Abstract
Penicillium digitatum is one of the most important phytopathogens. It causes deterioration and rotting of citrus fruits, generating significant economic losses worldwide. As a human pathogen, it is extremely rare. We present a case of pulmonary co-infection in a patient diagnosed with pneumonia due to SARS-CoV-2. A 20-year-old female patient, primigravid, 36 weeks of gestation, without comorbidities, and diagnosed with severe pneumonia due to the SARS-CoV-2, showed rapid lung deterioration for which their pregnancy was interrupted by surgery. The patient was hospitalized in the Intensive Care Unit (ICU), connected to mechanical ventilation and receiving corticosteroids and antibiotics. The diagnosis of pulmonary fungal infection was made through bronchoalveolar lavage (BAL) culture, and the species identification was performed by sequencing of β-tubulin. Phylogenetic analysis with related species was performed for the confirmation of species identification. Antifungal susceptibility tests were performed for itraconazole (4 µg/mL), voriconazole (2 µg/mL), and amphotericin B (2 µg/mL). The patient was successfully treated with itraconazole. This is the second worldwide report of pulmonary infection by P. digitatum and the first in Chile. Although it is a fungus that rarely infects humans, it could represent an emerging opportunistic fungal pathogen, with associated risk factors that should be considered in the differential diagnosis of Penicillium species isolated from infections in humans.
1. Introduction
Penicillium digitatum is a mesophilic fungus and one of the most devasting agents of deterioration and rotting of citrus fruits [1,2]. Together with Penicillium italicum, it causes about 90% of losses worldwide, mainly affecting the post-harvest stages. However, it can generate deleterious effects in all phases of production, from the cultivation of the fruits to collection, packaging, storage, transport, and market, or even after being acquired by the consumer [2]. Due to the production of ethylene, indole alkaloids, and the secretion of enzymes that soften the adjacent fruits, the infection in fruits spreads rapidly, generating complete rot in about 4–5 days, spreading easily through contact [3,4]. It acts as a necrotrophic organism generating infection through previous damage to the surface of the fruit and is recognized as an agent capable of producing a potential mycotoxin called citrinin, which is associated with nephrotoxic, embryotoxic, teratogenic, and carcinogenic effects in humans and animals [5,6,7].
Infections caused by P. digitatum in humans are extremely rare. Currently, there is only one clinical report developed in 2013 in Japan that corresponded to fatal pneumonia [8]. This study describes the second clinical report of P. digitatum worldwide and the first in Chile, in a patient who developed pneumonia associated with COVID-19 that was successfully treated with itraconazole.
Case Presentation
A 20-year-old woman from the rural area of La Araucanía region of Chile, primigravid, 36 weeks of gestation, without comorbidities, was admitted to a health center for presenting, in the prior 5 days, commitment of the general state, headache, myalgia, dry cough, progressive dyspnea, and anosmia with 10 days of evolution. On physical examination, she was found to be tachypneic, tachycardic, and saturating 88%; as the bilateral crepitus was auscultated, a chest radiograph was performed that showed bilateral interstitial compromise with basal predominance. According to clinical suspicion, SARS-CoV-2 virus detection tests were carried out, which were positive, so she was diagnosed with severe COVID-19 pneumonia. Due to the significant pulmonary compromise and thus need for oxygen therapy, and due to her advanced pregnancy condition, she was transferred to Dr. Hernán Henríquez Aravena Hospital (Temuco, Chile), where she underwent an emergency cesarean section and started antimicrobial treatment with ceftriaxone 2 g/iv/day, dexamethasone 6 mg/iv/day, enoxaparin 40 mg/sc/day. She was admitted to the ICU, where she was connected to a mechanical ventilator. Chest computed tomography (CT) on admission showed bilateral ground glass opacities and condensing opacities, consistent with organizing pneumonia. On the 10th day after surgery, she was successfully extubated and maintained with oxygen support through a High-Flow Nasal Cannula (HFNC) (50 L/min). However, she remained under surveillance, since she presented a dimer D elevation of 7.70 µg/mL, leukocytosis with 18,890 cells/µL, C-reactive protein (CRP) of 8.8 mg/L, and bilateral wheezing. Due to a worsening of her clinical condition with purulent secretions associated with poor respiratory mechanics, tachycardia, and hypertension, with a fever of 38 °C, she was reconnected to mechanical ventilation 2 days after being extubated and a new CT was performed, which showed consolidation of ground glass opacities and the appearance of new bilateral nodular opacities in the upper lobes; pulmonary embolism (PE) was ruled out (Figure 1). On the 13th day after admission, BAL was taken for a complete pathogen study that included bacterial culture, fungal culture, FilmArray (26 pathogens), galactomannan, and molecular detection of Pneumocystis jirovecii; peripheral blood and central venous catheter (CVC) blood cultures were also taken, and empiric therapy was started with vancomycin 2 g/iv/day for 4 days, meropenem 1 g every 8 h/iv for 7 days, and dexamethasone for 1 day. Bacterial culture was negative, molecular detection of P. jirovecii negative, galactomannan 0.15, Gram stain without germs and FilmArray (26 pathogens) negative. The patient persisted with fevers and pulmonary secretions, so on the 20th day after admission, a chest CT without contrast was performed, where multiple bilateral ground-glass opacities, predominantly at the bases and without pleural effusion or pneumothorax, were observed. Fungal growth was detected in fungal culture, identified by micromorphology as Penicillium sp. Therefore, treatment with itraconazole 400 mg for 10 days was started, followed by 200 mg/day for 28 days. After starting antifungal treatment, the patient began to evolve favorably, remaining stable, afebrile, and with decreasing inflammatory parameters. After completing 8 days with itraconazole, it was decided to disconnect her from mechanical ventilation, and the procedure was well-tolerated and uneventful. Finally, after 16 days of antifungal treatment and with good clinical and laboratory evolution, it was decided to continue antifungal treatment on an outpatient basis, and the patient was discharged from the hospital.
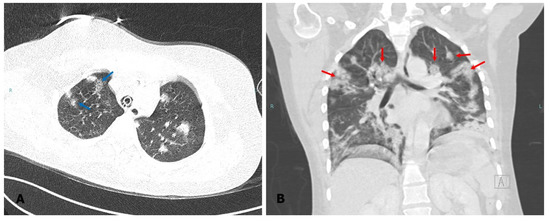
Figure 1.
Computed tomography of the chest. The image taken prior to the diagnosis of fungal infection shows consolidation of ground glass opacities ((A), blue arrow) and bilateral nodular opacities in the upper lobes ((B), red arrow).
2. Materials and Methods
2.1. Fungal Culture and Phenotypic Identification
The fungal culture of the BAL sample was performed on potato dextrose agar (PDA) (Biokar Diagnostics, France) in duplicate, incubating at 25 °C in darkness. The micromorphological study was carried out through the microscopic observation of preparations made with Lactophenol and with cotton blue Lactophenol solutions. The images were obtained with the TissueFAXS I PLUS Cytometer TissueGnostics Axio Observer 7 Carl Zeiss GmbH (TissueGnostics GmbH, Vienna, Austria). Images were obtained in the fields of view at 40× and 63× magnifications.
2.2. Molecular Identification and Phylogenetic Analysis
Species identification was performed through sequencing of the phylogenetic marker β-tubulin (BenA) according to the recommendations of Visagie et al. [9]. For this, the fungal genomic DNA was extracted using the commercial kit Mini kit QIAamp DNA (Qiagen), according to the manufacturer’s instructions. PCR for BenA amplification was performed using the primer pairs T10/bt2b following previously described protocols [10], and the PCR product was purified and stored at −20 °C until sequencing. The sequences were obtained using the same primer pairs at Austral-omics of the Universidad Austral de Chile using the ABI Prism 310 automated sequencer. Sequence editing was done with SeqMan v. 7.0.0 (DNAStar Lasergene, Madison, WI, USA), and the consensus sequence obtained was submitted for comparison in the National Center for Biotechnology Information (NCBI) database using the BLASTn tool. Subsequently, the phylogenetic analysis of the β-tubulin marker was performed with inclusion of the sequences of the strain CEMT 2 together with sequences of seven ex-type strains of species representatives of the series Clavigera, Sclerotigena, Italica, Penicillium, and Digitata. The alignment of the locus was performed in MEGA software (Molecular Evolutionary Genetics Analysis) v. 6.0 [11], through Clustal W algorithm [12] and refined with MUSCLE [13] or manually if necessary. The phylogenetic analysis was performed using the maximum-likelihood method (ML) under the same software. The best nucleotide substitution model for the ML was Kimura 2-parameter with gamma distribution (K2 + G). Bootstrap values ≥ 70% were considered significant. Sequences from Aspergillus fumigatus CBS 133.61 and Aspergillus clavatus CBS 513.65 were used as outgroup (Table 1).

Table 1.
Penicillium species included in the phylogenetic study, their respective origin, and GenBank accession number.
2.3. In Vitro Antifungals Susceptibility Tests
The broth microdilution method was performed according to the guidelines of the CLSI document M38-A2 [14]. Minimum inhibitory concentration (MIC) values were determined for amphotericin B (Sigma-Aldrich, St. Louis, MO, USA), voriconazole (Sigma-Aldrich), and itraconazole (Sigma-Aldrich). Fungal inoculum suspensions were prepared in sterile distilled water from 5-day-old fungal culture on PDA at 28 °C. The suspension was filtered through sterile gauze. Cell counts were determined with a hemocytometer and the inoculum size was adjusted to around 105 conidia/mL. Stock solutions of antifungals were prepared in 100% dimethyl sulfoxide (DMSO) to 100-fold the final concentration needed and further diluted in RPMI 1640 pH 7.0 (Sigma-Aldrich Co., St. Louis, MO, USA) to obtain the ×2 drug concentration. Dilutions of the fungicides were prepared and dispensed into 96-well microdilution plates. Each microdilution well containing 100 μL of the appropriate antifungal solution (2× final concentration) was inoculated with 100 μL of the conidial inoculum suspension, yielding final antifungal concentrations of 0.06, 0.12, 0.24, 0.5, 1, 2, 4, 8, 16, and 32 μg/mL. The growth control wells contained 100 μL of the inoculum suspension and 100 μL of RPMI medium. Sterility control wells contained 200 μL of RPMI medium. For determination of MIC, microdilution plates were incubated at 27 °C and visually examined using a stereoscopic magnifier (SZ61TR, OLYMPUS) from 0 up to 48 h from the time of inoculation. MIC was defined as the lowest concentration of each antifungal agent that causes a specified reduction in visible growth of the fungal strain on the broth dilution susceptibility test. The reference strains Fusarium oxysporum (ATCC 36031) and Fusarium keratoplasticum (ATCC 48112) were used as quality control for the test.
2.4. Ethics Statement
The present study has the approval of the Ethics Committee of Servicio de Salud Araucanía Sur (protocol code N° 26 approved on 25 January 2022). Informed consent was obtained from the patient involved in the study.
3. Results
3.1. Phenotypic Study and Preliminary Identification
On the PDA culture medium, fungal development was observed after 7 days of incubation at 25 °C with the development of velvety colonies with a greenish hue (Figure 2A). The microscopic characterization of the isolate (CEMT 2) was performed via observation of specimen mounted in Lactophenol and cotton blue Lactophenol. The observation of septate hyaline hyphae, from which conidiophores, metulae, phialides and oval conidia-forming chains originated, allowed us to identify the isolate as Penicillium sp. (Figure 2C–F)
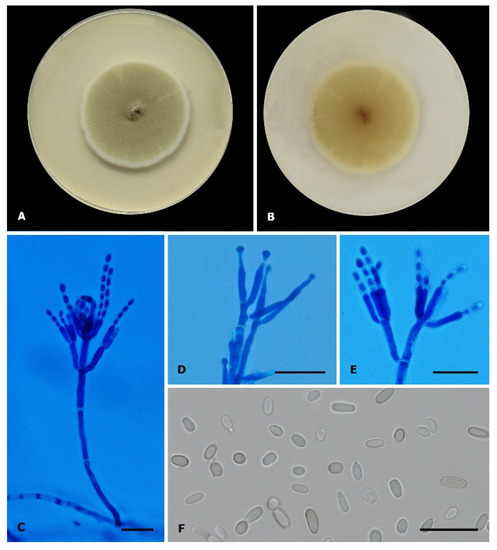
Figure 2.
Penicillium digitatum (CEMT-2). (A,B) Colony on PDA, in front and reverse respectively, incubated at 25 °C for 7 days. (C–E) Conidiophores, metulae, phialides, and conidia (Cotton blue Lactophenol mounting solution). (F) Conidia (Lactophenol mounting solution). Scale bars: 20 µm.
3.2. Molecular Identification and Phylogenetic Analysis
The molecular identification confirmed the preliminary identification and defined the strain as Penicillium digitatum, which presented 100% identity with several reference strain sequences of that species and 99.75% identity with the sequence of the type-strain (P. digitatum CBS 112082). The sequence data generated in the present study were deposited in GenBank (Table 1). The phylogenetic analysis carried out with the type-strain of the series Digitata (series that includes the identified species) and other phylogenetically related series (Penicillium, Italica, Sclerotigena and Clavigera) confirmed the identification, showing the formation of a well-supported clade (bootstrap of 99%) made up of the strain of the present study (CEMT 2) and the type-strain of P. digitatum (CBS 112082). The phylogenetic relationships between our isolate and other related species are shown in Figure 3.
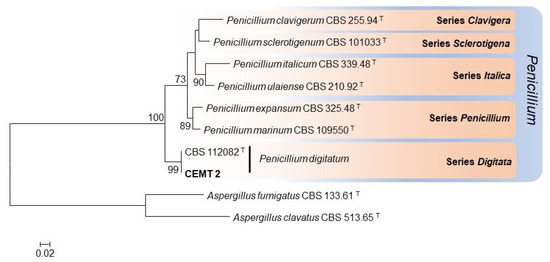
Figure 3.
Maximum-likelihood phylogenetic tree constructed with β-tubulin sequences from 7 representative species of the series Clavigera, Sclerotigena, Italica, Penicillium, and Digitata. Bootstrap values ≥ 70% are indicated in the nodes. The fungal strain of the present study is indicated in bold. Branch lengths are proportional to phylogenetic distance. T: Ex-type species.
3.3. Results of In Vitro Antifungal Susceptibility Test
The susceptibility profiles obtained showed an in vitro activity of the three antifungal drugs studied, with low MIC values ranging between 2 and 4 µg/mL, amphotericin B and voriconazole being the ones that showed better activity. MIC values for amphotericin B, voriconazole, and itraconazole are shown in Table 2.

Table 2.
Results of in vitro antifungal susceptibility test of the strain Penicillium digitatum CEMT-2.
4. Discussion
Invasive fungal infections (IFI) have shown a considerable increase in recent decades, constituting an emerging problem worldwide. Although they mainly affect patients with risk factors such as hematologic malignancies, solid organ and bone marrow transplants, acquired immunodeficiency syndrome (AIDS), corticosteroid treatment, extensive use of antibiotics, CVC, chemotherapy, or mechanical ventilation, among several others [15,16], serious fungal infections have also been described in immunocompetent individuals [17,18,19,20,21,22].
Aspergillosis undoubtedly constitutes the most frequent IFI, not only in patients with the aforementioned risk factors, but also in patients with pulmonary infection by SARS-CoV-2. In this last group, cases of co-infection have been reported worldwide, even establishing the concept of COVID-19-associated pulmonary aspergillosis (CAPA) [23]. Although COVID-19 patients are not immunocompromised patients as such, they present a series of risk factors that make them susceptible to the development of infections by other agents, whether bacterial or fungal. Among them, local pulmonary hypoxia, a product of the viral infection itself, and damage to the respiratory epithelium, mechanical ventilation, prolonged hospitalizations, and therapy with broad-spectrum antibiotics and immunosuppressive drugs, particularly systemic corticosteroids, are of great relevance, since together they contribute to the settlement and germination of external agents, facilitating colonization and invasion [24,25,26].
Infections by non-Aspergillus filamentous fungi are considerably less frequent and also constitute a challenge not only from the point of view of their treatment, due to the high levels of resistance that some species present, but also due to the difficulty in their correct identification, a product of the increasing number of cryptic species described in various fungal genera [27]. Regarding the genus Penicillium, until a few years ago, the species Penicillium marneffei was described as the most frequent in clinical infections in humans. However, this species is currently classified within the genus Talaromyces (T. marneffei), so the clinical reports caused by other Penicillium species are very rare. Lyratzopoulos et al. [28] conducted a review of 31 cases of invasive fungal infection by Penicillium species, among which they described P. chrysogenum, P. decumbens P. janthinellum, P. lilacinum, P. purporogenum, P. citrinum, and P. brevicompactum as the majority of them, isolated from invasive pulmonary infection [28].
Penicillium digitatum is a fungus with worldwide distribution, widely recognized as a phytopathogen in citrus fruits, with clinical isolates being extremely rare [2]. There has only been one previous report made in Japan, in which this species was described as an agent of pneumonia with a fatal outcome, in an elderly patient who was suffering from pulmonary emphysema and malnutrition [8]. Although the patient in the current case reported not having the habit of consuming citrus fruits very frequently, she could have acquired it from any other external source, even in her own home, precisely because it is an environmental fungus. This is why it is very difficult to establish the source of contagion in this type of organism with certainty. Although the patient was immunocompetent and without underlying diseases, she was suffering from a pulmonary infection due to SARS-CoV-2, requiring the administration of oxygen therapy, which conditioned the interruption of her pregnancy through surgery, hospitalization in ICU, installation of CVC, connection to mechanical ventilation, and treatment with corticosteroids and antibiotics—all of these being risk factors that facilitate the development of IFI. Co-infection between SARS-CoV-2 and fungal species has been reported in various studies worldwide [24,29,30,31,32,33]. The genus Aspergillus appears as the most frequent agent [23,34,35,36]; however, the present study highlights the pathogenic potential of other emerging fungal species in patients diagnosed with COVID-19.
Regarding the diagnostic tests recommended in COVID-19 patients for the diagnosis of CAPA, it is important to highlight the detection of galactomannan antigen, which constitutes an important component of the cell wall of Aspergillus spp., secreted in vivo during the growth of the fungus [37,38]. Therefore, its detection has high sensitivity and specificity for the diagnosis of this fungal infection; however, its usefulness in invasive infections by other genera is limited. Based on the consensus criteria established by ECMM/ISHAM for the diagnosis of CAPA, a patient with pulmonary infiltrates associated with an optical density index (ODI) value in BAL ≥ 1.0 could be diagnosed with a possible CAPA [39]. In this case, since the ODI in BAL was 0.15, it was possible to rule out CAPA, but not an invasive pulmonary infection caused by another type of fungus.
Unfortunately, in the report by Oshikata et al. [8], antifungal susceptibility tests were not carried out; however, they described P. digitatum as a species resistant to antifungals, an assertion based only on the clinical evolution of the patient due to a poor response despite the administration of itraconazole, micafungin, voriconazole, and amphotericin B. Our results disagree with what was described by Oshikata et al. [8], since the MIC values found were in general lower, 2 µg/mL for amphotericin B and voriconazole, or slightly higher, 4 µg/mL for itraconazole. Although amphotericin B is described as a good therapeutic option in cases of invasive fungal infection by Penicillium non-marneffei species [28], in this report and unlike that described by Oshikata et al. [8], only itraconazole was administered with good therapeutic success, showing high levels of susceptibility not only in vitro, but also in vivo. In any case, it is essential to analyze a larger number of isolates of clinical and environmental origin in order to establish an antifungal susceptibility profile in this species and determine if there is intra-species variability.
Although this species still has a low clinical frequency, it clearly implies a greater burden on the health system, generating higher costs, prolonged hospitalizations, and greater work absenteeism. In the present study, we describe the first clinical isolation of P. digitatum in Chile, being the second clinical report worldwide, postulating it as an emerging opportunistic fungal pathogen that should be considered in the differential diagnosis of Penicillium spp. isolated from infectious diseases in humans.
Author Contributions
Conceptualization, I.I.-G.; methodology, I.I.-G., C.S., M.S., F.C. and V.B.; software, I.I.-G.; writing—original draft preparation, I.I.-G.; writing—review and editing, I.I.-G., A.G., P.G.-M., F.V.; supervision, F.V., V.T., G.R., L.O. and A.S.M.; project administration, F.F.-S.; funding acquisition, R.S. All authors have read and agreed to the published version of the manuscript.
Funding
Study funded by Universidad de La Frontera, Project DFP21-0020.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Servicio de Salud Araucanía Sur (protocol code N°26 approved on 25 January 2022).
Informed Consent Statement
Informed consent was obtained from the patient involved in the study. Written informed consent has been obtained from the patient to publish this paper.
Acknowledgments
The authors wish to thank the professionals of the Universidad de La Frontera in Temuco, Chile for their help in obtaining the micromorphology images of P. digitatum incorporated in this article with the equipment TissueFAXS i PLUS Cytometer TissueGnostics Axio Observer 7 Carl Zeiss GmbH (TissueGnostics GmbH, Vienna, Austria), project ANID FONDEQUIP EQM200228. The authors also thank the Center of Excellence in Translational Medicine and Vicerrectoría Académica of the Universidad de La Frontera, Temuco, Chile, for their support and contributions to carrying out this study. C.S. and M.S. offer thanks for the partial funding from the Universidad de La Frontera through the research project FAPESP-UFRO 2020/07546-2.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Vilanova, L.; Viñas, I.; Torres, R.; Usall, J.; Jauset, A.; Teixidó, N. Infection capacities in the orange-pathogen relationship: Compatible (Penicillium digitatum) and incompatible (Penicillium expansum) interactions. Food Microbiol. 2012, 29, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Qian, X.; Dhanasekaran, S.; Boateng, N.A.S.; Yan, X.; Zhu, H.; He, F.; Zhang, H. Study on the Infection Mechanism of Penicillium digitatum on Postharvest Citrus (Citrus Reticulata Blanco) Based on Transcriptomics. Microorganisms 2019, 7, 672. [Google Scholar] [CrossRef] [PubMed]
- Ballester, A.R.; González-Candelas, L. EFE-Mediated Ethylene Synthesis Is the Major Pathway in the Citrus Postharvest Pathogen Penicillium digitatum during Fruit Infection. J. Fungi 2020, 6, 175. [Google Scholar] [CrossRef]
- Costa, J.H.; Bazioli, J.M.; Barbosa, L.D.; Júnior, P.L.T.D.S.; Reis, F.C.G.; Klimeck, T.; Crnkovic, C.M.; Berlinck, R.G.S.; Sussulini, A.; Rodrigues, M.L.; et al. Phytotoxic Tryptoquialanines Produced In Vivo by Penicillium digitatum Are Exported in Extracellular Vesicles. mBio 2021, 12, e03393-20. [Google Scholar] [CrossRef] [PubMed]
- Arai, M.; Hibino, T. Tumorigenicity of citrinin in male F344 rats. Cancer Lett. 1983, 17, 281–287. [Google Scholar] [CrossRef]
- Flajs, D.; Peraica, M. Toxicological properties of citrinin. Arh. Hig. Rada Toksikol. 2009, 60, 457–464. [Google Scholar] [CrossRef]
- Singh, N.D.; Sharma, A.K.; Patil, R.D.; Rahman, S.; Leishangthem, G.D.; Kumar, M. Effect of feeding graded doses of Citrinin on clinical and teratology in female Wistar rats. Indian J. Exp. Biol. 2014, 52, 159–167. [Google Scholar]
- Oshikata, C.; Tsurikisawa, N.; Saito, A.; Watanabe, M.; Kamata, Y.; Tanaka, M.; Tsuburai, T.; Mitomi, H.; Takatori, K.; Yasueda, H.; et al. Fatal Pneumonia Caused by Penicillium digitatum: A Case Report. BMC Pulm. Med. 2013, 13, 16. Available online: http://www.biomedcentral.com/1471-2466/13/16 (accessed on 23 March 2013). [CrossRef]
- Visagie, C.; Houbraken, J.; Frisvad, J.; Hong, S.-B.; Klaassen, C.; Perrone, G.; Seifert, K.; Varga, J.; Yaguchi, T.; Samson, R. Identification and nomenclature of the genus Penicillium. Stud. Mycol. 2014, 78, 343–371. [Google Scholar] [CrossRef]
- Houbraken, J.; Kocsubé, S.; Visagie, C.M.; Yilmaz, N.; Wang, X.-C.; Meijer, M.; Kraak, B.; Hubka, V.; Bensch, K.; Samson, R.A.; et al. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Stud. Mycol. 2020, 95, 5–169. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi; Approved Standard-Second Edition, CLSI document M38-A2; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2008. [Google Scholar]
- Low, C.Y.; Rotstein, C. Emerging fungal infections in immunocompromised patients. F1000 Med. Rep. 2011, 3, 14. [Google Scholar] [CrossRef] [PubMed]
- Enoch, D.A.; Yang, H.; Aliyu, S.H.; Micallef, C. The Changing Epidemiology of Invasive Fungal Infections. Methods Mol. Biol. 2017, 1508, 17–65. [Google Scholar] [PubMed]
- Denning, D.W.; Chakrabarti, A. Pulmonary and sinus fungal diseases in non-immunocompromised patients. Lancet Infect. Dis. 2017, 17, e357–e366. [Google Scholar] [CrossRef]
- Adulkar, N.G.; Radhakrishnan, S.; Vidhya, N.; Kim, U. Invasive sino-orbital fungal infections in immunocompetent patients: A clinico-pathological study. Eye 2019, 33, 988–994. [Google Scholar] [CrossRef]
- Rudramurthy, S.M.; Singh, S. Candida Infections in Immunocompetent Hosts: Pathogenesis and Diagnosis. Curr. Fungal Infect. Rep. 2020, 14, 233–245. [Google Scholar] [CrossRef]
- Cao, X.-G.; Yu, C.-W.; Zhou, S.-S.; Huang, Y.; Wang, C.-Y. Case Report: A Candida Meningitis in an Immunocompetent Patient Detected Through the Next-Generation Sequencing. Front. Med. 2021, 22, 656066. [Google Scholar] [CrossRef]
- He, J.; Sheng, G.; Yue, H.; Zhang, F.; Zhang, H.-L. Isolated pulmonary mucormycosis in an immunocompetent patient: A case report and systematic review of the literature. BMC Pulm. Med. 2021, 21, 138. [Google Scholar] [CrossRef]
- Kumar, S.; Muthu, V.; Bansal, Y.S.; Mehta, N.; Arora, V. Invasive pulmonary aspergillosis with brain dissemination in an immunocompetent host. Autops. Case Rep. 2021, 11, e2021280. [Google Scholar] [CrossRef] [PubMed]
- Arastehfar, A.; Carvalho, A.; Van De Veerdonk, F.L.; Jenks, J.D.; Koehler, P.; Krause, R.; Cornely, O.A.; Perlin, D.S.; Lass-Flörl, C.; Hoenigl, M. COVID-19 Associated Pulmonary Aspergillosis (CAPA)–From Immunology to Treatment. J. Fungi 2020, 6, 91. [Google Scholar] [CrossRef] [PubMed]
- Salmanton-García, J.; Sprute, R.; Stemler, J.; Bartoletti, M.; Dupont, D.; Valerio, M.; Garcia-Vidal, C.; Falces-Romero, I.; Machado, M.; de la Villa, S.; et al. COVID-19-Associated Pulmonary Aspergillosis, March-August 2020. Emerg. Infect. Dis. 2021, 27, 1077–1086. [Google Scholar] [CrossRef] [PubMed]
- Chaharom, F.E.; Pourafkari, L.; Chaharom, A.A.E.; Nader, N.D. Effects of corticosteroids on Covid-19 patients: A systematic review and meta-analysis on clinical outcomes. Pulm. Pharmacol. Ther. 2022, 72, 102107. [Google Scholar] [CrossRef]
- Närhi, F.; Moonesinghe, S.R.; Shenkin, S.D.; Drake, T.M.; Mulholland, R.H.; Donegan, C.; Dunning, J.; Fairfield, C.J.; Girvan, M.; Hardwick, H.E.; et al. Implementation of corticosteroids in treatment of COVID-19 in the ISARIC WHO Clinical Characterisation Protocol UK: Prospective, cohort study. Lancet Digit. Health 2022, 4, e220–e234. [Google Scholar] [CrossRef]
- Tan, Y.P.; Crous, P.W.; Shivas, R.G. Cryptic species of Curvularia in the culture collection of the Queensland Plant Pathology Herbarium. MycoKeys 2018, 35, 1–25. [Google Scholar] [CrossRef]
- Lyratzopoulos, G.; Ellis, M.; Nerringer, R.; Denning, D. Invasive infection due to Penicillium species other than P. marneffei. J. Infect. 2002, 45, 184–195. [Google Scholar] [CrossRef]
- Hughes, S.; Troise, O.; Donaldson, H.; Mughal, N.; Moore, L.S.P. Bacterial and fungal coinfection among hospitalized patients with COVID-19: A retrospective cohort study in a UK secondary-care setting. Clin. Microbiol. Infect. 2020, 26, 1395–1399. [Google Scholar] [CrossRef]
- Baskaran, V.; Lawrence, H.; Lansbury, L.E.; Webb, K.; Safavi, S.; Zainuddin, N.I.; Huq, T.; Eggleston, C.; Ellis, J.; Thakker, C.; et al. Co-infection in critically ill patients with COVID-19: An observational cohort study from England. J. Med. Microbiol. 2021, 70, 001350. [Google Scholar] [CrossRef]
- Peng, J.; Wang, Q.; Mei, H.; Zheng, H.; Liang, G.; She, X.; Liu, W. Fungal co-infection in COVID-19 patients: Evidence from a systematic review and meta-analysis. Aging 2021, 13, 7745–7757. [Google Scholar] [CrossRef]
- Yang, S.; Hua, M.; Liu, X.; Du, C.; Pu, L.; Xiang, P.; Wang, L.; Liu, J. Bacterial and fungal co-infections among COVID-19 patients in intensive care unit. Microbes Infect. 2021, 23, 104806. [Google Scholar] [CrossRef] [PubMed]
- Shafiekhani, M.; Shekari, Z.; Boorboor, A.; Zare, Z.; Arabsheybani, S.; Azadeh, N. Bacterial and fungal co-infections with SARS-CoV-2 in solid organ recipients: A retrospective study. Virol. J. 2022, 19, 35. [Google Scholar] [CrossRef] [PubMed]
- Koehler, P.; Cornely, O.A.; Böttiger, B.W.; Dusse, F.; Eichenauer, D.A.; Fuchs, F.; Hallek, M.; Jung, N.; Klein, F.; Persigehl, T.; et al. COVID-19 associated pulmonary aspergillosis. Mycoses 2020, 63, 528–534. [Google Scholar] [CrossRef] [PubMed]
- Nasir, N.; Farooqi, J.; Mahmood, S.F.; Jabeen, K. COVID-19-associated pulmonary aspergillosis (CAPA) in patients admitted with severe COVID-19 pneumonia: An observational study from Pakistan. Mycoses 2020, 63, 766–770. [Google Scholar] [CrossRef]
- van Arkel, A.L.; Rijpstra, T.A.; Belderbos, H.N.; Van Wijngaarden, P.; Verweij, P.E.; Bentvelsen, R.G. COVID-19-associated Pulmonary Aspergillosis. Am. J. Respir. Crit. Care Med. 2020, 202, 132–135. [Google Scholar] [CrossRef]
- Latgé, J.P.; Kobayashi, H.; Debeaupuis, J.P.; Diaquin, M.; Sarfati, J.; Wieruszeski, J.M.; Parra, E.; Bouchara, J.P.; Fournet, B. Chemical and immunological characterization of the extracellular galactomannan of Aspergillus fumigatus. Infect. Immun. 1994, 62, 5424–5433. [Google Scholar] [CrossRef]
- Mercier, T.; Guldentops, E.; Lagrou, K.; Maertens, J. Galactomannan, a Surrogate Marker for Outcome in Invasive Aspergillosis: Finally Coming of Age. Front. Microbiol. 2018, 9, 661. [Google Scholar] [CrossRef]
- Koehler, P.; Bassetti, M.; Chakrabarti, A.; Chen, S.C.A.; Colombo, A.L.; Hoenigl, M.; Klimko, N.; Lass-Flörl, C.; Oladele, R.O.; Vinh, D.C.; et al. Defining and managing COVID-19-associated pulmonary aspergillosis: The 2020 ECMM/ISHAM consensus criteria for research and clinical guidance. Lancet Infect. Dis. 2021, 21, e149–e162. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).