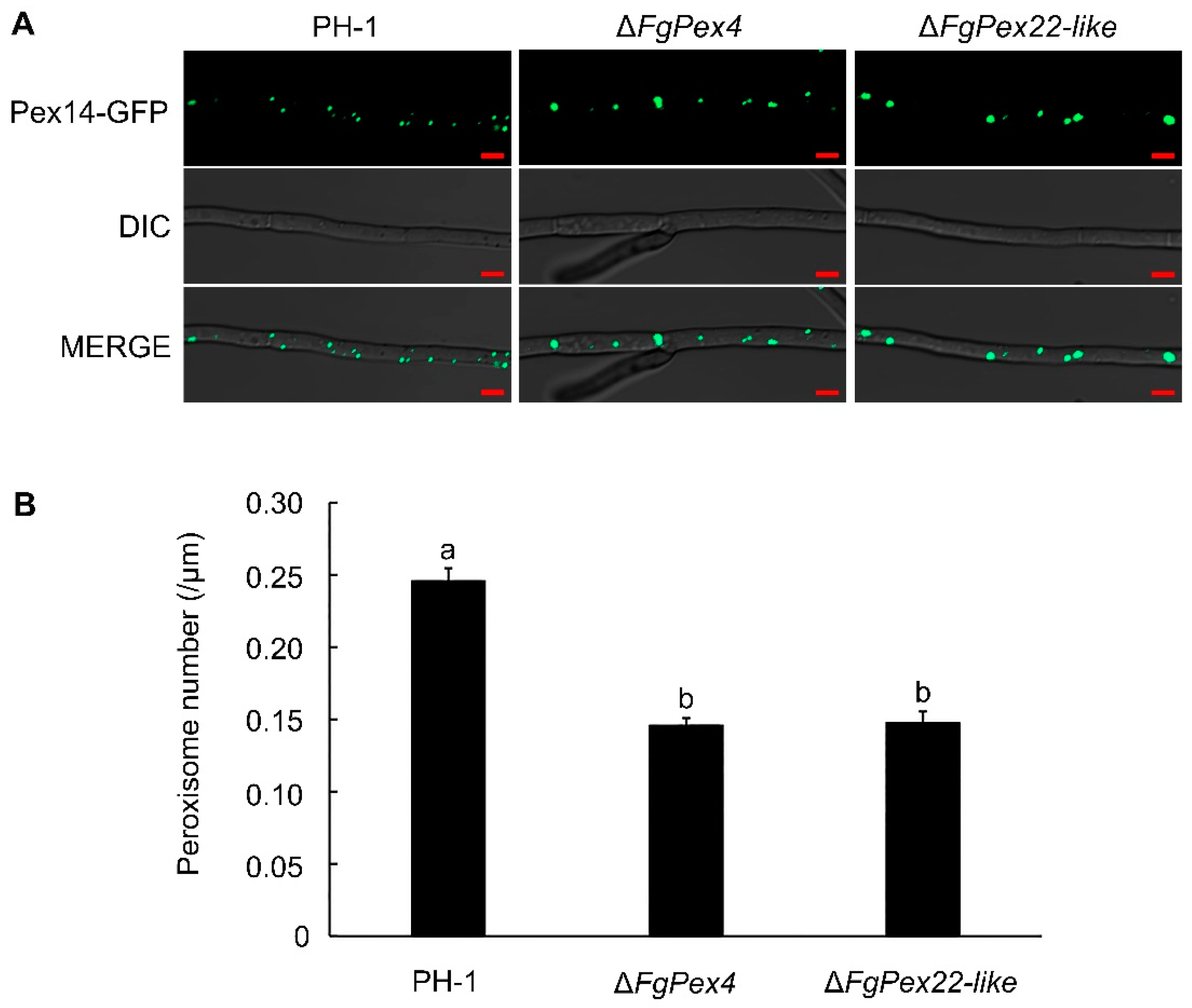
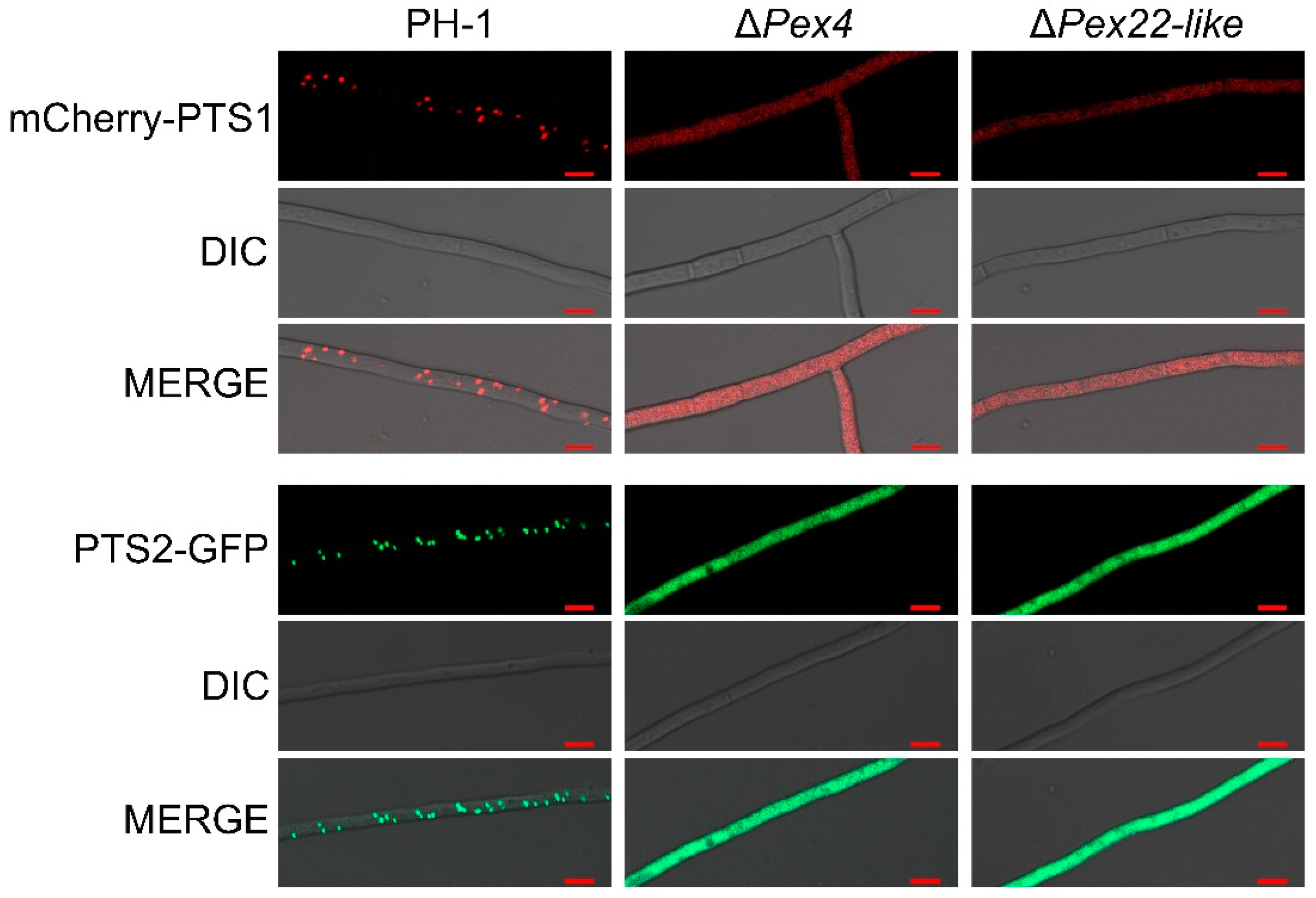
3.1. Deletion of FgPex4 and FgPex22-like Genes Reduced Number of Peroxisomes
3.2. Peroxisome Synthesis Affected by the Knockout of FgPex4 and FgPex22-like Genes
3.3. Knockout of FgPex4 and FgPex22-like Genes Affected Peroxisome Degradation
3.3.1. Effect of FgPex4 and FgPex22-like Deletions on Autophagy
3.3.2. Deletion of FgPex4 and FgPex22-like Aggravated the Pexophagy of F. graminearum
3.4. Effect of FgPex4 and FgPex22-like Genes Deletion on Polyubiquitination of Pex5p
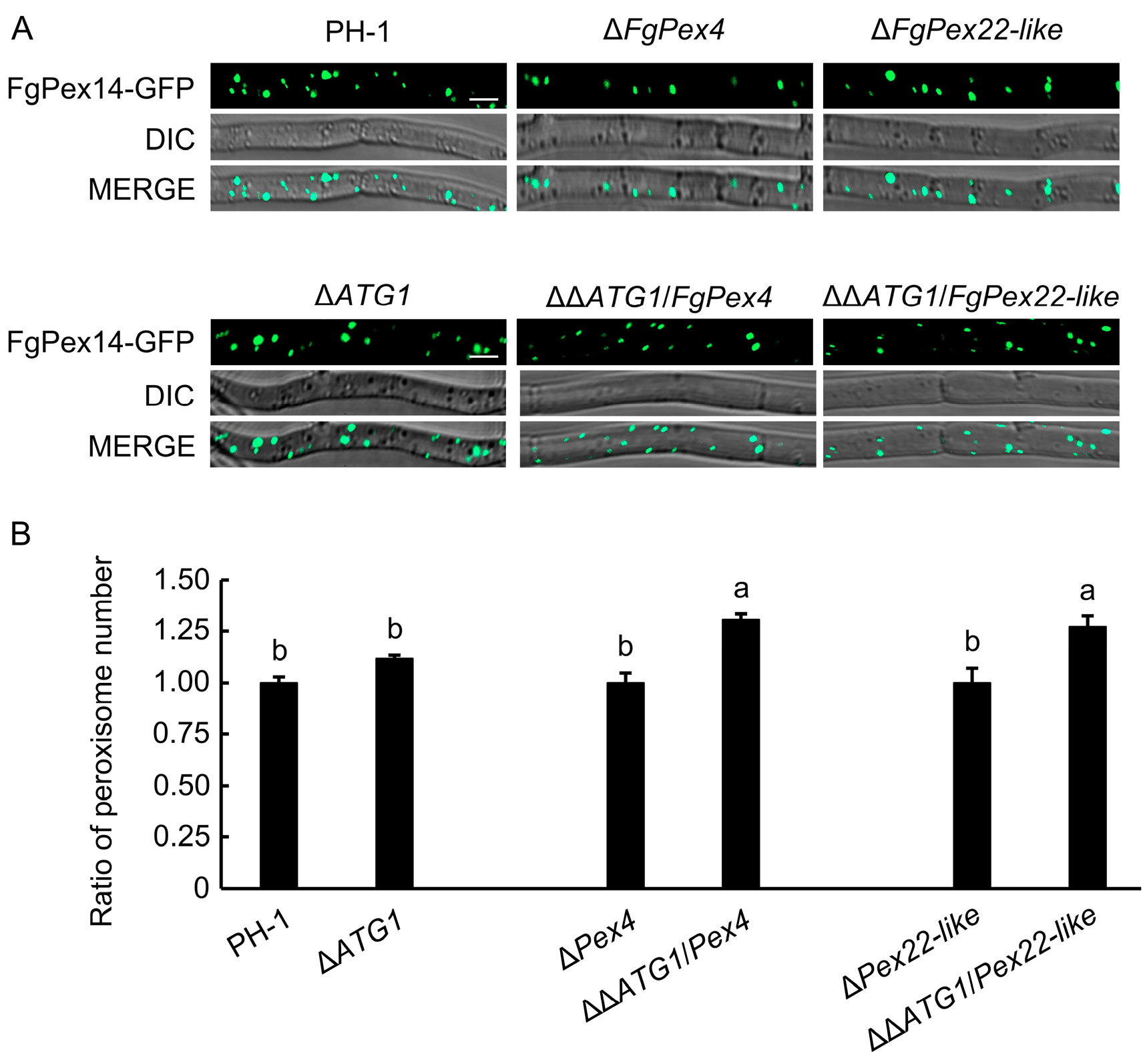
3.5. Effect of Blocking the Pexophagy Pathway on the Number of Peroxisomes
3.6. Effects of FgPex4 and FgPex22-like Deletions on Peroxisome Integrity
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, L.; Liu, C.; Wang, M.; Tao, Y.; Liang, Y.; Yu, J. Peroxin FgPEX22-Like Is Involved in FgPEX4 Tethering and Fusarium graminearum Pathogenicity. Front. Microbiol. 2021, 12, 756292. [Google Scholar] [CrossRef] [PubMed]
- Mano, S.; Hayashi, Y.; Hikino, K.; Otomo, M.; Kanai, M.; Nishimura, M. Ubiquitin-conjugating activity by PEX4 is required for efficient protein transport to peroxisomes in Arabidopsis thaliana. J. Biol. Chem. 2022, 298, 102038. [Google Scholar] [CrossRef]
- Suaste-Olmos, F.; Zirión-Martínez, C.; Takano-Rojas, H.; Peraza-Reyes, L. Meiotic development initiation in the fungus Podospora anserina requires the peroxisome receptor export machinery. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 572–586. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, L.; Liang, Y.; Yu, J. FgPEX4 is involved in development, pathogenicity, and cell wall integrity in Fusarium graminearum. Curr. Genet. 2019, 65, 747–758. [Google Scholar] [CrossRef]
- Skowyra, M.L.; Rapoport, T.A. PEX5 translocation into and out of peroxisomes drives matrix protein import. Mol. Cell 2022, 82, 3209–3225.e7. [Google Scholar] [CrossRef]
- Gould, S.J.; Keller, G.A.; Hosken, N.; Wilkinson, J.; Subramani, S. A conserved tripeptide sorts proteins to peroxisomes. J. Cell Biol. 1989, 108, 1657–1664. [Google Scholar] [CrossRef]
- Miura, S.; Kasuya-Arai, I.; Mori, H.; Miyazawa, S.; Osumi, T.; Hashimoto, T.; Fujiki, Y. Carboxyl-terminal consensus Ser-Lys-Leu-related tripeptide of peroxisomal proteins functions in vitro as a minimal peroxisome-targeting signal. J. Biol. Chem. 1992, 267, 14405–14411. [Google Scholar] [CrossRef]
- Miyazawa, S.; Osumi, T.; Hashimoto, T.; Ohno, K.; Miura, S.; Fujiki, Y. Peroxisome targeting signal of rat liver acyl-coenzyme A oxidase resides at the carboxy terminus. Mol. Cell Biol. 1989, 9, 83–91. [Google Scholar]
- Williams, C.; van den Berg, M.; Sprenger, R.R.; Distel, B. A conserved cysteine is essential for Pex4p-dependent ubiquitination of the peroxisomal import receptor Pex5p. J. Biol. Chem. 2007, 282, 22534–22543. [Google Scholar] [CrossRef]
- Platta, H.W.; El Magraoui, F.; Schlee, D.; Grunau, S.; Girzalsky, W.; Erdmann, R. Ubiquitination of the peroxisomal import receptor Pex5p is required for its recycling. J. Cell Biol. 2007, 177, 197–204. [Google Scholar] [CrossRef]
- Carvalho, A.F.; Pinto, M.P.; Grou, C.P.; Alencastre, I.S.; Fransen, M.; Sa-Miranda, C.; Azevedo, J.E. Ubiquitination of mammalian Pex5p, the peroxisomal import receptor. J. Biol. Chem. 2007, 282, 31267–31272. [Google Scholar] [CrossRef] [PubMed]
- Okumoto, K.; Misono, S.; Miyata, N.; Matsumoto, Y.; Mukai, S.; Fujiki, Y. Cysteine ubiquitination of PTS1 receptor Pex5p regulates Pex5p recycling. Traffic 2011, 12, 1067–1083. [Google Scholar] [CrossRef] [PubMed]
- Kiel, J.A.; Emmrich, K.; Meyer, H.E.; Kunau, W.H. Ubiquitination of the peroxisomal targeting signal type 1 receptor, Pex5p, suggests the presence of a quality control mechanism during peroxisomal matrix protein import. J. Biol. Chem. 2005, 280, 1921–1930. [Google Scholar] [CrossRef] [PubMed]
- Kragt, A.; Voorn-Brouwer, T.; van den Berg, M.; Distel, B. The Saccharomyces cerevisiae peroxisomal import receptor Pex5p is monoubiquitinated in wild type cells. J. Biol. Chem. 2005, 280, 7867–7874. [Google Scholar] [CrossRef]
- Haan, G.J.; Baerends, R.J.; Krikken, A.M.; Otzen, M.; Veenhuis, M.; van der Klei, I.J. Reassembly of peroxisomes in Hansenula polymorpha pex3 cells on reintroduction of Pex3p involves the nuclear envelope. FEMS Yeast Res. 2006, 6, 186–194. [Google Scholar] [CrossRef]
- Hoepfner, D.; Schildknegt, D.; Braakman, I.; Philippsen, P.; Tabak, H.F. Contribution of the Endoplasmic Reticulum to Peroxisome Formation. Cell 2005, 122, 85–95. [Google Scholar] [CrossRef]
- Kragt, A.; Voorn-Brouwer, T.; van den Berg, M.; Distel, B. Endoplasmic Reticulum-directed Pex3p Routes to Peroxisomes and Restores Peroxisome Formation in a Saccharomyces cerevisiae pex3Δ Strain. J. Biol. Chem. 2005, 280, 34350–34357. [Google Scholar] [CrossRef]
- Tam, Y.Y.C.; Fagarasanu, A.; Fagarasanu, M.; Rachubinski, R.A. Pex3p Initiates the Formation of a Preperoxisomal Compartment from a Subdomain of the Endoplasmic Reticulum in Saccharomyces cerevisiae. J. Biol. Chem. 2005, 280, 34933–34939. [Google Scholar] [CrossRef]
- Hoepfner, D.; van den Berg, M.; Philippsen, P.; Tabak, H.F.; Hettema, E.H. A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae. J. Cell Biol. 2001, 155, 979–990. [Google Scholar] [CrossRef]
- Koch, A.; Yoon, Y.; Bonekamp, N.A.; McNiven, M.A.; Schrader, M. A role for Fis1 in both mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell 2005, 16, 5077–5086. [Google Scholar] [CrossRef]
- Kuravi, K.; Nagotu, S.; Krikken, A.M.; Sjollema, K.; Deckers, M.; Erdmann, R.; Veenhuis, M.; van der Klei, I.J. Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae. J. Cell Sci. 2006, 119, 3994–4001. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gould, S.J. The Dynamin-like GTPase DLP1 Is Essential for Peroxisome Division and Is Recruited to Peroxisomes in Part by PEX11. J. Biol. Chem. 2003, 278, 17012–17020. [Google Scholar] [CrossRef] [PubMed]
- Koch, J.; Brocard, C. PEX11 proteins attract Mff and human Fis1 to coordinate peroxisomal fission. J. Cell Sci. 2012, 125 Pt 16, 3813–3826. [Google Scholar]
- Fagarasanu, M.; Fagarasanu, A.; Tam, Y.Y.; Aitchison, J.D.; Rachubinski, R.A. Inp1p is a peroxisomal membrane protein required for peroxisome inheritance in Saccharomyces cerevisiae. J. Cell Biol. 2005, 169, 765–775. [Google Scholar] [CrossRef] [PubMed]
- Fagarasanu, M.; Fagarasanu, A.; Rachubinski, R.A. Sharing the wealth: Peroxisome inheritance in budding yeast. Biochim. Biophys. Acta 2006, 1763, 1669–1677. [Google Scholar] [CrossRef]
- Fagarasanu, A.; Fagarasanu, M.; Rachubinski, R.A. Maintaining peroxisome populations: A story of division and inheritance. Annu. Rev. Cell Dev. Biol. 2007, 23, 321–344. [Google Scholar] [CrossRef]
- Otzen, M.; Rucktaschel, R.; Thoms, S.; Emmrich, K.; Krikken, A.M.; Erdmann, R.; van der Klei, I.J. Pex19p contributes to peroxisome inheritance in the association of peroxisomes to Myo2p. Traffic 2012, 13, 947–959. [Google Scholar] [CrossRef]
- Fagarasanu, A.; Fagarasanu, M.; Eitzen, G.A.; Aitchison, J.D.; Rachubinski, R.A. The peroxisomal membrane protein Inp2p is the peroxisome-specific receptor for the myosin V motor Myo2p of Saccharomyces cerevisiae. Dev. Cell 2006, 10, 587–600. [Google Scholar] [CrossRef]
- Fagarasanu, A.; Mast, F.D.; Knoblach, B.; Jin, Y.; Brunner, M.J.; Logan, M.R.; Glover, J.N.; Eitzen, G.A.; Aitchison, J.D.; Weisman, L.S.; et al. Myosin-driven peroxisome partitioning in S. cerevisiae. J. Cell Biol. 2009, 186, 541–554. [Google Scholar] [CrossRef]
- Marelli, M.; Smith, J.J.; Jung, S.; Yi, E.; Nesvizhskii, A.I.; Christmas, R.H.; Saleem, R.A.; Tam, Y.Y.C.; Fagarasanu, A.; Goodlett, D.R.; et al. Quantitative mass spectrometry reveals a role for the GTPase Rho1p in actin organization on the peroxisome membrane. J. Cell Biol. 2004, 167, 1099–1112. [Google Scholar] [CrossRef]
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of cells and tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef] [PubMed]
- Green, D.R.; Levine, B. To be or not to be? How selective autophagy and cell death govern cell fate. Cell 2014, 157, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Lamb, C.A.; Yoshimori, T.; Tooze, S.A. The autophagosome: Origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 2013, 14, 759–774. [Google Scholar] [CrossRef]
- Mochida, K.; Oikawa, Y.; Kimura, Y.; Kirisako, H.; Hirano, H.; Ohsumi, Y.; Nakatogawa, H. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature 2015, 522, 359–362. [Google Scholar] [CrossRef]
- Ohsumi, Y. Molecular dissection of autophagy: Two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2001, 2, 211–216. [Google Scholar] [CrossRef]
- Suzuki, K.; Kubota, Y.; Sekito, T.; Ohsumi, Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 2007, 12, 209–218. [Google Scholar] [CrossRef]
- Nakatogawa, H.; Ichimura, Y.; Ohsumi, Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 2007, 130, 165–178. [Google Scholar] [CrossRef]
- Walker, C.L.; Pomatto, L.C.D.; Tripathi, D.N.; Davies, K.J.A. Redox Regulation of Homeostasis and Proteostasis in Peroxisomes. Physiol. Rev. 2018, 98, 89–115. [Google Scholar] [CrossRef]
- Oku, M.; Sakai, Y. Pexophagy in yeasts. Biochim. Biophys. Acta 2016, 1863, 992–998. [Google Scholar] [CrossRef]
- Germain, K.; Kim, P.K. Pexophagy: A Model for Selective Autophagy. Int. J. Mol. Sci. 2020, 21, 578. [Google Scholar] [CrossRef]
- Eberhart, T.; Kovacs, W.J. Pexophagy in yeast and mammals: An update on mysteries. Histochem. Cell Biol. 2018, 150, 473–488. [Google Scholar] [CrossRef] [PubMed]
- Bellu, A.R.; Salomons, F.A.; Kiel, J.A.; Veenhuis, M.; Van Der Klei, I.J. Removal of Pex3p is an important initial stage in selective peroxisome degradation in Hansenula polymorpha. J. Biol. Chem. 2002, 277, 42875–42880. [Google Scholar] [CrossRef] [PubMed]
- de Vries, B.; Todde, V.; Stevens, P.; Salomons, F.; van der Klei, I.J.; Veenhuis, M. Pex14p is not required for N-starvation induced microautophagy and in catalytic amounts for macropexophagy in Hansenula polymorpha. Autophagy 2006, 2, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Bellu, A.R.; Komori, M.; van der Klei, I.J.; Kiel, J.A.; Veenhuis, M. Peroxisome biogenesis and selective degradation converge at Pex14p. J. Biol. Chem. 2001, 276, 44570–44574. [Google Scholar] [CrossRef]
- Nuttall, J.M.; Motley, A.M.; Hettema, E.H. Deficiency of the exportomer components Pex1, Pex6, and Pex15 causes enhanced pexophagy in Saccharomyces cerevisiae. Autophagy 2014, 10, 835–845. [Google Scholar] [CrossRef]
- Demers, N.D.; Riccio, V.; Jo, D.S.; Bhandari, S.; Law, K.B.; Liao, W.; Kim, C.; McQuibban, G.A.; Choe, S.K.; Cho, D.H.; et al. PEX13 prevents pexophagy by regulating ubiquitinated PEX5 and peroxisomal ROS. Autophagy 2023, 19, 1781–1802. [Google Scholar] [CrossRef]
- Chen, Y.; Zheng, S.; Ju, Z.; Zhang, C.; Tang, G.; Wang, J.; Wen, Z.; Chen, W.; Ma, Z. Contribution of peroxisomal docking machinery to mycotoxin biosynthesis, pathogenicity and pexophagy in the plant pathogenic fungus Fusarium graminearum. Environ. Microbiol. 2018, 20, 3224–3245. [Google Scholar] [CrossRef]
- Tam, Y.Y.; Torres-Guzman, J.C.; Vizeacoumar, F.J.; Smith, J.J.; Marelli, M.; Aitchison, J.D.; Rachubinski, R.A. Pex11-related proteins in peroxisome dynamics: A role for the novel peroxin Pex27p in controlling peroxisome size and number in Saccharomyces cerevisiae. Mol. Biol. Cell 2003, 14, 4089–4102. [Google Scholar] [CrossRef]
- Vizeacoumar, F.J.; Torres-Guzman, J.C.; Tam, Y.Y.C.; Aitchison, J.D.; Rachubinski, R.A. YHR150w and YDR479c encode peroxisomal integral membrane proteins involved in the regulation of peroxisome number, size, and distribution in Saccharomyces cerevisiae. J. Cell Biol. 2003, 161, 321–332. [Google Scholar] [CrossRef]
- Li, L.; Wang, J.; Chen, H.; Chai, R.; Zhang, Z.; Mao, X.; Qiu, H.; Jiang, H.; Wang, Y.; Sun, G. Pex14/17, a filamentous fungus-specific peroxin, is required for the import of peroxisomal matrix proteins and full virulence of Magnaporthe oryzae. Mol. Plant Pathol. 2017, 18, 1238–1252. [Google Scholar] [CrossRef]
- Wróblewska, J.P.; van der Klei, I.J. Peroxisome Maintenance Depends on De Novo Peroxisome Formation in Yeast Mutants Defective in Peroxisome Fission and Inheritance. Int. J. Mol. Sci. 2019, 20, 4023. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, C.; Wang, L.; Sun, S.; Liu, A.; Liang, Y.; Yu, J.; Dong, H. FgPEX1 and FgPEX10 are required for the maintenance of Woronin bodies and full virulence of Fusarium graminearum. Curr. Genet. 2019, 65, 1383–1396. [Google Scholar] [CrossRef] [PubMed]
- Goswami, R.S. Targeted gene replacement in fungi using a split-marker approach. Methods Mol. Biol. 2012, 835, 255–269. [Google Scholar] [PubMed]
- Gravelat, F.N.; Askew, D.S.; Sheppard, D.C. Targeted gene deletion in Aspergillus fumigatus using the hygromycin-resistance split-marker approach. Methods Mol. Biol. 2012, 845, 119–130. [Google Scholar]
- Yun, Y.; Liu, Z.; Zhang, J.; Shim, W.B.; Chen, Y.; Ma, Z. The MAPKK FgMkk1 of Fusarium graminearum regulates vegetative differentiation, multiple stress response, and virulence via the cell wall integrity and high-osmolarity glycerol signaling pathways. Environ. Microbiol. 2014, 16, 2023–2037. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- He, D.; Li, M.; Damaris, R.N.; Bu, C.; Xue, J.; Yang, P. Quantitative ubiquitylomics approach for characterizing the dynamic change and extensive modulation of ubiquitylation in rice seed germination. Plant J. 2019, 101, 1430–1447. [Google Scholar] [CrossRef]
- Zhu, L.; Cheng, H.; Peng, G.; Wang, S.; Zhang, Z.; Ni, E.; Fu, X.; Zhuang, C.; Liu, Z.; Zhou, H. Ubiquitinome Profiling Reveals the Landscape of Ubiquitination Regulation in Rice Young Panicles. Genom. Proteom. Bioinform. 2020, 18, 305–320. [Google Scholar] [CrossRef]
- Managadze, D.; Wurtz, C.; Wiese, S.; Schneider, M.; Girzalsky, W.; Meyer, H.E.; Erdmann, R.; Warscheid, B.; Rottensteiner, H. Identification of PEX33, a novel component of the peroxisomal docking complex in the filamentous fungus Neurospora crassa. Eur. J. Cell Biol. 2010, 89, 955–964. [Google Scholar] [CrossRef]
- Vizeacoumar, F.J.; Torres-Guzman, J.C.; Bouard, D.; Aitchison, J.D.; Rachubinski, R.A. Pex30p, Pex31p, and Pex32p form a family of peroxisomal integral membrane proteins regulating peroxisome size and number in Saccharomyces cerevisiae. Mol. Biol. Cell 2004, 15, 665–677. [Google Scholar] [CrossRef]
- Zutphen, T.; Veenhuis, M.; van der Klei, I.J. Pex14 is the sole component of the peroxisomal translocon that is required for pexophagy. Autophagy 2008, 4, 63–66. [Google Scholar] [CrossRef] [PubMed]
- Kim, P.K.; Hailey, D.W.; Mullen, R.T.; Lippincott-Schwartz, J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc. Natl. Acad. Sci. USA 2008, 105, 20567–20574. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, S.; Abe, K.; Tatemichi, Y.; Fujiki, Y. The membrane peroxin PEX3 induces peroxisome-ubiquitination-linked pexophagy. Autophagy 2014, 10, 1549–1564. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Tripathi, D.N.; Jing, J.; Alexander, A.; Kim, J.; Powell, R.T.; Dere, R.; Tait-Mulder, J.; Lee, J.H.; Paull, T.T.; et al. ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat. Cell Biol. 2015, 17, 1259–1269. [Google Scholar] [CrossRef]
- Deosaran, E.; Larsen, K.B.; Hua, R.; Sargent, G.; Wang, Y.; Kim, S.; Lamark, T.; Jauregui, M.; Law, K.; Lippincott-Schwartz, J.; et al. NBR1 acts as an autophagy receptor for peroxisomes. J. Cell Sci. 2013, 126 Pt 4, 939–952. [Google Scholar] [CrossRef]
- Law, K.B.; Bronte-Tinkew, D.; Di Pietro, E.; Snowden, A.; Jones, R.O.; Moser, A.; Brumell, J.H.; Braverman, N.; Kim, P.K. The peroxisomal AAA ATPase complex prevents pexophagy and development of peroxisome biogenesis disorders. Autophagy 2017, 13, 868–884. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Bi, Z.; Xu, H.; Zhang, R.; Wang, J.; Liang, Y.; Zhang, L.; Yu, J. Regulatory Mechanism of Peroxisome Number Reduction Caused by FgPex4 and FgPex22-like Deletion in Fusarium graminearum. J. Fungi 2023, 9, 1083. https://doi.org/10.3390/jof9111083
Liu C, Bi Z, Xu H, Zhang R, Wang J, Liang Y, Zhang L, Yu J. Regulatory Mechanism of Peroxisome Number Reduction Caused by FgPex4 and FgPex22-like Deletion in Fusarium graminearum. Journal of Fungi. 2023; 9(11):1083. https://doi.org/10.3390/jof9111083
Chicago/Turabian StyleLiu, Chunjie, Zhuoyu Bi, Hao Xu, Renjie Zhang, Jiayi Wang, Yuancun Liang, Li Zhang, and Jinfeng Yu. 2023. "Regulatory Mechanism of Peroxisome Number Reduction Caused by FgPex4 and FgPex22-like Deletion in Fusarium graminearum" Journal of Fungi 9, no. 11: 1083. https://doi.org/10.3390/jof9111083
APA StyleLiu, C., Bi, Z., Xu, H., Zhang, R., Wang, J., Liang, Y., Zhang, L., & Yu, J. (2023). Regulatory Mechanism of Peroxisome Number Reduction Caused by FgPex4 and FgPex22-like Deletion in Fusarium graminearum. Journal of Fungi, 9(11), 1083. https://doi.org/10.3390/jof9111083





