Unveiling the Core Effector Proteins of Oil Palm Pathogen Ganoderma boninense via Pan-Secretome Analysis
Abstract
:1. Introduction
2. Results and Discussion
2.1. Whole-Genome Sequencing, Assembly, and Functional Annotation of G. boninense Genomes
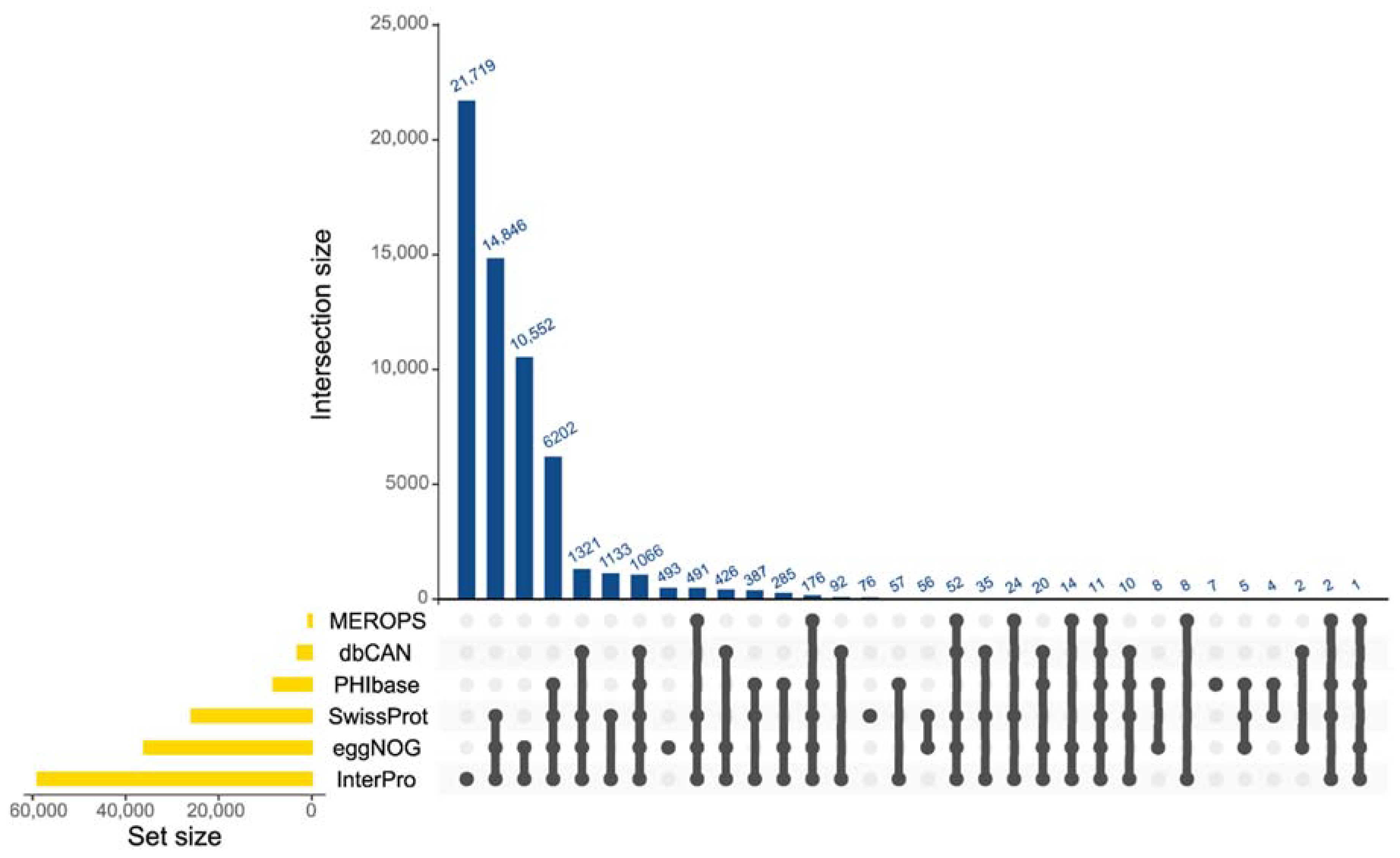
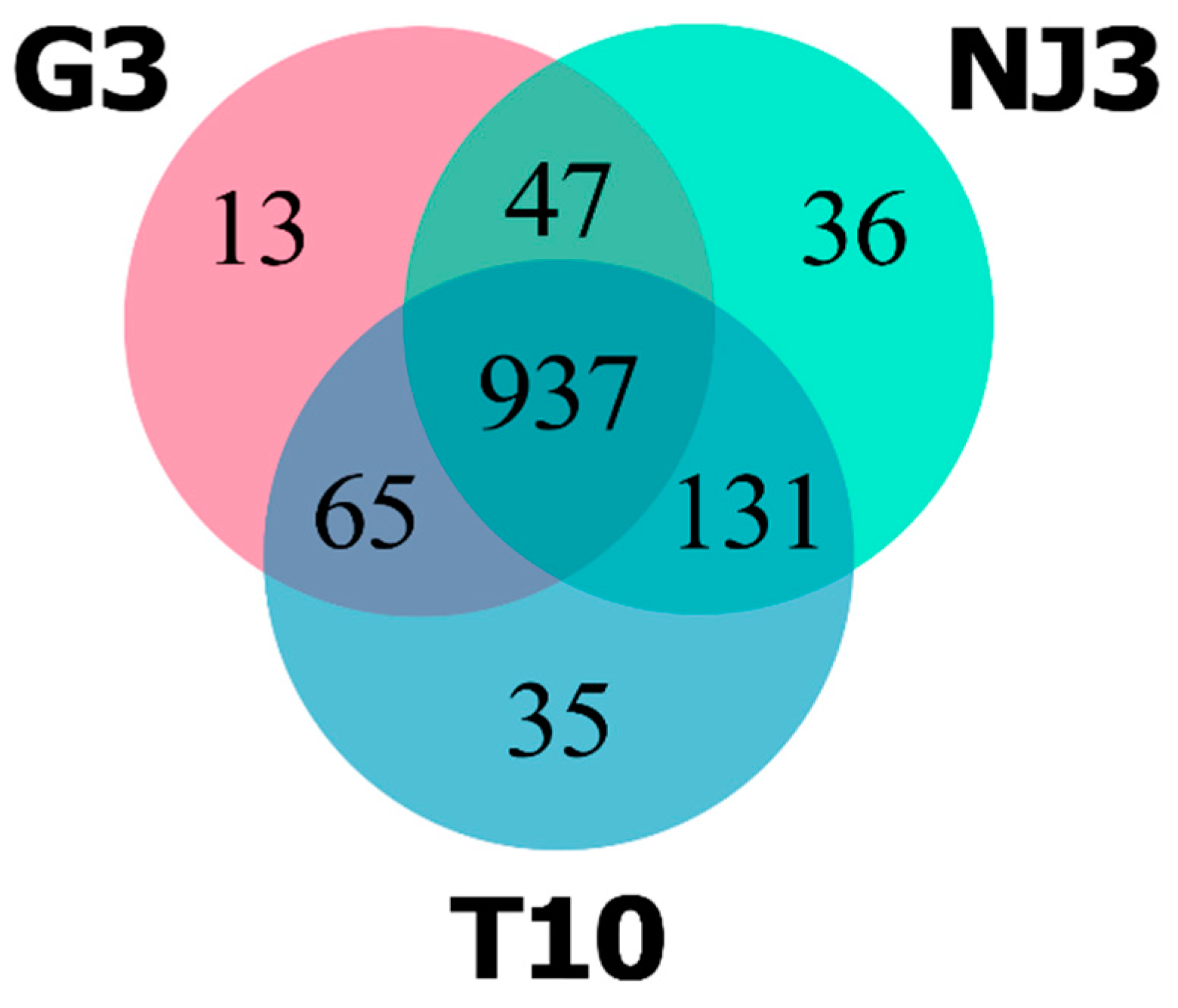
2.2. The Pan-Secretome of Ganoderma boninense
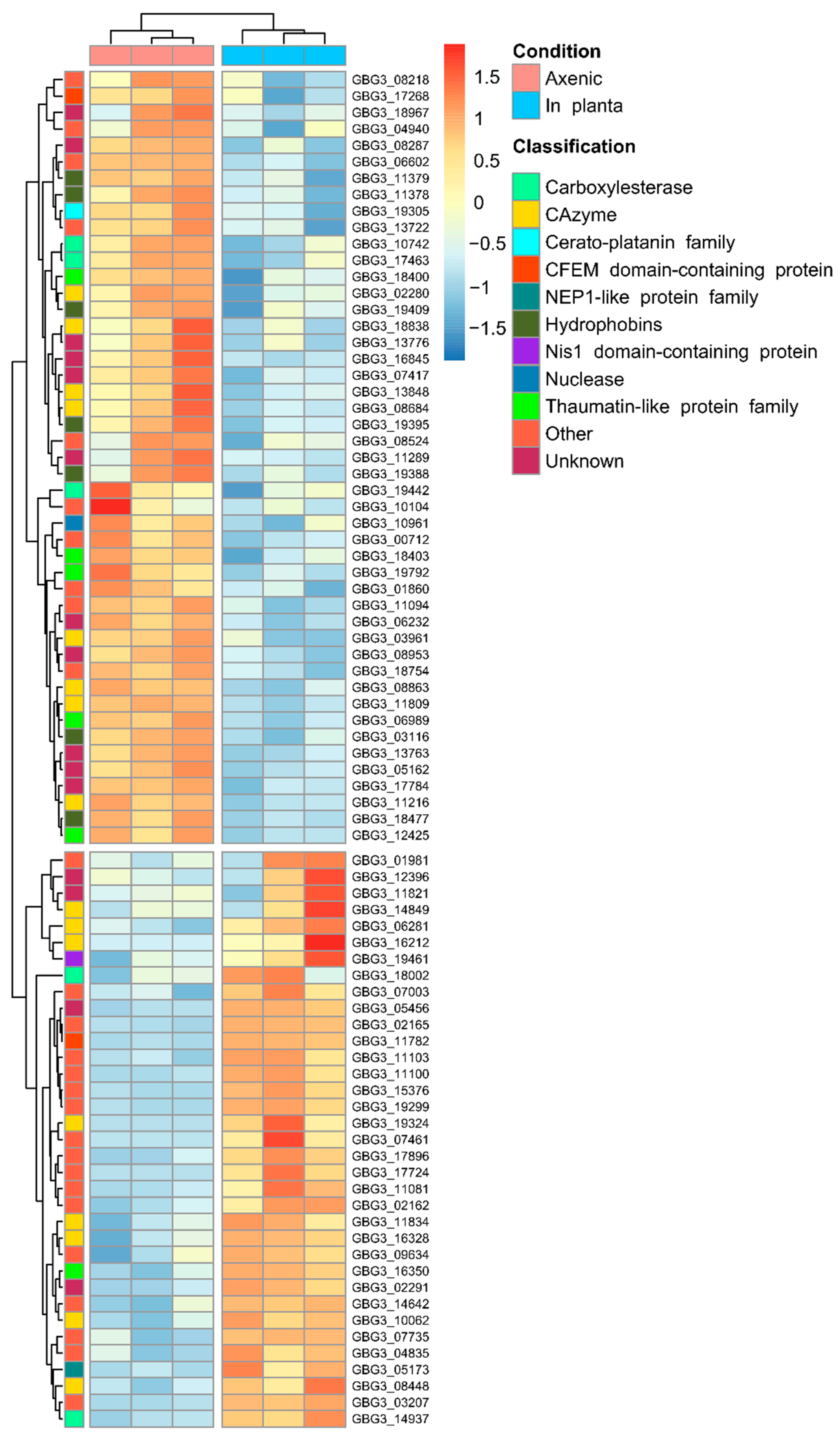
2.3. Differential Expression of Candidate Effector Genes in G. boninense
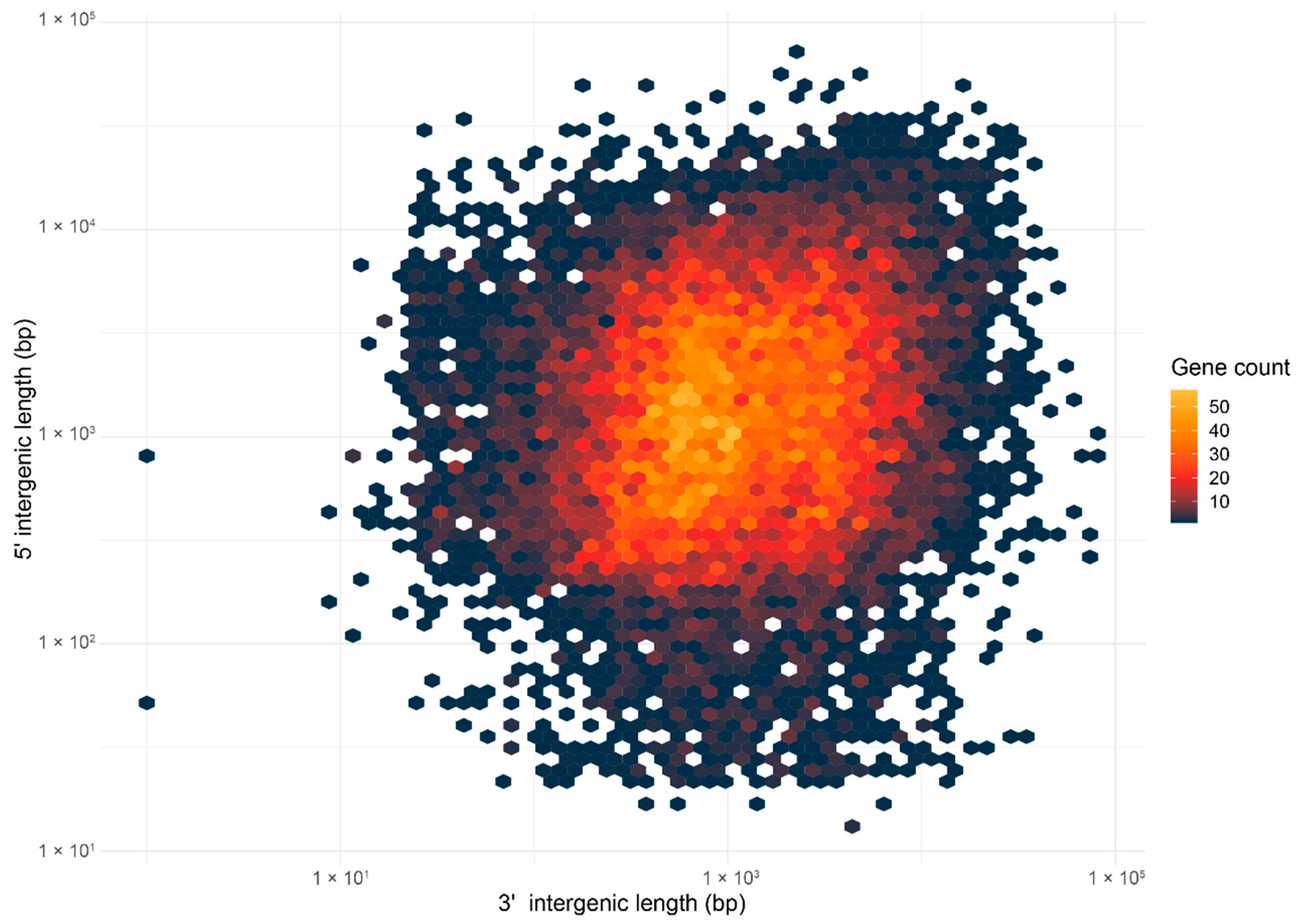
2.4. Ganoderma boninense Possesses the One-Speed Genome Architecture
3. Materials and Methods
3.1. Data Retrieval
3.2. Ganoderma Boninense Strain T10
3.3. Genomic DNA Extraction and Whole-Genome Sequencing
3.4. Data Pre-Processing and Genome Assembly
3.5. Annotation of Repetitive Elements
3.6. Gene Prediction and Annotation
3.7. Differential Expression Analysis
3.8. Genome Architecture Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Corley, R.H.V.; Tinker, P.B. The Oil Palm, 4th ed.; John Wiley & Sons: Hoboken, NJ, USA, 2008. [Google Scholar]
- Siddiqui, Y.; Surendran, A.; Paterson, R.R.M.; Ali, A.; Ahmad, K. Current strategies and perspectives in detection and control of basal stem rot of oil palm. Saudi J. Biol. Sci. 2021, 28, 2840–2849. [Google Scholar] [CrossRef] [PubMed]
- Olaniyi, O.N.; Szulczyk, K.R. Estimating the economic damage and treatment cost of basal stem rot striking the Malaysian oil palms. For. Policy Econ. 2020, 116, 102163. [Google Scholar] [CrossRef]
- Paterson, R.R.M. Ganoderma boninense Disease of Oil Palm to Significantly Reduce Production After 2050 in Sumatra if Projected Climate Change Occurs. Microorganisms 2019, 7, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Govender, N.; Wong, M.Y. Detection of Oil Palm Root Penetration by Agrobacterium-Mediated Transformed Ganoderma boninense, Expressing Green Fluorescent Protein. Phytopathology 2017, 107, 483–490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rees, R.W.; Flood, J.; Hasan, Y.; Potter, U.; Cooper, R.M. Basal stem rot of oil palm (Elaeis guineensis); mode of root infection and lower stem invasion by Ganoderma boninense. Plant Pathol. 2009, 58, 982–989. [Google Scholar] [CrossRef]
- Ramzi, A.B.; Che Me, M.L.; Ruslan, U.S.; Baharum, S.N.; Nor Muhammad, N.A. Insight into plant cell wall degradation and pathogenesis of Ganoderma boninense via comparative genome analysis. PeerJ 2019, 7, e8065. [Google Scholar] [CrossRef] [Green Version]
- Chong, K.P.; Dayou, J.; Alexander, A. Detection and Control of Ganoderma boninense in Oil Palm Crop; Springer: Berlin/Heidelberg, Germany, 2017; p. 50. [Google Scholar]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [Green Version]
- Kanyuka, K.; Rudd, J.J. Cell surface immune receptors: The guardians of the plant’s extracellular spaces. Curr. Opin. Plant Biol. 2019, 50, 1–8. [Google Scholar] [CrossRef]
- Abdul Malik, N.A.; Kumar, I.S.; Nadarajah, K. Elicitor and Receptor Molecules: Orchestrators of Plant Defense and Immunity. Int. J. Mol. Sci. 2020, 21, 963. [Google Scholar] [CrossRef] [Green Version]
- Uhse, S.; Djamei, A. Effectors of plant-colonizing fungi and beyond. PLoS Pathog. 2018, 14, e1006992. [Google Scholar] [CrossRef] [Green Version]
- Ho, C.L.; Tan, Y.C.; Yeoh, K.A.; Ghazali, A.K.; Yee, W.Y.; Hoh, C.C. De novo transcriptome analyses of host-fungal interactions in oil palm (Elaeis guineensis Jacq.). BMC Genom. 2016, 17, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shokrollahi, N.; Ho, C.-L.; Zainudin, N.A.I.M.; Wahab, M.A.w.B.A.; Wong, M.-Y. Identification of non-ribosomal peptide synthetase in Ganoderma boninense Pat. that was expressed during the interaction with oil palm. Sci. Rep. 2021, 11, 16330. [Google Scholar] [CrossRef] [PubMed]
- Teh, C.-Y.; Pang, C.-L.; Tor, X.-Y.; Ho, P.-Y.; Lim, Y.-Y.; Namasivayam, P.; Ho, C.-L. Molecular cloning and functional analysis of a necrosis and ethylene inducing protein (NEP) from Ganoderma boninense. Physiol. Mol. Plant Pathol. 2019, 106, 42–48. [Google Scholar] [CrossRef]
- Yoshida, K.; Saunders, D.G.; Mitsuoka, C.; Natsume, S.; Kosugi, S.; Saitoh, H.; Inoue, Y.; Chuma, I.; Tosa, Y.; Cano, L.M.; et al. Host specialization of the blast fungus Magnaporthe oryzae is associated with dynamic gain and loss of genes linked to transposable elements. BMC Genom. 2016, 17, 370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, Z.; Oliveira-Garcia, E.; Lin, G.; Hu, Y.; Dalby, M.; Migeon, P.; Tang, H.; Farman, M.; Cook, D.; White, F.F.; et al. Effector gene reshuffling involves dispensable mini-chromosomes in the wheat blast fungus. PLoS Genet. 2019, 15, e1008272. [Google Scholar] [CrossRef] [Green Version]
- Fouche, S.; Plissonneau, C.; Croll, D. The birth and death of effectors in rapidly evolving filamentous pathogen genomes. Curr. Opin. Microbiol. 2018, 46, 34–42. [Google Scholar] [CrossRef]
- Dong, S.; Raffaele, S.; Kamoun, S. The two-speed genomes of filamentous pathogens: Waltz with plants. Curr. Opin. Genet. Dev. 2015, 35, 57–65. [Google Scholar] [CrossRef]
- Sanchez-Vallet, A.; Fouche, S.; Fudal, I.; Hartmann, F.E.; Soyer, J.L.; Tellier, A.; Croll, D. The Genome Biology of Effector Gene Evolution in Filamentous Plant Pathogens. Annu. Rev. Phytopathol. 2018, 56, 21–40. [Google Scholar] [CrossRef]
- Kok, S.M.; Goh, Y.K.; Jiat, T.; Goh, K.; Wei Chee, W.; Goh, Y.K. In vitro growth of Ganoderma boninense isolates on novel palm extract medium and virulence on oil palm (Elaeis guineensis) seedlings. Malays. J. Microbiol. 2013, 9, 33–42. [Google Scholar] [CrossRef]
- Lex, A.; Gehlenborg, N.; Strobelt, H.; Vuillemot, R.; Pfister, H. UpSet: Visualization of Intersecting Sets. IEEE Trans. Vis. Comput. Graph. 2014, 20, 1983–1992. [Google Scholar] [CrossRef]
- Urban, M.; Cuzick, A.; Seager, J.; Wood, V.; Rutherford, K.; Venkatesh, S.Y.; De Silva, N.; Martinez, M.C.; Pedro, H.; Yates, A.D.; et al. PHI-base: The pathogen–host interactions database. Nucleic Acids Res. 2019, 48, D613–D620. [Google Scholar] [CrossRef] [PubMed]
- Kubicek, C.P.; Starr, T.L.; Glass, N.L. Plant cell wall-degrading enzymes and their secretion in plant-pathogenic fungi. Annu. Rev. Phytopathol. 2014, 52, 427–451. [Google Scholar] [CrossRef] [PubMed]
- Leuthner, B.; Aichinger, C.; Oehmen, E.; Koopmann, E.; Müller, O.; Müller, P.; Kahmann, R.; Bölker, M.; Schreier, P.H. A H2O2-producing glyoxal oxidase is required for filamentous growth and pathogenicity in Ustilago maydis. Mol. Genet. Genom. 2005, 272, 639–650. [Google Scholar] [CrossRef] [Green Version]
- Song, X.S.; Xing, S.; Li, H.P.; Zhang, J.B.; Qu, B.; Jiang, J.H.; Fan, C.; Yang, P.; Liu, J.L.; Hu, Z.Q.; et al. An antibody that confers plant disease resistance targets a membrane-bound glyoxal oxidase in Fusarium. New Phytol. 2016, 210, 997–1010. [Google Scholar] [CrossRef] [PubMed]
- Daou, M.; Faulds, C.B. Glyoxal oxidases: Their nature and properties. World J. Microbiol. Biotechnol. 2017, 33, 87. [Google Scholar] [CrossRef] [PubMed]
- Gardiner, D.M.; Kazan, K.; Manners, J.M. Novel Genes of Fusarium graminearum That Negatively Regulate Deoxynivalenol Production and Virulence. Mol. Plant-Microbe Interact. 2009, 22, 1588–1600. [Google Scholar] [CrossRef] [Green Version]
- Mercière, M.; Boulord, R.; Carasco-Lacombe, C.; Klopp, C.; Lee, Y.P.; Tan, J.S.; Syed Alwee, S.S.R.; Zaremski, A.; De Franqueville, H.; Breton, F.; et al. About Ganoderma boninense in oil palm plantations of Sumatra and peninsular Malaysia: Ancient population expansion, extensive gene flow and large scale dispersion ability. Fungal. Biol. 2017, 121, 529–540. [Google Scholar] [CrossRef]
- Wong, W.C.; Tung, H.J.; Fadhilah, M.N.; Midot, F.; Lau, S.Y.L.; Melling, L.; Astari, S.; Hadziabdic, Đ.; Trigiano, R.N.; Goh, K.J.; et al. Genetic diversity and gene flow amongst admixed populations of Ganoderma boninense, causal agent of basal stem rot in African oil palm (Elaeis guineensis Jacq.) in Sarawak (Malaysia), Peninsular Malaysia, and Sumatra (Indonesia). Mycologia 2021, 113, 902–917. [Google Scholar] [CrossRef]
- Midot, F.; Lau, S.Y.L.; Wong, W.C.; Tung, H.J.; Yap, M.L.; Lo, M.L.; Jee, M.S.; Dom, S.P.; Melling, L. Genetic Diversity and Demographic History of Ganoderma boninense in Oil Palm Plantations of Sarawak, Malaysia Inferred from ITS Regions. Microorganisms 2019, 7, 464. [Google Scholar] [CrossRef] [Green Version]
- Sulaiman, S.; Yusoff, N.; Tan, J.S.; Lee, Y.P. Deciphering the pan-genome of Ganoderma sp. to depict potential genomic components that contribute to Ganoderma boninense pathogenicity. Malays. Appl. Biol. 2018, 47, 71–80. [Google Scholar]
- Gui, Y.-J.; Chen, J.-Y.; Zhang, D.-D.; Li, N.-Y.; Li, T.-G.; Zhang, W.-Q.; Wang, X.-Y.; Short, D.P.G.; Li, L.; Guo, W.; et al. Verticillium dahliae manipulates plant immunity by glycoside hydrolase 12 proteins in conjunction with carbohydrate-binding module 1. Environ. Microbiol. 2017, 19, 1914–1932. [Google Scholar] [CrossRef] [Green Version]
- Ma, Z.; Song, T.; Zhu, L.; Ye, W.; Wang, Y.; Shao, Y.; Dong, S.; Zhang, Z.; Dou, D.; Zheng, X.; et al. A Phytophthora sojae Glycoside Hydrolase 12 Protein Is a Major Virulence Factor during Soybean Infection and Is Recognized as a PAMP. Plant Cell 2015, 27, 2057–2072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ökmen, B.; Bachmann, D.; de Wit, P.J.G.M. A conserved GH17 glycosyl hydrolase from plant pathogenic Dothideomycetes releases a DAMP causing cell death in tomato. Mol. Plant Pathol. 2019, 20, 1710–1721. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Zhu, L.; Song, T.; Wang, Y.; Zhang, Q.; Xia, Y.; Qiu, M.; Lin, Y.; Li, H.; Kong, L.; et al. A paralogous decoy protects Phytophthora sojae apoplastic effector PsXEG1 from a host inhibitor. Science 2017, 355, 710–714. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Zhong, X.; Shi, Y.; Liu, Z.; Jiang, N.; Liu, J.; Ding, B.; Li, Z.; Kang, H.; Ning, Y.; et al. A fungal effector targets a heat shock–dynamin protein complex to modulate mitochondrial dynamics and reduce plant immunity. Sci. Adv. 2020, 6, eabb7719. [Google Scholar] [CrossRef]
- Li, S.; Peng, X.; Wang, Y.; Hua, K.; Xing, F.; Zheng, Y.; Liu, W.; Sun, W.; Wei, S. The Effector AGLIP1 in Rhizoctonia solani AG1 IA Triggers Cell Death in Plants and Promotes Disease Development Through Inhibiting PAMP-Triggered Immunity in Arabidopsis thaliana. Front. Microbiol. 2019, 10, 2228. [Google Scholar] [CrossRef]
- Blumke, A.; Falter, C.; Herrfurth, C.; Sode, B.; Bode, R.; Schafer, W.; Feussner, I.; Voigt, C.A. Secreted fungal effector lipase releases free fatty acids to inhibit innate immunity-related callose formation during wheat head infection. Plant Physiol. 2014, 165, 346–358. [Google Scholar] [CrossRef] [Green Version]
- He, M.; Ding, N.-Z. Plant Unsaturated Fatty Acids: Multiple Roles in Stress Response. Front. Plant Sci. 2020, 11, 562785. [Google Scholar] [CrossRef]
- Alizadeh, F.; Abdullah, S.N.A.; Chong, P.P.; Selamat, A.B. Expression Analysis of Fatty Acid Biosynthetic Pathway Genes during Interactions of Oil Palm (Elaeis guineensis Jacq.) with the Pathogenic Ganoderma boninense and Symbiotic Trichoderma harzianum Fungal Organisms. Plant Mol. Biol. Rep. 2014, 32, 70–81. [Google Scholar] [CrossRef]
- Isha, A.; Yusof, N.A.; Shaari, K.; Osman, R.; Abdullah, S.N.A.; Wong, M.-Y. Metabolites identification of oil palm roots infected with Ganoderma boninense using GC–MS-based metabolomics. Arab. J. Chem. 2020, 13, 6191–6200. [Google Scholar] [CrossRef]
- Alexander, A.; Dayou, J.; Abdullah, S.; Chong, K.P. The changes of oil palm roots cell wall lipids during pathogenesis of Ganoderma boninense. IOP Conf. Ser. Earth Environ. Sci. 2017, 77, 012014. [Google Scholar] [CrossRef]
- Ahmad, R.; Lim, C.K.; Marzuki, N.F.; Goh, Y.K.; Azizan, K.A.; Goh, Y.K.; Goh, K.J.; Ramzi, A.B.; Baharum, S.N. Metabolic Profile of Scytalidium parasiticum-Ganoderma boninense Co-Cultures Revealed the Alkaloids, Flavonoids and Fatty Acids that Contribute to Anti-Ganoderma Activity. Molecules 2020, 25, 5965. [Google Scholar] [CrossRef] [PubMed]
- Choquer, M.; Rascle, C.; Gonçalves, I.R.; de Vallée, A.; Ribot, C.; Loisel, E.; Smilevski, P.; Ferria, J.; Savadogo, M.; Souibgui, E.; et al. The infection cushion of Botrytis cinerea: A fungal ‘weapon’ of plant-biomass destruction. Environ. Microbiol. 2021, 23, 2293–2314. [Google Scholar] [CrossRef] [PubMed]
- Jashni, M.K.; Dols, I.H.M.; Iida, Y.; Boeren, S.; Beenen, H.G.; Mehrabi, R.; Collemare, J.; de Wit, P.J.G.M. Synergistic Action of a Metalloprotease and a Serine Protease from Fusarium oxysporum f. sp. lycopersici Cleaves Chitin-Binding Tomato Chitinases, Reduces Their Antifungal Activity, and Enhances Fungal Virulence. Mol. Plant-Microbe Interact. 2015, 28, 996–1008. [Google Scholar] [CrossRef] [Green Version]
- Naumann, T.A.; Wicklow, D.T.; Price, N.P.J. Identification of a Chitinase-modifying protein from Fusarium verticillioides: Truncation of a host resistance protein by a fungalysin metalloprotease. J. Biol. Chem. 2011, 286, 35358–35366. [Google Scholar] [CrossRef] [Green Version]
- Ökmen, B.; Kemmerich, B.; Hilbig, D.; Wemhöner, R.; Aschenbroich, J.; Perrar, A.; Huesgen, P.F.; Schipper, K.; Doehlemann, G. Dual function of a secreted fungalysin metalloprotease in Ustilago maydis. New Phytol. 2018, 220, 249–261. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; He, J.; Jia, L.-J.; Yuan, T.-L.; Zhang, D.; Guo, Y.; Wang, Y.; Tang, W.-H. Cellular Tracking and Gene Profiling of Fusarium graminearum during Maize Stalk Rot Disease Development Elucidates Its Strategies in Confronting Phosphorus Limitation in the Host Apoplast. PLoS Pathog. 2016, 12, e1005485. [Google Scholar] [CrossRef] [Green Version]
- Yun, Y.; Liu, Z.; Yin, Y.; Jiang, J.; Chen, Y.; Xu, J.-R.; Ma, Z. Functional analysis of the Fusarium graminearum phosphatome. New Phytol. 2015, 207, 119–134. [Google Scholar] [CrossRef]
- Tran, T.M.; MacIntyre, A.; Hawes, M.; Allen, C. Escaping Underground Nets: Extracellular DNases Degrade Plant Extracellular Traps and Contribute to Virulence of the Plant Pathogenic Bacterium Ralstonia solanacearum. PLoS Pathog. 2016, 12, e1005686. [Google Scholar] [CrossRef]
- Hawes, M.C.; Curlango-Rivera, G.; Wen, F.; White, G.J.; Vanetten, H.D.; Xiong, Z. Extracellular DNA: The tip of root defenses? Plant Sci. 2011, 180, 741–745. [Google Scholar] [CrossRef]
- Luti, S.; Sella, L.; Quarantin, A.; Pazzagli, L.; Baccelli, I. Twenty years of research on cerato-platanin family proteins: Clues, conclusions, and unsolved issues. Fungal Biol. Rev. 2020, 34, 13–24. [Google Scholar] [CrossRef]
- Li, S.; Dong, Y.; Li, L.; Zhang, Y.; Yang, X.; Zeng, H.; Shi, M.; Pei, X.; Qiu, D.; Yuan, Q. The Novel Cerato-Platanin-Like Protein FocCP1 from Fusarium oxysporum Triggers an Immune Response in Plants. Int. J. Mol. Sci. 2019, 20, 2849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, Y.; Wei, J.; Yao, C.; Reng, H.; Gao, Z. SsSm1, a Cerato-platanin family protein, is involved in the hyphal development and pathogenic process of Sclerotinia sclerotiorum. Plant Sci. 2018, 270, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; An, B.; Feng, L.; He, C.; Luo, H. A Colletotrichum gloeosporioides cerato-platanin protein, CgCP1, contributes to conidiation and plays roles in the interaction with rubber tree. Can. J. Microbiol. 2018, 64, 826–834. [Google Scholar] [CrossRef]
- Baccelli, I.; Luti, S.; Bernardi, R.; Scala, A.; Pazzagli, L. Cerato-platanin shows expansin-like activity on cellulosic materials. Appl. Microbiol. Biotechnol. 2014, 98, 175–184. [Google Scholar] [CrossRef]
- Baccelli, I.; Gonthier, P.; Bernardi, R. Gene expression analyses reveal a relationship between conidiation and cerato-platanin in homokaryotic and heterokaryotic strains of the fungal plant pathogen Heterobasidion irregulare. Mycol. Prog. 2015, 14, 40. [Google Scholar] [CrossRef]
- Quarantin, A.; Castiglioni, C.; Schäfer, W.; Favaron, F.; Sella, L. The Fusarium graminearum cerato-platanins loosen cellulose substrates enhancing fungal cellulase activity as expansin-like proteins. Plant Physiol. Biochem. 2019, 139, 229–238. [Google Scholar] [CrossRef]
- Quarantin, A.; Glasenapp, A.; Schäfer, W.; Favaron, F.; Sella, L. Involvement of the Fusarium graminearum cerato-platanin proteins in fungal growth and plant infection. Plant Physiol. Biochem. 2016, 109, 220–229. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, Y.; Liang, Y.; Dong, Y.; Yang, X.; Yuan, J.; Qiu, D. The Verticillium dahliae SnodProt1-Like Protein VdCP1 Contributes to Virulence and Triggers the Plant Immune System. Front. Plant Sci. 2017, 8, 1880. [Google Scholar] [CrossRef] [Green Version]
- Catanzariti, A.-M.; Dodds, P.N.; Lawrence, G.J.; Ayliffe, M.A.; Ellis, J.G. Haustorially Expressed Secreted Proteins from Flax Rust Are Highly Enriched for Avirulence Elicitors. Plant Cell 2006, 18, 243–256. [Google Scholar] [CrossRef] [Green Version]
- Zhu, W.; Wei, W.; Wu, Y.; Zhou, Y.; Peng, F.; Zhang, S.; Chen, P.; Xu, X. BcCFEM1, a CFEM Domain-Containing Protein with Putative GPI-Anchored Site, Is Involved in Pathogenicity, Conidial Production, and Stress Tolerance in Botrytis cinerea. Front. Microbiol. 2017, 8, 1807. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Zhong, D.; Xie, W.; He, Y.; Zheng, Y.; Lin, Y.; Chen, Z.; Han, Y.; Tian, D.; Liu, W.; et al. Functional Identification of Novel Cell Death-inducing Effector Proteins from Magnaporthe oryzae. Rice 2019, 12, 59. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Songkumarn, P.; Venu, R.C.; Gowda, M.; Bellizzi, M.; Hu, J.; Liu, W.; Ebbole, D.; Meyers, B.; Mitchell, T.; et al. Identification and Characterization of In planta–Expressed Secreted Effector Proteins from Magnaporthe oryzae That Induce Cell Death in Rice. Mol. Plant-Microbe Interact. 2012, 26, 191–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.-x.; Long, F.; Zhu, H.; Zhang, Y.; Wu, J.-y.; Shen, S.; Dong, J.-g.; Hao, Z.-m. Bioinformatic analysis and functional characterization of CFEM proteins in Setosphaeria turcica. J. Integr. Agric. 2021, 20, 2438–2449. [Google Scholar] [CrossRef]
- Gong, A.-d.; Jing, Z.-y.; Zhang, K.; Tan, Q.-q.; Wang, G.-l.; Liu, W.-d. Bioinformatic analysis and functional characterization of the CFEM proteins in maize anthracnose fungus Colletotrichum graminicola. J. Integr. Agric. 2020, 19, 541–550. [Google Scholar] [CrossRef]
- Tian, H.; MacKenzie, C.I.; Rodriguez-Moreno, L.; van den Berg, G.C.M.; Chen, H.; Rudd, J.J.; Mesters, J.R.; Thomma, B.P.H.J. Three LysM effectors of Zymoseptoria tritici collectively disarm chitin-triggered plant immunity. Mol. Plant Pathol. 2021, 22, 683–693. [Google Scholar] [CrossRef]
- Suarez-Fernandez, M.; Aragon-Perez, A.; Lopez-Llorca, L.V.; Lopez-Moya, F. Putative LysM Effectors Contribute to Fungal Lifestyle. Int. J. Mol. Sci. 2021, 22, 3147. [Google Scholar] [CrossRef]
- Sánchez-Vallet, A.; Tian, H.; Rodriguez-Moreno, L.; Valkenburg, D.-J.; Saleem-Batcha, R.; Wawra, S.; Kombrink, A.; Verhage, L.; de Jonge, R.; van Esse, H.P.; et al. A secreted LysM effector protects fungal hyphae through chitin-dependent homodimer polymerization. PLoS Pathog. 2020, 16, e1008652. [Google Scholar] [CrossRef]
- Irieda, H.; Inoue, Y.; Mori, M.; Yamada, K.; Oshikawa, Y.; Saitoh, H.; Uemura, A.; Terauchi, R.; Kitakura, S.; Kosaka, A.; et al. Conserved fungal effector suppresses PAMP-triggered immunity by targeting plant immune kinases. Proc. Natl. Acad. Sci. USA 2019, 116, 496–505. [Google Scholar] [CrossRef] [Green Version]
- An, B.; Wang, W.; Guo, Y.; Wang, Q.; Luo, H.; He, C. BAS2 Is Required for Conidiation and Pathogenicity of Colletotrichum gloeosporioides from Hevea brasiliensis. Int. J. Mol. Sci. 2018, 19, 1860. [Google Scholar] [CrossRef] [Green Version]
- Motteram, J.; Küfner, I.; Deller, S.; Brunner, F.; Hammond-Kosack, K.E.; Nürnberger, T.; Rudd, J.J. Molecular characterization and functional analysis of MgNLP, the sole NPP1 domain-containing protein, from the fungal wheat leaf pathogen Mycosphaerella graminicola. Mol. Plant Microbe Interact. 2009, 22, 790–799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Staats, M.; Van Baarlen, P.; Schouten, A.; Van Kan, J.A.L. Functional analysis of NLP genes from Botrytis elliptica. Mol. Plant Pathol. 2007, 8, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.L.; Peng, Y.L.; Fan, J. The Nep1-like protein family of Magnaporthe oryzae is dispensable for the infection of rice plants. Sci. Rep. 2017, 7, 4372. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Han, C.; Chen, J.; Li, H.; He, K.; Liu, A.; Li, D. Fungal cellulase is an elicitor but its enzymatic activity is not required for its elicitor activity. Mol. Plant Pathol. 2015, 16, 14–26. [Google Scholar] [CrossRef] [PubMed]
- Santhanam, P.; van Esse, H.P.; Albert, I.; Faino, L.; Nürnberger, T.; Thomma, B.P. Evidence for functional diversification within a fungal NEP1-like protein family. Mol. Plant Microbe Interact. 2013, 26, 278–286. [Google Scholar] [CrossRef] [Green Version]
- Zhou, B.J.; Jia, P.S.; Gao, F.; Guo, H.S. Molecular characterization and functional analysis of a necrosis- and ethylene-inducing, protein-encoding gene family from Verticillium dahliae. Mol. Plant Microbe Interact. 2012, 25, 964–975. [Google Scholar] [CrossRef] [Green Version]
- Mattinen, L.; Tshuikina, M.; Mäe, A.; Pirhonen, M. Identification and characterization of Nip, necrosis-inducing virulence protein of Erwinia carotovora subsp. carotovora. Mol. Plant Microbe Interact. 2004, 17, 1366–1375. [Google Scholar] [CrossRef] [Green Version]
- Wong, M.Y.; Govender, N.T.; Ong, C.S. RNA-seq data of Ganoderma boninense at axenic culture condition and under in planta pathogen-oil palm (Elaeis guineensis Jacq.) interaction. BMC Res. Notes 2019, 12, 631. [Google Scholar] [CrossRef] [Green Version]
- Dhillon, B.; Hamelin, R.C.; Rollins, J.A. Transcriptional profile of oil palm pathogen, Ganoderma boninense, reveals activation of lignin degradation machinery and possible evasion of host immune response. BMC Genom. 2021, 22, 326. [Google Scholar] [CrossRef]
- Meyer, M.; Bourras, S.; Gervais, J.; Labadie, K.; Cruaud, C.; Balesdent, M.H.; Rouxel, T. Impact of biotic and abiotic factors on the expression of fungal effector-encoding genes in axenic growth conditions. Fungal Genet. Biol. 2017, 99, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Liu, X.; Liu, M.; Wang, Y.; Zou, Y.; You, Y.; Yang, L.; Hu, J.; Zhang, H.; Zheng, X.; et al. Magnaporthe oryzae Auxiliary Activity Protein MoAa91 Functions as Chitin-Binding Protein To Induce Appressorium Formation on Artificial Inductive Surfaces and Suppress Plant Immunity. mBio 2020, 11, e03304–e03319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bi, K.; Scalschi, L.; Jaiswal, N.; Mengiste, T.; Fried, R.; Sanz, A.B.; Arroyo, J.; Zhu, W.; Masrati, G.; Sharon, A. The Botrytis cinerea Crh1 transglycosylase is a cytoplasmic effector triggering plant cell death and defense response. Nat. Commun. 2021, 12, 2166. [Google Scholar] [CrossRef] [PubMed]
- Charova, S.N.; Dölfors, F.; Holmquist, L.; Moschou, P.N.; Dixelius, C.; Tzelepis, G. The RsRlpA Effector Is a Protease Inhibitor Promoting Rhizoctonia solani Virulence through Suppression of the Hypersensitive Response. Int. J. Mol. Sci. 2020, 21, 8070. [Google Scholar] [CrossRef]
- Pennington, H.G.; Jones, R.; Kwon, S.; Bonciani, G.; Thieron, H.; Chandler, T.; Luong, P.; Morgan, S.N.; Przydacz, M.; Bozkurt, T.; et al. The fungal ribonuclease-like effector protein CSEP0064/BEC1054 represses plant immunity and interferes with degradation of host ribosomal RNA. PLoS Pathog. 2019, 15, e1007620. [Google Scholar] [CrossRef] [PubMed]
- Grenier, J.; Potvin, C.; Asselin, A. Some Fungi Express β-1,3-Glucanases Similar to Thaumatin-like Proteins. Mycologia 2000, 92, 841–848. [Google Scholar] [CrossRef]
- Sakamoto, Y.; Watanabe, H.; Nagai, M.; Nakade, K.; Takahashi, M.; Sato, T. Lentinula edodes tlg1 encodes a thaumatin-like protein that is involved in lentinan degradation and fruiting body senescence. Plant Physiol. 2006, 141, 793–801. [Google Scholar] [CrossRef] [Green Version]
- Cai, F.; Gao, R.; Zhao, Z.; Ding, M.; Jiang, S.; Yagtu, C.; Zhu, H.; Zhang, J.; Ebner, T.; Mayrhofer-Reinhartshuber, M.; et al. Evolutionary compromises in fungal fitness: Hydrophobins can hinder the adverse dispersal of conidiospores and challenge their survival. ISME J. 2020, 14, 2610–2624. [Google Scholar] [CrossRef]
- Dubey, M.K.; Jensen, D.F.; Karlsson, M. Hydrophobins are required for conidial hydrophobicity and plant root colonization in the fungal biocontrol agent Clonostachys rosea. BMC Microbiol. 2014, 14, 18. [Google Scholar] [CrossRef] [Green Version]
- Frantzeskakis, L.; Kracher, B.; Kusch, S.; Yoshikawa-Maekawa, M.; Bauer, S.; Pedersen, C.; Spanu, P.D.; Maekawa, T.; Schulze-Lefert, P.; Panstruga, R. Signatures of host specialization and a recent transposable element burst in the dynamic one-speed genome of the fungal barley powdery mildew pathogen. BMC Genom. 2018, 19, 381. [Google Scholar] [CrossRef] [Green Version]
- Stam, R.; Münsterkötter, M.; Pophaly, S.D.; Fokkens, L.; Sghyer, H.; Güldener, U.; Hückelhoven, R.; Hess, M. A New Reference Genome Shows the One-Speed Genome Structure of the Barley Pathogen Ramularia collo-cygni. Genome Biol. Evol. 2018, 10, 3243–3249. [Google Scholar] [CrossRef] [Green Version]
- Schwessinger, B.; Sperschneider, J.; Cuddy, W.S.; Garnica, D.P.; Miller, M.E.; Taylor, J.M.; Dodds, P.N.; Figueroa, M.; Park, R.F.; Rathjen, J.P.; et al. A Near-Complete Haplotype-Phased Genome of the Dikaryotic Wheat Stripe Rust Fungus Puccinia striiformis f. sp. tritici Reveals High Interhaplotype Diversity. mBio 2018, 9, e02275-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torres, D.E.; Oggenfuss, U.; Croll, D.; Seidl, M.F. Genome evolution in fungal plant pathogens: Looking beyond the two-speed genome model. Fungal Biol. Rev. 2020, 34, 136–143. [Google Scholar] [CrossRef]
- Utomo, C.; Tanjung, Z.A.; Aditama, R.; Buana, R.F.N.; Pratomo, A.D.M.; Tryono, R.; Liwang, T. Draft Genome Sequence of the Phytopathogenic Fungus Ganoderma boninense, the Causal Agent of Basal Stem Rot Disease on Oil Palm. Genome Announc. 2018, 6, e00122-18. [Google Scholar] [CrossRef] [Green Version]
- Mercière, M.; Laybats, A.; Carasco-Lacombe, C.; Tan, J.S.; Klopp, C.; Durand-Gasselin, T.; Alwee, S.S.R.S.; Camus-Kulandaivelu, L.; Breton, F. Identification and development of new polymorphic microsatellite markers using genome assembly for Ganoderma boninense, causal agent of oil palm basal stem rot disease. Mycol. Prog. 2015, 14, 103. [Google Scholar] [CrossRef] [Green Version]
- Jackman, S.D.; Vandervalk, B.P.; Mohamadi, H.; Chu, J.; Yeo, S.; Hammond, S.A.; Jahesh, G.; Khan, H.; Coombe, L.; Warren, R.L.; et al. ABySS 2.0: Resource-efficient assembly of large genomes using a Bloom filter. Genome Res. 2017, 27, 768–777. [Google Scholar] [CrossRef] [Green Version]
- Chikhi, R.; Medvedev, P. Informed and automated k-mer size selection for genome assembly. Bioinformatics 2013, 30, 31–37. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [Green Version]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Simão, F.A.; Zdobnov, E.M. BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes. Mol. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef]
- Smit, A.; Hubley, R.; Green, P. RepeatMasker Open-4.0. 2013–2015. Available online: http://www.repeatmasker.org (accessed on 21 October 2021).
- Storer, J.; Hubley, R.; Rosen, J.; Wheeler, T.J.; Smit, A.F. The Dfam community resource of transposable element families, sequence models, and genome annotations. Mob. DNA 2021, 12, 2. [Google Scholar] [CrossRef]
- Bao, W.; Kojima, K.K.; Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 2015, 6, 11. [Google Scholar] [CrossRef] [Green Version]
- Wicker, T.; Matthews, D.E.; Keller, B. TREP: A database for Triticeae repetitive elements. Trends Plant Sci. 2002, 7, 561–562. [Google Scholar] [CrossRef]
- Ou, S.; Su, W.; Liao, Y.; Chougule, K.; Agda, J.R.A.; Hellinga, A.J.; Lugo, C.S.B.; Elliott, T.A.; Ware, D.; Peterson, T.; et al. Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biol. 2019, 20, 275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smit, A.F.; Hubley, R. RepeatModeler Open-1.0. 2008–2015. Available online: http://www.repeatmasker.org (accessed on 21 October 2021).
- Brůna, T.; Hoff, K.J.; Lomsadze, A.; Stanke, M.; Borodovsky, M. BRAKER2: Automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR Genom. Bioinform. 2021, 3, lqaa108. [Google Scholar] [CrossRef] [PubMed]
- Isaac, I.L.; Walter, A.W.C.Y.; Bakar, M.F.A.; Idris, A.S.; Bakar, F.D.A.; Bharudin, I.; Murad, A.M.A. Transcriptome datasets of oil palm pathogen Ganoderma boninense. Data Brief 2018, 17, 1108–1111. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Zdobnov, E.M.; Kuznetsov, D.; Tegenfeldt, F.; Manni, M.; Berkeley, M.; Kriventseva, E.V. OrthoDB in 2020: Evolutionary and functional annotations of orthologs. Nucleic Acids Res. 2020, 49, D389–D393. [Google Scholar] [CrossRef]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2020, 49, D344–D354. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [Green Version]
- Consortium, T.U. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2020, 49, D480–D489. [Google Scholar] [CrossRef]
- Coordinators, N.R. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018, 46, D8–D13. [Google Scholar] [CrossRef] [Green Version]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2018, 47, D309–D314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rawlings, N.D.; Barrett, A.J.; Thomas, P.D.; Huang, X.; Bateman, A.; Finn, R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2017, 46, D624–D632. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef] [Green Version]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Käll, L.; Krogh, A.; Sonnhammer, E.L. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004, 338, 1027–1036. [Google Scholar] [CrossRef]
- Käll, L.; Krogh, A.; Sonnhammer, E.L. An HMM posterior decoder for sequence feature prediction that includes homology information. Bioinformatics 2005, 21 (Suppl. S1), i251–i257. [Google Scholar] [CrossRef]
- Käll, L.; Krogh, A.; Sonnhammer, E.L. Advantages of combined transmembrane topology and signal peptide prediction—The Phobius web server. Nucleic Acids Res. 2007, 35, W429–W432. [Google Scholar] [CrossRef] [Green Version]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [Green Version]
- Sonnhammer, E.L.; von Heijne, G.; Krogh, A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998, 6, 175–182. [Google Scholar]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchfink, B.; Reuter, K.; Drost, H.-G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 2021, 18, 366–368. [Google Scholar] [CrossRef] [PubMed]
- Sperschneider, J.; Gardiner, D.M.; Dodds, P.N.; Tini, F.; Covarelli, L.; Singh, K.B.; Manners, J.M.; Taylor, J.M. EffectorP: Predicting fungal effector proteins from secretomes using machine learning. New Phytol. 2016, 210, 743–761. [Google Scholar] [CrossRef] [Green Version]
- Sperschneider, J.; Dodds, P.N.; Gardiner, D.M.; Singh, K.B.; Taylor, J.M. Improved prediction of fungal effector proteins from secretomes with EffectorP 2.0. Mol. Plant Pathol. 2018, 19, 2094–2110. [Google Scholar] [CrossRef] [Green Version]
- Sperschneider, J.; Dodds, P.N. EffectorP 3.0: Prediction of apoplastic and cytoplasmic effectors in fungi and oomycetes. Mol Plant Microbe Interact. 2022, 35, 146–156. [Google Scholar] [CrossRef]
- Sundram, S.; Meon, S.; Seman, I.A.; Othman, R. Symbiotic interaction of endophytic bacteria with arbuscular mycorrhizal fungi and its antagonistic effect on Ganoderma boninense. J. Microbiol. 2011, 49, 551. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Kolde, R. pheatmap: Pretty Heatmaps. R package version 1.0. 12. Available online: https://cran.r-project.org/web/packages/pheatmap/index.html (accessed on 16 February 2022).
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [Green Version]

| Strain | G3 | NJ3 | T10 |
|---|---|---|---|
| Assembly Statistics | |||
| BioProject Accession | PRJNA421251 | PRJNA287769 | PRJNA789134 |
| Sequencing platform | Illumina HiSeq 4000 and PacBio RSII | Illumina HiSeq 2000 and 454 GS FLX | Illumina HiSeq 4000 |
| Number of contigs (≥1000 base) | 494 | 12,643 | 9678 |
| Total length | 79,188,463 | 60,324,849 | 64,965,238 |
| N50 | 272,644 | 6116 | 8987 |
| GC (%) | 56 | 56 | 56 |
| Repetitive elements (%) | 8.99 | 6.98 | 11.50 |
| Predicted genes | 19,978 | 25,745 | 25,220 |
| BUSCO | |||
| Complete (%) | 1606 (91.0%) | 1341 (76.0%) | 1588 (90.0%) |
| Single-copy (%) | 1413 (80.1%) | 1286 (72.9%) | 1336 (75.7%) |
| Duplicated (%) | 193 (10.9%) | 55 (3.1%) | 252 (14.3%) |
| Fragmented (%) | 64 (3.6%) | 200 (11.3%) | 35 (2.0%) |
| Missing (%) | 94 (5.3%) | 223 (12.6%) | 141 (8.0%) |
| Class | Core | Accessory | G3-Specific | NJ3-Specific | T10-Specific |
|---|---|---|---|---|---|
| Carboxylesterase | 118 | 26 | 4 | 5 | |
| CAZyme | |||||
| Cell wall-degrading enzyme (CWDE) | 672 | 110 | 12 | 12 | 10 |
| Other CAZyme | 253 | 41 | 6 | 5 | 11 |
| Peptidase | |||||
| Aspartic peptidase | 129 | 19 | 2 | ||
| Glutamic peptidase | 40 | 2 | 2 | ||
| Metallo peptidase | 65 | 15 | |||
| Serine peptidase | 162 | 11 | 9 | ||
| Effector-associated | |||||
| CP family protein | 37 | 2 | |||
| CFEM domain-containing protein | 34 | 9 | |||
| Nis1 domain-containing protein | 21 | 8 | |||
| NEP1-like protein family | 6 | ||||
| LysM domain-containing protein | 9 | 3 | |||
| Biotrophy-associated secreted protein 2 | 8 | 1 | 2 | ||
| Other classes | |||||
| Isomerase | 20 | 2 | |||
| Nuclease | 18 | 2 | |||
| Phosphatase | 73 | 8 | 2 | ||
| Chaperone | 19 | 3 | 1 | ||
| Hydrophobins | 79 | 18 | |||
| Thaumatin-like protein family | 34 | ||||
| Other | 767 | 160 | 4 | 14 | 12 |
| Unannotated | 349 | 73 | 5 | 18 | 22 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khairi, M.H.F.; Nor Muhammad, N.A.; Bunawan, H.; Abdul Murad, A.M.; Ramzi, A.B. Unveiling the Core Effector Proteins of Oil Palm Pathogen Ganoderma boninense via Pan-Secretome Analysis. J. Fungi 2022, 8, 793. https://doi.org/10.3390/jof8080793
Khairi MHF, Nor Muhammad NA, Bunawan H, Abdul Murad AM, Ramzi AB. Unveiling the Core Effector Proteins of Oil Palm Pathogen Ganoderma boninense via Pan-Secretome Analysis. Journal of Fungi. 2022; 8(8):793. https://doi.org/10.3390/jof8080793
Chicago/Turabian StyleKhairi, Mohamad Hazwan Fikri, Nor Azlan Nor Muhammad, Hamidun Bunawan, Abdul Munir Abdul Murad, and Ahmad Bazli Ramzi. 2022. "Unveiling the Core Effector Proteins of Oil Palm Pathogen Ganoderma boninense via Pan-Secretome Analysis" Journal of Fungi 8, no. 8: 793. https://doi.org/10.3390/jof8080793
APA StyleKhairi, M. H. F., Nor Muhammad, N. A., Bunawan, H., Abdul Murad, A. M., & Ramzi, A. B. (2022). Unveiling the Core Effector Proteins of Oil Palm Pathogen Ganoderma boninense via Pan-Secretome Analysis. Journal of Fungi, 8(8), 793. https://doi.org/10.3390/jof8080793






