Abstract
Penicillium rubens is a filamentous fungus of great biotechnological importance due to its role as an industrial producer of the antibiotic penicillin. However, despite its significance, our understanding of the regulatory mechanisms governing biological processes in this fungus is still limited. In fungi, zinc finger proteins containing a Zn(II)2Cys6 domain are particularly interesting regulators. Although the P. rubens genome harbors many genes encoding proteins with this domain, only two of them have been investigated thus far. In this study, we employed CRISPR-Cas9 technology to disrupt the pcz1 gene, which encodes a Zn(II)2Cys6 protein in P. rubens. The disruption of pcz1 resulted in a decrease in the production of penicillin in P. rubens. This decrease in penicillin production was accompanied by the downregulation of the expression of pcbAB, pcbC and penDE genes, which form the biosynthetic gene cluster responsible for penicillin production. Moreover, the disruption of pcz1 also impacts on asexual development, leading to decreased growth and conidiation, as well as enhanced conidial germination. Collectively, our results indicate that pcz1 acts as a positive regulator of penicillin production, growth, and conidiation, while functioning as a negative regulator of conidial germination in P. rubens. To the best of our knowledge, this is the first report involving a gene encoding a Zn(II)2Cys6 protein in the regulation of penicillin biosynthesis in P. rubens.
1. Introduction
The genus Penicillium is one of the largest and most diverse in the fungal kingdom. Most species in this genus are saprophytic, displaying high catabolic activity and the ability to produce a wide range of specialized metabolites [1]. Moreover, several species within the Penicillium genus are economically important. One of these species is Penicillium rubens. This filamentous fungal species is an important industrial producer of β-lactam antibiotics, specifically penicillins, and it comprises historically important penicillin-producing strains formerly classified as Penicillium chrysogenum [2]. Penicillins are of great economic importance, serving as both direct therapeutic agents against certain diseases and as a scaffold for the development of semi-synthetic antibiotics [3].
At the molecular level, the biosynthesis pathway of penicillin in P. rubens was elucidated many years ago and has been extensively documented in recent reviews [3,4,5]. The production of this specialized metabolite involves the sequential action of three enzymes encoded by the genes pcbAB, pcbC, and penDE, which form a biosynthetic gene cluster (BGC) [3,4]. These enzymes, namely δ(α-aminoadipyl)-cysteinyl-valine (ACV) synthetase, isopenicillin N synthase, and isopenicillin N acyltransferase, synthesize penicillin from the precursor amino acids α-aminoadipate, cysteine, and valine [3,4].
The regulation of penicillin biosynthesis in P. rubens involves a complex network of regulatory signals, transduction pathways, and effector proteins [3,4,6,7]. Interestingly, the penicillin BGC lacks a gene encoding for a specific transcription factor [8,9]. Thereby, the production of this metabolite in P. rubens is governed by well-known global regulators in fungi, including CreA, AreA, PacC, LaeA, Velvet proteins, PcRFX1, and PcFKH1 [3,4,6].
Zinc finger proteins constitute one of the largest families of regulators in eukaryotes. These proteins are classified into different groups based on the type of “zinc fingers” they possess [10,11]. Among these proteins, those containing a Zn(II)2Cys6 domain, in which six cysteines coordinate two zinc atoms, are particularly interesting in fungi, where they are exclusively found [10,11]. Fungal genomes can contain hundreds of genes encoding Zn(II)2Cys6 proteins, and several of them have been functionally characterized in diverse genera such as Aspergillus, Bipolaris, Fusarium, Trichoderma, and Penicillium [10].
In the genome of P. rubens, numerous genes encoding proteins with Zn(II)2Cys6 domains were identified [12]. However, to date, only two of these genes have been functionally characterized in this fungus. One of these genes, sorR1, is part of the sorbicillin BGC in P. rubens. Guzmán-Chávez et al. [13] demonstrated that sorR1 positively regulates the expression of all the genes within this BGC. The second characterized gene is nirA, a well-known regulatory gene involved in nitrate assimilation in fungi [14]. Espeso et al. [15] examined a P. rubens strain with a natural mutation in nirA, resulting in a truncation of the NirA protein and an impaired nitrate assimilation. The subsequent complementation of this strain with a functional nirA gene led to an increase in the transcriptional levels of genes related to nitrate assimilation and improved growth of the fungus on nitrate-containing medium. To the best of our knowledge, no other gene encoding Zn(II)2Cys6 proteins has been functionally studied in P. rubens.
A few years ago, a gene named pcz1 (for Penicillium C6 zinc domain protein 1), encoding a protein with a Zn(II)2Cys6 domain, was characterized in the cheese-ripening fungus Penicillium roqueforti [16]. In P. roqueforti, Pcz1 exerts significant effects on asexual development, including promotion of apical growth and conidiation, and repression of conidial germination [16,17]. Furthermore, pcz1 exerts important effects on the production of the main specialized metabolites of P. roqueforti [17]. It is worth noting that orthologues of pcz1 are widely distributed in fungi of the Ascomycota phylum, including P. rubens [16]. However, the functional role of this gene has only been experimentally investigated in P. roqueforti and remains unexplored in other fungi.
Recently, Pohl et al. [18] developed CRISPR/Cas9 tools for genetic modification in P. rubens, thus enabling the application of this technique in the fungus [19,20,21]. The authors established two CRISPR/Cas9 systems for P. rubens. One approach involved direct introduction of Cas9 protein and an in vitro synthesized sgRNA-ribonucleoprotein complex into fungal cells through transformation. The other approach involved transient transformation of the fungus with an AMA-based plasmid carrying cassettes for sgRNA and Cas9 expression within the fungal cells. The simplicity of the second approach makes it particularly attractive for gene disruption in the fungus. Consequently, in this study, we employed the second approach to disrupt the pcz1 gene in P. rubens using CRISPR/Cas9. The functional characterization of the pcz1-disrupted mutants indicated that the gene acts as a positive regulator of penicillin BGC expression and penicillin production in P. rubens. Furthermore, pcz1 exerts an important influence on the asexual development processes in the fungus, specifically controlling growth, conidiation, and germinal conidiation.
2. Materials and Methods
2.1. Fungal Strain and Culture Media
In this work, we used P. rubens Wis 54-1255, a strain widely used as a reference in molecular biology and OMICs studies [3]. The fungus was routinely kept on potato dextrose agar (PDA, Difco, Sparks, MD, USA) and grown at 28 °C. Various culture media were employed for phenotypic characterization, including Czapek, CYA, YES [22], Power [23], and CM [16]. The specific media utilized for penicillin production are described in the corresponding protocol (see below).
2.2. Selection of a Target Site for CRISPR/Cas9-Mediated Disruption of the pcz1 Gene
The sequence of the pcz1 gene from P. rubens Wis 54-1255 (see Results) was used to select a target site using the CRISPR/Cas9 target prediction tool CCTop (https://cctop.cos.uni-heidelberg.de:8043, accessed on 18 August 2021) [24], which contains the genome sequence of P. rubens as a reference. CCTop was utilized with default parameters, except for mismatches considered during the prediction in the core sequence and the total number of mismatches, which were both set to 0. A predicted target sequence (5′ GGCCGGGCATGAGATCGACG 3′) with a high efficacy score (0.76) and located at the 5′ end of the coding sequence of the gene, was selected. To ensure specificity to the target sequence of interest and the absence of off-target effects, the selected sequence was aligned against the complete genome of P. rubens Wis 54-1255 using BLASTN.
2.3. Construction of Plasmid pFC333-Pcpcz1 for CRISPR/Cas9-Mediated Disruption of pcz1 in P. rubens Wis 54-1255
To achieve the disruption of pcz1 in P. rubens Wis 54-1255 using CRISPR/Cas9, a plasmid named pFC333-Pcpcz1 was constructed. The plasmid construction involved the previous synthesis of an expression cassette by Integrated DNA Technologies (IDT, Coralville, IA, USA). The cassette was designed based on an expression cassette utilized in previous studies conducted on Aspergillus by Song et al. [25] and van Leewe et al. [26]. It comprised the promoter and gene sequence of the proline-tRNA (tRNAPro1), the sgRNA (including the target sequence), and the terminator of tRNAPro1. Additionally, a hammerhead ribozyme sequence was included between the tRNAPro1 gene sequence and sgRNA, and PacI restriction sites were added at the ends of the cassette.
The cassette synthesized was then digested with PacI and cloned into plasmid pFC333 [27], previously digested with PacI, thus giving rise to the final plasmid pFC333-Pcpcz1. Plasmid pFC333 contains the gene encoding Cas9 under the control of tef1-promoter from A. nidulans, the AMA region enabling autonomous replication of the plasmid, and the phleomycin resistance marker for selection of fungal transformants [27].
2.4. Transformation of P. rubens Wis 54-1255 and Obtainment of Transformants
The plasmid pFC333-Pcpcz1 was introduced into P. rubens Wis 54-1255 through protoplast transformation. Briefly, protoplasts were obtained using lytic enzymes (Sigma, St. Louis, MO, USA) and the transformation process followed the protocol established by Chávez et al. [28]. Transformants were selected on Czapek-sorbitol medium containing phleomycin, as described by Gil-Durán et al. [16]. Subsequently, conidia of the transformants underwent serial dilutions and were seeded onto phleomycin-containing Czapek medium to obtain homokaryotic strains. To induce the loss of phleomycin resistance, the homokaryotic strains were transferred four times to PDA medium without phleomycin (non-selective growth conditions). Confirmation of the loss of phleomycin resistance was achieved by observing the absence of growth of the transformants when plated again on selective media.
DNA was extracted following the method described by Gil-Durán et al. [16]. This DNA was used as a template for amplifying the target region using appropriate primers (Table 1). The resulting amplicons were then cloned in pGEM-T Easy and sent to Macrogen Inc. (Seoul, Republic of Korea) for sequencing.

Table 1.
Primers used in this work.
2.5. Growth, Conidiation, and Conidial Germination Analyses
Apical extension rates were determined following the method described by Ivey et al. [29]. Briefly, 0.2 μL of a conidia suspension (106 conidia/mL) was inoculated at the center of a Petri dish containing PDA, Czapek, CYA, YES, or Power medium. The plates were then incubated at 28 °C for 7 days. The diameters of the colonies were initially measured after 48 h and subsequently at 24 h intervals, with the outer white edge of the colonies serving as the reference point. To calculate the apical extension rate, a linear regression analysis was performed using the colony diameters plotted against time.
The production of conidia was measured following the methodology outlined by García-Rico et al. [30]. In brief, 100 μL of a conidial suspension (5 × 105 conidia/mL) was spread onto Petri dishes containing either Czapek or Power media. The dishes were incubated at 28 °C for 3, 5, or 7 days, and the resulting conidia were collected by adding NT solution (0.9% NaCl, 0.05% Triton) and gently scraping the plate’s surface using an inverted Pasteur pipette. This procedure was repeated once, and the collected conidia were quantified using a Neubauer chamber. The conidia counts were expressed as conidia/mm2 of surface area.
Finally, conidial germination analysis was performed following the method described by Gil-Duran et al. [16]. Three replicate flasks containing CM or Czapek medium were inoculated with 2 × 105 conidia/mL. The flasks were incubated at 28 °C for 12 h, and at regular intervals, 10 μL samples were collected from each flask. These samples were observed under a microscope, and the number of germinated and non-germinated conidia was counted in 10 randomly selected fields. This process was repeated twice for each flask to ensure technical replication. Conidia were classified as germinated when the length of their germ tubes equaled or exceeded the diameter of the conidia. The resulting data were plotted as the percentage of germination over time.
2.6. Production of Benzylpenicillin and HPLC Analyses
To produce benzylpenicillin, P. rubens was grown on Power medium agar [23] at 28 °C for 5 days. All the spores from a plate were collected and inoculated into 50 mL of Penicillium seed medium (MCIP) (sucrose 20 g/L, corn steep solids 20 g/L, yeast extract 10 g/L, CaCO3 5 g/L, pH 5.7) [31]. The culture was incubated at 25 °C and 250 rpm for 24 h. Subsequently, 10 mL of this culture was inoculated into 100 mL of complex medium for fermentation of Penicillium (MCFP) (lactose 55 g/L, corn steep solids 35 g/L, CaCO3 10 g/L, KH2PO4 7 g/L, MgSO4·7H2O 3 g/L, potassium phenylacetate 4 g/L, pH 6.8) [31] and incubated under the same conditions for 48 and 72 h.
The extraction and quantification of benzylpenicillin from these broths were conducted as described by García-Estrada et al. [32]. In brief, 5 mL of the culture broth was centrifuged at 9000 rpm to separate the broth from the mycelium. The mycelium was stored for subsequent use (dry weight determination, see below), while the clarified broth (approximately 3 mL) was acidified to pH 2.0 using 0.1 N HCl. Benzylpenicillin was extracted from the broth three times by adding 2 mL of n-butyl acetate, and then the organic phase was re-extracted three times with 2 mL of 10 mM phosphate buffer at pH 7.5. The aqueous phase was lyophilized, resuspended in 1 mL of Milli-Q water, and analyzed using HPLC.
HPLC analysis was performed using a Waters system comprising a Waters 1525 Binary HPLC pump, a Waters 2996 Photodiode Array Detector, and an analytical 4.6 × 250 mm (5 μm) RPC18 SunFire® 100 column (Waters, Wexford, Ireland). A flow rate of 1 mL/min and a detector wavelength of 214 nm were employed. Samples (50 μL) were injected and eluted using a mobile phase consisting of buffer A (30 mM ammonium formate pH 5.0 and 5% acetonitrile) and buffer B (buffer A plus acetonitrile 20:80, v/v) with an isocratic method (85% of A). In all analyses, pure commercial benzylpenicillin was used as standard.
Penicillin production was normalized based on the dry weight of the mycelium. For this purpose, the remaining mycelium was washed with 2 mL of a 1 M HCl solution, followed by centrifugation at 9000 g for 5 min. The supernatant was discarded, and the washed mycelium was then lyophilized, and its dry weight was determined.
2.7. qRT-PCR Analysis of the Expression of Penicillin Biosynthesis Genes
To isolate RNA, the mycelia were frozen in liquid nitrogen and ground in a mortar. Total RNA extraction was performed using the Plant/FungiTotal RNA Purification Kit (Norgen Biotek Corp., Thorold, ON, Canada) according to the manufacturer’s instructions. To eliminate any contaminating DNA, the extracted RNA was treated with the RNase-Free DNase I Kit (Norgen Biotek Corp, Thorold, ON, Canada). The concentration of RNA was determined using a MultiSkan GO quantification system with a μDrop plate (Thermo Fischer Scientific, Braunschweig, Germany). Next, 1 μg of total RNA was used to synthesize cDNA using All-In-One 5× RT MasterMix (Applied Biological Materials, Richmond, BC, Canada) according to the manufacturer’s instructions.
For gene expression analysis, qRT-PCR was conducted using the primers described in Table 1. Reactions were prepared in 20 μL volumes, containing 10 μL of KAPA SYBR Fast qRT-PCR Master Mix 2× (Kapa Biosystems, Wilmington, MA, USA), 0.4 μL of each primer (at a concentration of 10 μM each), 0.4 μL de 50× ROX High/Low, 6.8 μL of water, and 2 μL of cDNA. The quantification was performed on a StepOne Real-Time PCR System (Applied Biosystems, Waltham, MA, USA) with the following conditions: 30 s at 95 °C and 40 cycles of 3 s at 95 °C and 30 s at 60 °C. Negative controls were included in the experiments. Three replicates were performed for each gene expression analysis. Data were analyzed using the comparative Ct (2−ΔΔCt) method and were normalized to β-tubulin gene expression in each sample.
2.8. Phylogenetic Analysis of Pcz1
The Pcz1 protein sequence from P. rubens Wis 54-1255 was deduced from the corresponding gene sequence and subjected to BlastP analysis to identify sequences with high similarity. Multiple sequence alignment was performed using Clustal Omega, and a phylogenetic tree was constructed using MEGA version 7.0 [33]. The neighbor-joining method was employed to reconstruct the tree, and evolutionary distances were calculated using the Poisson correction model. To assess the robustness of the tree topology, bootstrap analyses with 1000 replications were conducted to estimate the support of internal nodes.
3. Results
3.1. Analysis of the pcz1 Gene and Deduced Protein from P. rubens Wis 54-1255
The pcz1 gene from P. roqueforti [16] was utilized to scan the genome of P. rubens Wis 54-1255 using BLASTN and BLASTX. Both analyses yielded redundant results, confirming that the pcz1 gene corresponds to the gene Pc22g12400 in the P. rubens Wis 54-1255 genome. The pcz1 gene in P. rubens Wis 54-1255 spans 2446 bp and contains a single intron of 70 bp located at the 3′ end of the gene (Figure 1). This gene encodes a protein consisting of 791 amino acids. Analyses of conserved domains were performed using the HMMER tool at Interpro, and CDD at NCBI. Both analyses indicated the presence of a Zn(II)2Cys6 fungal-type domain spanning positions 394–431 of the Pcz1 protein, which contains the conserved six cysteines characteristic of such domains (Figure 1).
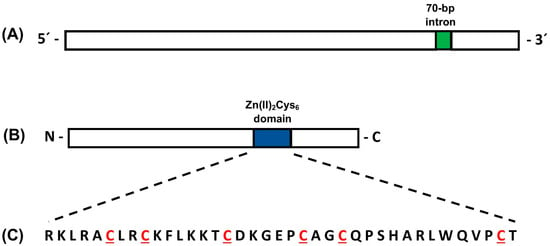
Figure 1.
Schematic representations of the pcz1 gene (A) and Pcz1 protein (B) from P. rubens Wis 54-1255. In panel (A), the single 70 bp intron is depicted in green. Panel (B) highlights the position of the Zn(II)2Cys6 domain in blue. Panel (C) shows the amino acid sequence of the Zn(II)2Cys6 domain, with the six conserved cysteines highlighted in red. The drawings are not to scale.
BlastP analysis of Pcz1 revealed the presence of closely related orthologues in various fungal species belonging to the phylum Ascomycota, particularly within the genus Penicillium. The evolutionary relationships between Pcz1 from P. rubens Wis 54-1255 and its closest orthologous proteins are depicted in Figure 2. This analysis showed that Pcz1 clustered with orthologues within section Chrysogena in Penicillium, in agreement with the established species phylogeny [1].
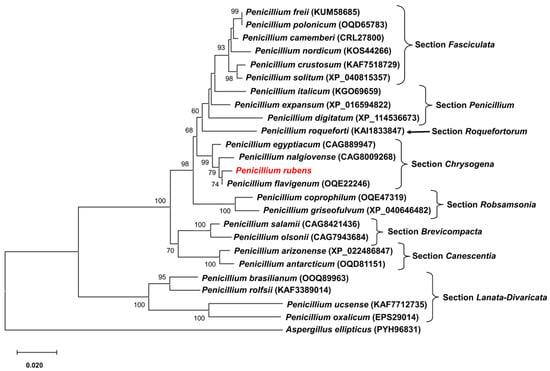
Figure 2.
Phylogenetic tree depicting the relationships among Pcz1 proteins from P. rubens Wis 54-1255 and other fungal species. Pcz1 protein sequences from various species were obtained through BlastP analysis. The sequences included representatives from different sections within the Penicillium genus. The placement of the Pcz1 protein from P. rubens is highlighted in red. The GenBank accession number of each sequence is indicated in parentheses. The Pcz1 protein from Aspergillus ellipticus was used as an outgroup. Bootstrap values of 50% or higher are displayed at each node.
3.2. Generation of pcz1-Disrupted Strains of P. rubens Wis 54-1255 by CRISPR-Cas9
After the transformation of P. rubens Wis 54-1255 with plasmid pFC333-Pcpcz1, thirty-five transformants were obtained. These transformants underwent passages in Czapek media containing phleomycin, followed by passages in phleomycin-free Czapek media. Ten transformants lost phleomycin resistance, and their target region was amplified and sequenced. Notably, four of these transformants exhibited insertions in the pcz1 gene, resulting in gene disruption due to the generation of multiple stop codons (Figure 3). Specifically, three transformants (T12, T14, and T25) shared the same 1 bp insertion, while transformant T6 displayed a large insertion of 336 bp, encompassing various fragments from the AMA region of plasmid pFC333 (Figure 3). Based on these findings, transformants T6 and T14, along with the wild-type strain, were selected for subsequent experiments.
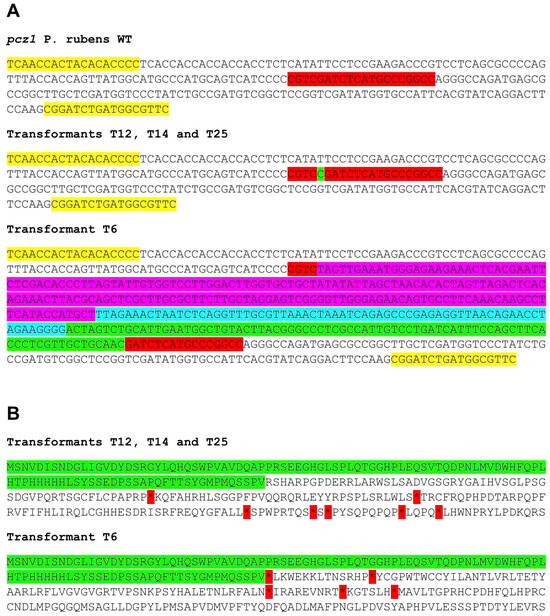
Figure 3.
(A) Sequences of the target region in the pcz1 gene from P. rubens wild type (WT) and transformants T6, T12, T14, and T25. The target sequence is highlighted in red, while the yellow region represents the primers used for amplification (see Table 1). Transformants T12, T14, and T25 exhibit the same one-nucleotide insertion (highlighted in green), while T6 displays different inserted segments from plasmid pFC333 (highlighted in fuchsia, turquoise, and green). (B) Deduced protein sequence of the target region from transformants T6, T12, T14, and T25. The unaltered protein sequence is highlighted in green. After this region, frameshifts occur in transformants, resulting in changes to the protein sequence and the generation of multiple stop codons (asterisks highlighted in red).
3.3. Morphological Features of the pcz1-Disrupted Strains of P. rubens Wis 54-1255
As an initial approach to assess the phenotypic characterization of the transformants T6 and T14, they were subjected to examination in five different media and compared to P. rubens Wis 54-1255 grown under the same conditions (Figure 4A). In general, two discernible morphological changes were noted. Firstly, the characteristic green pigmentation associated with normal sporulation in P. rubens Wis 54-1255 was attenuated in transformants T6 and T14 in certain media, resulting in a visibly paler or even white aspect. This phenomenon was particularly pronounced in PDA, CYA, and Power media (Figure 4A). Additionally, the transformants displayed subtle differences in growth compared to the wild-type strain (Figure 4A), suggesting a slight reduction in their growth rate of the transformants. Both conidiation and growth rates will be examined in detail in subsequent experiments.
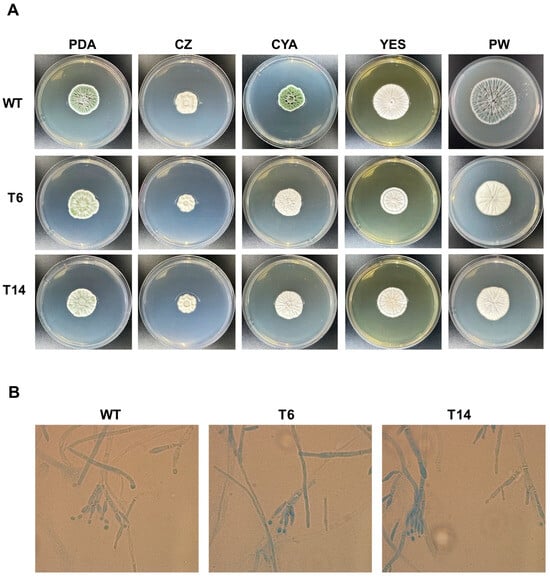
Figure 4.
Phenotypic characteristics of P. rubens strains. (A) Colony morphology of P. rubens Wis 54-1255 (WT) and transformants T6 and T14 after 7 days of growth at 28 °C on PDA, Czapek (CZ), CYA, YES and Power (PW) media. (B) Microscopic observation of P. rubens Wis 54-1255 (WT) and transformants T6 and T14. Fungal samples were obtained from colonies grown on PDA agar for 7 days at 28 °C. Samples were stained with lactophenol cotton blue and observed under bright-field microscopy at 100× magnification.
Furthermore, a microscopic examination of the strains was conducted (Figure 4B). All strains exhibited similar morphological characteristics under microscopic analysis, with no evidence of major morphological alterations. Thereby, hyphae displayed their typical elongated, branch-like structures characteristic of hyphal growth, while conidiophores exhibited the distinctive brush-like appearance typical of Penicillium.
3.4. The Disruption of pcz1 Reduces Growth and Conidiation, but Promotes Conidial Germination in P. rubens Wis 54-1255
The apical growth of fungal strains was evaluated in five different media. In all cases, the pcz1-disrupted strains T6 and T14 exhibited a slight delay in apical growth compared to the wild-type strain of P. rubens (Figure 5). Depending on the specific media used, transformants displayed a growth rate ranging between 77.6% and 89.7% of the wild-type fungus, indicating that pcz1 acts as a positive regulator of growth in P. rubens Wis 54-1255.
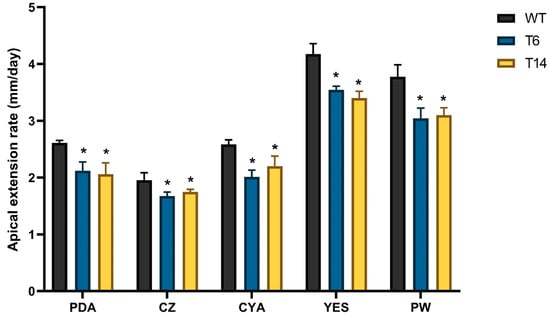
Figure 5.
Apical extension rates of P. rubens Wis 54-1255 (WT) and transformants T6 and T14 on PDA, Czapek (CZ), CYA, YES, and Power (PW) media. Strains were incubated at 28 °C for 7 days. Error bars represent the standard deviation of three replicates in three different experiments. The symbol * indicates statistically significant differences (p < 0.05 using Student’s t-test) in apical extension rates compared to the wild-type strain in the respective medium.
Regarding conidiation, it was assessed in two different media: the minimal medium Czapek, and the nutrient-rich medium Power, specifically designed to enhance conidiation [23]. In both media, the pcz1-disrupted strains T6 and T14 displayed a significant decrease in conidia production compared to the wild-type strain (Figure 6). Depending on the specific media used and day of measurement, the transformants produced between 44.8% and 63.2% of the conidia produced by the wild-type fungus. These findings support the role of pcz1 as a positive regulator of conidiation in P. rubens Wis 54-1255.
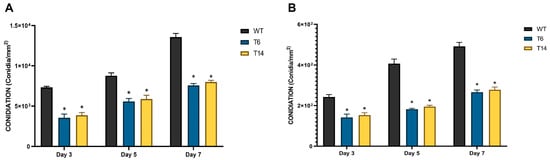
Figure 6.
Conidia production of P. rubens Wis 54-1255 (WT) and transformants T6 and T14 in Power (A) and Czapek (B) media grown at 28 °C for 3, 5, and 7 days. Error bars represent the standard deviation of three replicates in three independent experiments. The asterisk (*) indicates statistically significant differences (p < 0.05 using Student’s t-test) in conidia production between the transformants and the wild-type strain.
Finally, conidial germination was assessed. As shown in Figure 7, the pcz1-disrupted strains T6 and T14 exhibited earlier conidial germination compared to the wild-type strain. For instance, after 7, 8, and 9 h in CM medium, the transformants displayed approximately 56%, 80%, and 90% of conidia germinated, respectively. In contrast, the wild-type P. rubens Wis 54-1255 exhibited significantly lower values of 35%, 56%, and 69% of conidia germinated at the same time points and in the same medium. A similar trend was observed in the minimal medium Czapek, although the differences were less pronounced. These findings indicate that pcz1 functions as a negative regulator of conidial germination in P. rubens Wis 54-1255.
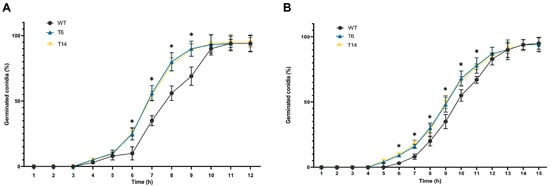
Figure 7.
Germination rates of P. rubens Wis 54-1255 (WT) and transformants T6 and T14 represented as the percentage of germinated conidia versus hours of incubation in CM-rich medium (A) and Czapek minimal medium (B). Error bars represent the standard deviation of three replicates from three independent experiments. The asterisks (*) indicate points on the curves where statistically significant differences (p < 0.05 using Student’s t-test) were observed between the transformants and the wild-type strain.
3.5. The Inactivation of pcz1 Reduces the Production of Penicillin and the Expression of the Penicillin Gene Cluster in P. rubens Wis 54-1255
Penicillin is the most important specialized metabolite produced by P. rubens; therefore, we studied its production in the transformants. As depicted in Figure 8, the production of benzylpenicillin was significantly reduced in the pcz1-disrupted strains. While the wild-type strain of P. rubens Wis 54-1255 produced an average of 2.4 and 3.9 μg/mg of penicillin at 48 and 72 h, respectively, transformant T6 produced 1.2 μg/mg of penicillin, and transformant T14 produced 0.6 and 0.5 μg/mg of penicillin at the same time points (Figure 8). These results indicate that the inactivation of pcz1 decreases the production of benzylpenicillin, suggesting that this gene exerts a positive regulation on the production of this important specialized metabolite in P. rubens Wis 54-1255.
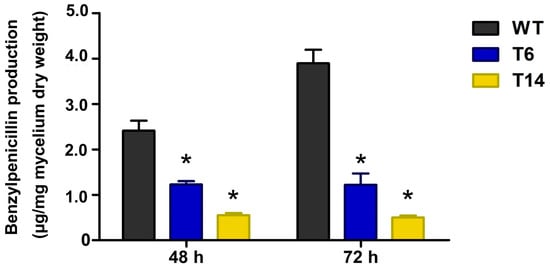
Figure 8.
Production of penicillin by P. rubens Wis 54-1255 (WT) and transformants T6 and T14 in MCFP medium at 48 and 72 h. Data are average of three replicas from three different experiments. Error bars indicate the standard deviation of the mean value. The symbol * indicates statistically significant differences of transformants (p < 0.05 using Student’s t-test) as compared to the wild-type strain at the same times.
Finally, the impact of pcz1 disruption on the expression of the genes in the penicillin BGC was investigated. For this purpose, the relative expression of pcbAB, pcbC, and penDE genes was measured. As shown in Figure 9, the relative expression of these three genes in the pcz1-disrupted strains was significantly lower compared to the wild-type strain. Transformants T6 and T14 exhibited an important reduction in pcbAB transcripts, with a 2.4- and 3.2-fold decrease at 48 h, and a 2.5- and 3.0-fold decrease at 72 h, respectively, compared to P. rubens Wis 54-1255. Similarly, pcbC transcripts showed a 2.3- and 3.0-fold decrease at 48 h, and a 2.4- and 3.2-fold decrease at 72 h, respectively, in the disrupted strains. And in the case of penDE, transcripts displayed a 2.3- and 3.1-fold decrease at 48 h, and a 2.4- and 3.1-fold decrease at 72 h, respectively. These findings confirm that Pcz1 positively regulates the expression of penicillin biosynthetic genes.
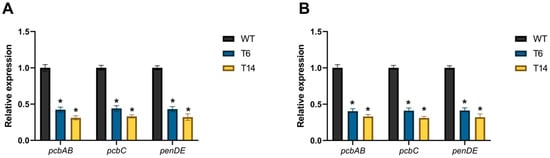
Figure 9.
Relative expression of pcbAB, pcbC, and penDE in pcz1-disrupted transformants at 48 h (A) and 72 h (B) compared to the control P. rubens wild-type strain (WT). The data represent the mean values from three separate experiments, each consisting of three replicates. The error bars indicate the standard deviation of the mean. The symbol * indicates statistically significant differences between transformants and the wild-type strain (p < 0.05, assessed using Student’s t-test).
4. Discussion
To date, the role of the pcz1 gene has only been investigated in the cheese-ripening fungus P. roqueforti. In this organism, it has been observed that pcz1 acts as a positive regulator of growth and conidiation, while exerting a negative regulation on conidial germination [16]. The results obtained in our study indicate that the role of pcz1 in the regulation of these processes is conserved in P. rubens. Taken together, these findings suggest an emerging role of pcz1 in asexual development across Penicillium species, possibly through conserved mechanisms, and underscore the importance of further exploring the regulatory role of pcz1 in fungi beyond the Penicillium genus.
In addition to its impact on asexual development, the inactivation of pcz1 in P. rubens Wis 54-1255 has an important effect on penicillin production. Specifically, the pcz1-disrupted strains T6 and T14 exhibited a significant reduction in benzylpenicillin production compared to the wild-type strain. The decrease in penicillin production ranged from approximately 50% to 87% in comparison to the wild-type strain at 48 and 72 h, respectively (Figure 8). These results provide clear evidence that the inactivation of pcz1 diminishes the production of penicillin, indicating a positive regulatory role of pcz1 in the biosynthesis of this important specialized metabolite in P. rubens Wis 54-1255. To the best of our knowledge, this is the first study implicating a gene encoding a Zn(II)2Cys6 protein in the regulation of penicillin biosynthesis in P. rubens.
The positive regulatory role of pcz1 in penicillin production suggests that the overexpression of this regulator could potentially serve to increase penicillin production in P. rubens. This hypothesis finds support in recent research demonstrating that overexpression of pcz1 in P. roqueforti increased the production of mycophenolic acid, an important immunosuppressive compound [17]. In P. rubens, various efforts have been made to increase penicillin production through the overexpression of global regulators, yielding varied results. For instance, the overexpression of regulators like PcLaeA, PcRFX1, PcYap1, PcRsmA, and PcFKH1 led to an increased production of penicillin, although in some cases (for example, PcFKH1) the increase was modest [34,35,36,37]. Conversely, the overexpression of the positive regulators of penicillin production StuA and MAT1-1-1 had no impact on penicillin titers in P. rubens [38,39]. Thus, the effectiveness of employing the overexpression of global regulators to enhance penicillin production in P. rubens varies on a case-by-case basis. In the future, it will be interesting to investigate whether the overexpression of pcz1 effectively increases penicillin production in P. rubens.
In P. roqueforti, Rojas-Aedo et al. [17] employed RNAi-mediated gene silencing to downregulate the expression of pcz1 and measured the effect of this genetic manipulation on the production of three specialized metabolites: roquefortine C, andrastin A, and mycophenolic acid. They observed reduced production of the three specialized metabolites in the RNAi-downregulated strains, which correlated with decreased transcription of genes within the biosynthetic gene clusters responsible for their biosynthesis [17]. Collectively, our results and these previous findings in P. roqueforti suggest a potential general role for pcz1 in regulating specialized metabolite production in fungi.
The precise molecular mechanism by which Pcz1 exerts its regulatory role remains unknown. As mentioned in the Introduction, Pcz1 is a protein containing a Zn(II)2Cys6 domain, a domain unique to fungi that is commonly found in “zinc finger” proteins [10,40]. Notable examples of fungal proteins containing this kind of domain include Gal4, a transcription factor involved in galactose metabolism in yeast [41], and AflR, a regulator of aflatoxin biosynthesis in Aspergillus [42]. These proteins typically bind specific DNA promoter sequences of their target genes [41,42]. Therefore, it is plausible to hypothesize that Pcz1 exerts its regulatory function by directly or indirectly interacting with promoter sequences of one or more target genes. Further investigations, such as chromatin immunoprecipitation coupled with DNA sequencing, are necessary to elucidate the putative binding sequences of Pcz1 in promoter regions. These experimental approaches are clearly beyond the scope of the present study.
A recent review proposed a regulatory model of pcz1 based on findings in P. roqueforti [43]. According to this model, the expression of pcz1 is negatively influenced by pga1, a gene that encodes a heterotrimeric G protein alpha subunit [43]. The effects of Pga1 on asexual development in P. rubens have been previously investigated [30], and these findings match well with the proposed model for P. roqueforti and the results obtained in our study. However, regarding penicillin production, the regulatory model of Pga1 on Pcz1 proposed for P. roqueforti does not agree with the experimental evidence in P. rubens. According to the model, a negative regulatory role of Pga1 on Pcz1 in P. rubens would imply a negative regulation of penicillin production by Pga1, contradicting previous experimental data that indicate a positive effect of Pga1 on penicillin production in P. rubens [44]. Thus, the relationship between Pga1 and Pcz1 appears to be more complex than the model proposed for P. roqueforti, and it is possible that the relationship between Pga1 and Pcz1 differs between P. rubens and P. roqueforti. This highlights the importance of conducting investigations to understand the regulation of fungal specialized metabolism on a case-by-case basis, as the functional relationships observed in one fungal species may not necessarily apply to others.
In conclusion, our study provides evidence for the significant role of Pcz1, a protein containing a Zn(II)2Cys6 domain, in governing asexual development and functioning as a regulator of penicillin production in P. rubens. It is important to note that penicillin production in this fungus is controlled by a complex network of regulatory circuits that are not yet fully understood [3]. Pcz1 emerges as a novel component within this regulatory network, and the future study of its mechanisms will contribute to improving our understanding of the regulation of penicillin production.
Author Contributions
Conceptualization, R.C. and I.V.; methodology, C.G.-D., D.P., Y.M., J.F.R.-A., R.C. and I.V.; formal analysis, C.G.-D. and R.C.; investigation, C.G.-D., D.P., Y.M. and J.F.R.-A.; resources, R.C., I.V., J.-L.P., C.M. and G.L.; data curation, R.C.; writing—original draft preparation, R.C. and I.V.; writing—review and editing, R.C. and I.V.; visualization, C.G.-D. and R.C.; supervision, R.C., I.V., J.-L.P., C.M. and G.L.; project administration, R.C., I.V., C.G.-D. and G.L.; funding acquisition, R.C., I.V. and G.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by “Universidad de Santiago de Chile, Usach, Proyecto DICYT 021943CHR, Vicerrectoría de Investigación, Desarrollo e Innovación”.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank Martin E. Kogle and Uffe H. Mortensen of the Technical University of Denmark for providing the plasmid pFC333. R.C. and I.V. would like to dedicate this work to Francisco “Paco” Fierro, mentor and friend who recently passed away.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Houbraken, J.; Kocsubé, S.; Visagie, C.M.; Yilmaz, N.; Wang, X.C.; Meijer, M.; Kraak, B.; Hubka, V.; Bensch, K.; Samson, R.A.; et al. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Stud. Mycol. 2020, 95, 5–169. [Google Scholar] [CrossRef] [PubMed]
- Houbraken, J.; Frisvad, J.C.; Samson, R.A. Fleming’s penicillin producing strain is not Penicillium chrysogenum but P. rubens. IMA Fungus 2011, 2, 87–95. [Google Scholar] [CrossRef]
- Fierro, F.; Vaca, I.; Castillo, N.I.; García-Rico, R.O.; Chávez, R. Penicillium chrysogenum, a vintage model with a cutting-edge profile in biotechnology. Microorganisms 2022, 10, 573. [Google Scholar] [CrossRef]
- García-Estrada, C.; Martín, J.F.; Cueto, L.; Barreiro, C. Omics approaches applied to Penicillium chrysogenum and penicillin production: Revealing the secrets of improved productivity. Genes 2020, 11, 712. [Google Scholar] [CrossRef] [PubMed]
- Sawant, A.M.; Vamkudoth, K.R. Biosynthetic process and strain improvement approaches for industrial penicillin production. Biotechnol. Lett. 2022, 44, 179–192. [Google Scholar] [CrossRef]
- El Hajj Assaf, C.; Zetina-Serrano, C.; Tahtah, N.; Khoury, A.E.; Atoui, A.; Oswald, I.P.; Puel, O.; Lorber, S. Regulation of secondary metabolism in the Penicillium genus. Int. J. Mol. Sci. 2020, 21, 9462. [Google Scholar] [CrossRef] [PubMed]
- Martín, J.F. Vacuolal and peroxisomal calcium ion transporters in yeasts and fungi: Key role in the translocation of intermediates in the biosynthesis of fungal metabolites. Genes 2022, 13, 1450. [Google Scholar] [CrossRef] [PubMed]
- Fierro, F.; García-Estrada, C.; Castillo, N.I.; Rodríguez, R.; Velasco-Conde, T.; Martin, J.F. Transcriptional and bioinformatics analysis of the 58.6 kb DNA region amplified in tandem repeats containing the penicillin gene cluster in Penicillium chrysogenum. Fungal Genet. Biol. 2006, 43, 618–629. [Google Scholar] [CrossRef]
- van den Berg, M.A.; Westerlaken, I.; Leeflang, C.; Kerkman, R.; Bovenberg, R.A.L. Functional characterization of the penicillin biosynthetic gene cluster of Penicillium chrysogenum Wisconsin 54-1255. Fungal Genet. Biol. 2007, 44, 830–844. [Google Scholar] [CrossRef]
- García-Estrada, C.; Domínguez-Santos, R.; Kosalková, K.; Martín, J.-F. Transcription factors controlling primary and secondary metabolism in filamentous fungi: The β-lactam paradigm. Fermentation 2018, 4, 47. [Google Scholar] [CrossRef]
- MacPherson, S.; Larochelle, M.; Turcotte, B. A fungal family of transcriptional regulators: The zinc cluster proteins. Microbiol. Mol. Biol. Rev. 2006, 70, 583–604. [Google Scholar] [CrossRef]
- van den Berg, M.A.; Albang, R.; Albermann, K.; Badger, J.H.; Daran, J.M.; Driessen, A.J.; Garcia-Estrada, C.; Fedorova, N.D.; Harris, D.M.; Heijne, W.H.; et al. Genome sequencing and analysis of the filamentous fungus Penicillium chrysogenum. Nat. Biotechnol. 2008, 26, 1161–1168. [Google Scholar] [CrossRef]
- Guzmán-Chávez, F.; Salo, O.; Nygård, Y.; Lankhorst, P.P.; Bovenberg, R.A.L.; Driessen, A.J.M. Mechanism and regulation of sorbicillin biosynthesis by Penicillium chrysogenum. Microb. Biotechnol. 2017, 10, 958–968. [Google Scholar] [CrossRef] [PubMed]
- Tudzynski, B. Nitrogen regulation of fungal secondary metabolism in fungi. Front. Microbiol. 2014, 5, 656. [Google Scholar] [CrossRef] [PubMed]
- Espeso, E.A.; Villarino, M.; Carreras, M.; Alonso-Guirado, L.; Alonso, J.M.; Melgarejo, P.; Larena, I. Altered nitrogen metabolism in biocontrol strains of Penicillium rubens. Fungal Genet. Biol. 2019, 132, 103263. [Google Scholar] [CrossRef]
- Gil-Durán, C.; Rojas-Aedo, J.F.; Medina, E.; Vaca, I.; García-Rico, R.O.; Villagrán, S.; Levicán, G.; Chávez, R. The pcz1 gene, which encodes a Zn(II)2Cys6 protein, is involved in the control of growth, conidiation, and conidial germination in the filamentous fungus Penicillium roqueforti. PLoS ONE 2015, 10, e0120740. [Google Scholar] [CrossRef]
- Rojas-Aedo, J.F.; Gil-Durán, C.; Goity, A.; Vaca, I.; Levicán, G.; Larrondo, L.F.; Chávez, R. The developmental regulator Pcz1 affects the production of secondary metabolites in the filamentous fungus Penicillium roqueforti. Microbiol. Res. 2018, 212–213, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Pohl, C.; Kiel, J.A.K.W.; Driessen, A.J.M.; Bovenberg, R.A.L.; Nygård, Y. CRISPR/Cas9 based genome editing of Penicillium chrysogenum. ACS Synth. Biol. 2016, 5, 754–764. [Google Scholar] [CrossRef]
- Pohl, C.; Polli, F.; Schütze, T.; Viggiano, A.; Mózsik, L.; Jung, S.; de Vries, M.; Bovenberg, R.A.L.; Meyer, V.; Driessen, A.J.M. A Penicillium rubens platform strain for secondary metabolite production. Sci. Rep. 2020, 10, 7630. [Google Scholar] [CrossRef]
- Mózsik, L.; Büttel, Z.; Bovenberg, R.A.L.; Driessen, A.J.M.; Nygård, Y. Synthetic control devices for gene regulation in Penicillium chrysogenum. Microb. Cell Fact. 2019, 18, 203. [Google Scholar] [CrossRef]
- Mózsik, L.; Hoekzema, M.; de Kok, N.A.W.; Bovenberg, R.A.L.; Nygård, Y.; Driessen, A.J.M. CRISPR-based transcriptional activation tool for silent genes in filamentous fungi. Sci. Rep. 2021, 11, 1118. [Google Scholar] [CrossRef]
- Del-Cid, A.; Gil-Durán, C.; Vaca, I.; Rojas-Aedo, J.F.; García-Rico, R.O.; Levicán, G.; Chávez, R. Identification and functional analysis of the mycophenolic acid gene cluster of Penicillium roqueforti. PLoS ONE 2016, 11, e0147047. [Google Scholar] [CrossRef]
- Fierro, F.; Montenegro, E.; Gutiérrez, S.; Martín, J.F. Mutants blocked in penicillin biosynthesis show a deletion of the entire penicillin gene cluster at a specific site within a conserved hexanucleotide sequence. Appl. Microbiol. Biotechnol. 1996, 44, 597–604. [Google Scholar] [CrossRef]
- Stemmer, M.; Thumberger, T.; Del Sol Keyer, M.; Wittbrodt, J.; Mateo, J.L. CCTop: An intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS ONE 2015, 10, e0124633. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Ouedraogo, J.-P.; Kolbusz, M.; Nguyen, T.T.M.; Tsang, A. Efficient genome editing using tRNA promoter-driven CRISPR/Cas9 gRNA in Aspergillus niger. PLoS ONE 2018, 13, e0202868. [Google Scholar] [CrossRef] [PubMed]
- van Leeuwe, T.M.; Arentshorst, M.; Ernst, T.; Alazi, E.; Punt, P.J.; Ram, A.F.J. Efficient marker free CRISPR/Cas9 genome editing for functional analysis of gene families in filamentous fungi. Fungal Biol. Biotechnol. 2019, 6, 13. [Google Scholar] [CrossRef] [PubMed]
- Nødvig, C.S.; Nielsen, J.B.; Kogle, M.E.; Mortensen, U.H. A CRISPR-Cas9 system for genetic engineering of filamentous fungi. PLoS ONE 2015, 10, e0133085. [Google Scholar] [CrossRef]
- Chávez, R.; Roa, A.; Navarrete, K.; Trebotich, J.; Espinosa, Y.; Vaca, I. Evaluation of properties of several cheese-ripening fungi for potential biotechnological applications. Mycoscience 2010, 51, 84–87. [Google Scholar] [CrossRef]
- Ivey, F.D.; Hodge, P.N.; Turner, G.E.; Borkovich, K.A. The Gαi homologue gna-1 controls multiple differentiation pathways in Neurospora crassa. Mol. Biol. Cell. 1996, 7, 1283–1297. [Google Scholar] [CrossRef]
- García-Rico, R.O.; Martín, J.F.; Fierro, F. The pga1 gene of Penicillium chrysogenum NRRL 1951 encodes a heterotrimeric G protein alpha subunit that controls growth and development. Res. Microbiol. 2007, 158, 437–446. [Google Scholar] [CrossRef]
- Pérez, E.A.; Fernández, F.J.; Fierro, F.; Mejía, A.; Marcos, A.T.; Martín, J.F.; Barrios-González, J. Yeast HXK2 gene reverts glucose regulation mutation of penicillin biosynthesis in P. chrysogenum. Braz. J. Microbiol. 2014, 45, 873–883. [Google Scholar] [CrossRef] [PubMed]
- García-Estrada, C.; Vaca, I.; Lamas-Maceiras, M.; Martín, J.F. In vivo transport of the intermediates of the penicillin biosynthetic pathway in tailored strains of Penicillium chrysogenum. Appl. Microbiol. Biotechnol. 2007, 76, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Kosalková, K.; García-Estrada, C.; Ullán, R.V.; Godio, R.P.; Feltrer, R.; Teijeira, F.; Mauriz, E.; Martín, J.F. The global regulator LaeA controls penicillin biosynthesis, pigmentation and sporulation, but not roquefortine C synthesis in Penicillium chrysogenum. Biochimie 2009, 91, 214–225. [Google Scholar] [CrossRef]
- Domínguez-Santos, R.; Martín, J.F.; Kosalková, K.; Prieto, C.; Ullán, R.V.; García-Estrada, C. The regulatory factor PcRFX1 controls the expression of the three genes of β-lactam biosynthesis in Penicillium chrysogenum. Fungal Genet. Biol. 2012, 49, 866–881. [Google Scholar] [CrossRef]
- Pérez-Pérez, W.D.; Carrasco-Navarro, U.; García-Estrada, C.; Kosalková, K.; Gutiérrez-Ruíz, M.C.; Barrios-González, J.; Fierro, F. bZIP transcription factors PcYap1 and PcRsmA link oxidative stress response to secondary metabolism and development in Penicillium chrysogenum. Microb. Cell Fact. 2022, 21, 50. [Google Scholar] [CrossRef] [PubMed]
- Domínguez-Santos, R.; García-Estrada, C.; Kosalková, K.; Prieto, C.; Santamarta, I.; Martín, J.F. PcFKH1, a novel regulatory factor from the forkhead family, controls the biosynthesis of penicillin in Penicillium chrysogenum. Biochimie 2015, 115, 162–176. [Google Scholar] [CrossRef] [PubMed]
- Böhm, J.; Hoff, B.; O’Gorman, C.M.; Wolfers, S.; Klix, V.; Binger, D.; Zadra, I.; Kürnsteiner, H.; Pöggeler, S.; Dyer, P.S.; et al. Sexual reproduction and mating-type-mediated strain development in the penicillin-producing fungus Penicillium chrysogenum. Proc. Natl. Acad. Sci. USA 2013, 110, 1476–1481. [Google Scholar] [CrossRef]
- Sigl, C.; Haas, H.; Specht, T.; Pfaller, K.; Kürnsteiner, H.; Zadra, I. Among developmental regulators, StuA but not BrlA is essential for penicillin V production in Penicillium chrysogenum. Appl. Environ. Microbiol. 2011, 77, 972–982. [Google Scholar] [CrossRef]
- Chang, P.K.; Ehrlich, K.C. Genome-wide analysis of the Zn(II)2Cys6 zinc cluster-encoding gene family in Aspergillus flavus. Appl. Microbiol. Biotechnol. 2013, 97, 4289–4300. [Google Scholar] [CrossRef]
- Choudhury, B.I.; Whiteway, M. Evolutionary transition of GAL regulatory circuit from generalist to specialist function in Ascomycetes. Trends Microbiol. 2018, 26, 692–702. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.; Ghazali, F.M.; Mahyudin, N.A.; Samsudin, N.I.P. Aflatoxin biosynthesis, genetic regulation, toxicity, and control strategies: A review. J. Fungi 2021, 7, 606. [Google Scholar] [CrossRef] [PubMed]
- Coton, E.; Coton, M.; Hymery, N.; Mounier, J.; Jany, J.-L. Penicillium roqueforti: An overview of its genetics, physiology, metabolism and biotechnological applications. Fungal Biol. Rev. 2020, 34, 59–73. [Google Scholar] [CrossRef]
- García-Rico, R.O.; Fierro, F.; Mauriz, E.; Gómez, A.; Fernández-Bodega, M.A.; Martín, J.F. The heterotrimeric Gα protein Pga1 regulates biosynthesis of penicillin, chrysogenin and roquefortine in Penicillium chrysogenum. Microbiology 2008, 154, 3567–3578. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).