Past, Present, and Future Perspectives on Whey as a Promising Feedstock for Bioethanol Production by Yeast
Abstract
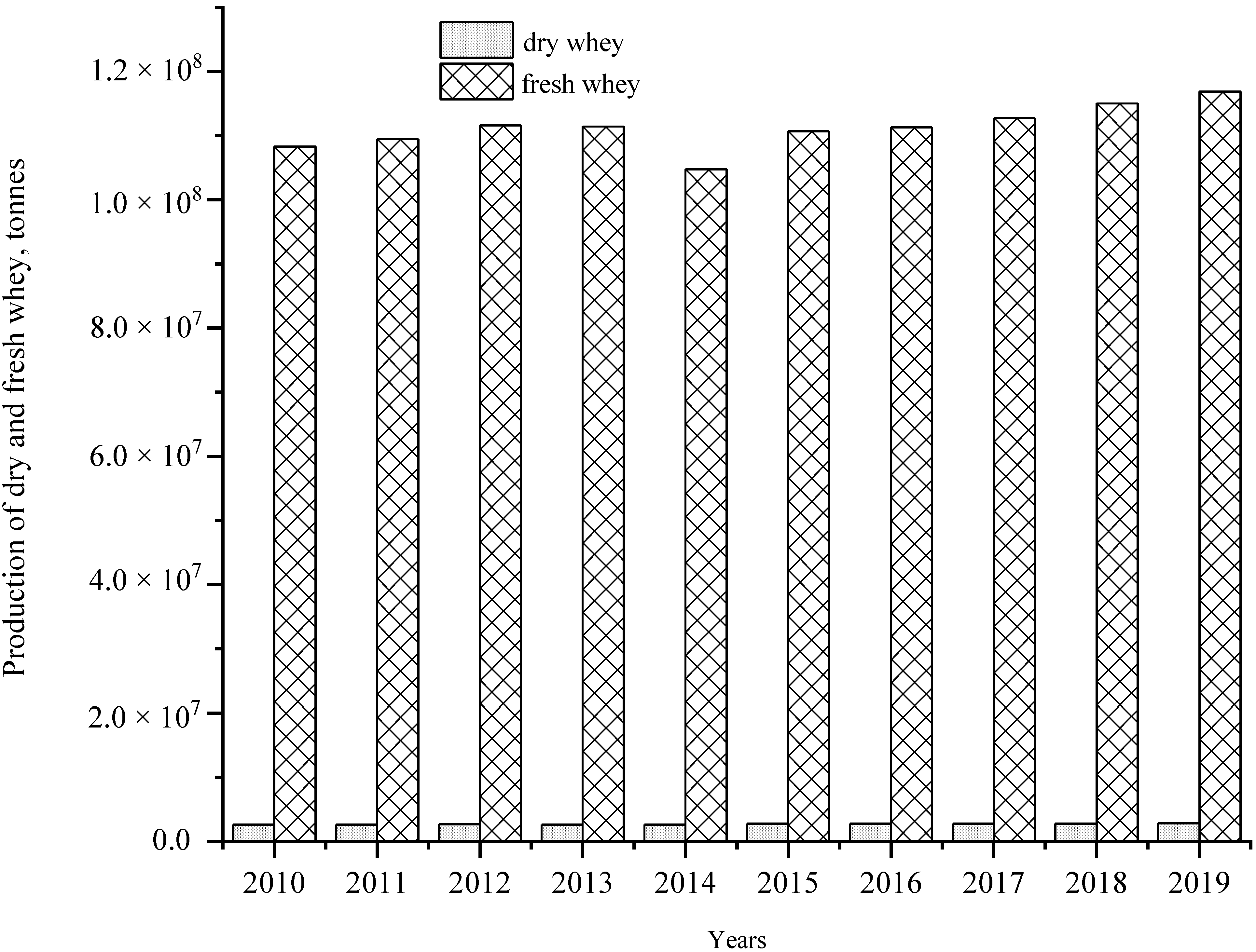
1. Overview of Bioethanol Production and Whey as a Feedstock
2. The Lactose/Galactose Metabolic Pathway and Its Regulation in Fungi
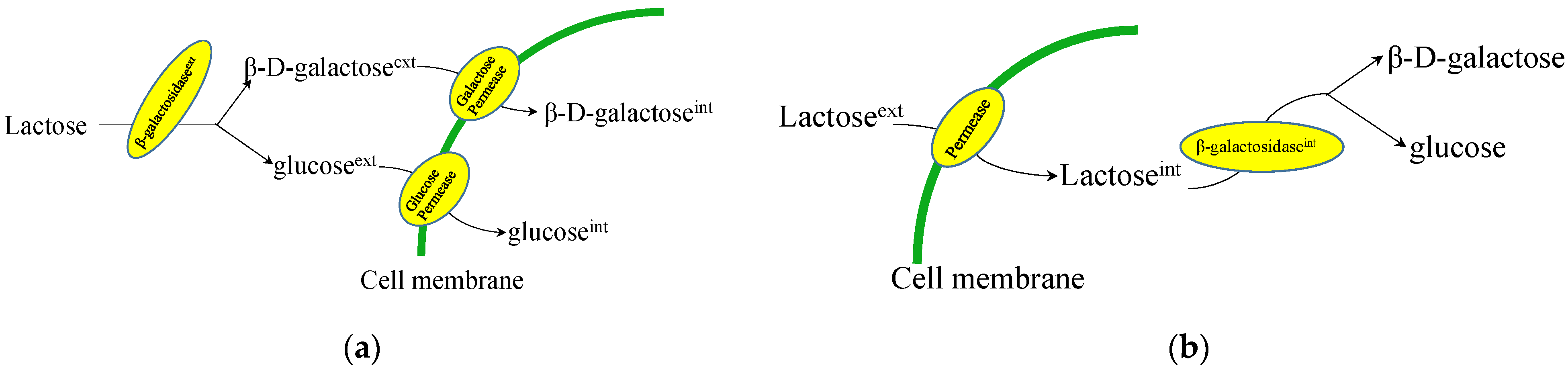
2.1. Lactose/Galactose Consumption in Fungi
2.2. GAL Gene Regulation in S. cerevisiae and K. lactis
3. Bioethanol Production from Whey/Lactose by Kluyveromyces
4. Strategies for Conferring Lactose Utilization to S. cerevisiae
4.1. Pre-Hydrolysis of Extracellular Lactose for S. cerevisiae Utilization
4.2. Protoplast Fusions of S. cerevisiae and Kluyveromyces spp.
4.3. Exogenous Expression of the Lactose Hydrolase Gene in S. cerevisiae
4.3.1. Lactose Metabolism Genes from E. coli
4.3.2. The Lactose Metabolism Genes from A. niger
4.3.3. The Lactose Metabolism Genes from Kluyveromyces spp.
5. Approaches for Enhancing Lactose Utilization Rate
5.1. Optimizing Culture Conditions and Media Components
5.2. Metabolic Engineering
5.3. Evolutionary Engineering
6. The Future of Ethanol Production from Whey or Lactose by Yeast
6.1. Whey/Cheese Whey Mixed with Other High Carbon Feedstocks for Bioethanol
6.2. Approaches for Efficient Generation of Lactose-Consuming Strains
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zeppini, P.; Van Den Bergh, J.C. Global competition dynamics of fossil fuels and renewable energy under climate policies and peak oil: A behavioural model. Energy Policy 2020, 136, 110907. [Google Scholar] [CrossRef]
- Franta, B. Early oil industry knowledge of CO2 and global warming. Nat. Clim. Chang. 2018, 8, 1024–1025. [Google Scholar] [CrossRef]
- Franta, B. Early oil industry disinformation on global warming. Environ. Polit. 2021, 30, 663–668. [Google Scholar] [CrossRef]
- Yoro, K.O.; Daramola, M.O. CO2 emission sources, greenhouse gases, and the global warming effect. In Advances in Carbon Capture; Rahimpour, M.R., Farsi, M., Makarem, M.A., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; Section 1; pp. 3–28. [Google Scholar]
- Uslu, S.; Celik, M.B. Combustion and emission characteristics of isoamyl alcohol-gasoline blends in spark ignition engine. Fuel 2020, 262, 116496. [Google Scholar] [CrossRef]
- Simsek, S.; Uslu, S. Experimental study of the performance and emissions characteristics of fusel oil/gasoline blends in spark ignited engine using response surface methodology. Fuel 2020, 277, 118182. [Google Scholar] [CrossRef]
- Iodice, P.; Cardone, M. Ethanol/gasoline blends as alternative fuel in last generation spark-ignition engines: A review on CO and HC engine out emissions. Energies 2021, 14, 4034. [Google Scholar] [CrossRef]
- Iodice, P.; Amoresano, A.; Langella, G. A review on the effects of ethanol/gasoline fuel blends on NOX emissions in spark-ignition engines. Biofuel Res. J. 2021, 8, 1465–1480. [Google Scholar] [CrossRef]
- Lynd, L.R. Overview and evaluation of fuel ethanol from cellulosic biomass: Technology, economics, the environment, and policy. Annu. Rev. Energy Environ. 1996, 21, 403–465. [Google Scholar] [CrossRef]
- Tyner, W.E. US ethanol policy—Possibilities for the future. In Historical Documents of the Purdue Cooperative Extension Service; Purdue University: West Lafayette, IN, USA, 2015. [Google Scholar]
- Nwufo, O.; Nwafor, O.; Igbokwe, J. Effects of blends on the physical properties of bioethanol produced from selected Nigerian crops. Int. J. Ambient Energy 2016, 37, 10–15. [Google Scholar] [CrossRef]
- Sriputorn, B.; Laopaiboon, P.; Phukoetphim, N.; Polsokchuak, N.; Butkun, K.; Laopaiboon, L. Enhancement of ethanol production efficiency in repeated-batch fermentation from sweet sorghum stem juice: Effect of initial sugar, nitrogen and aeration. Electron. J. Biotechnol. 2020, 46, 55–64. [Google Scholar] [CrossRef]
- Ayodele, B.V.; Alsaffar, M.A.; Mustapa, S.I. An overview of integration opportunities for sustainable bioethanol production from first-and second-generation sugar-based feedstocks. J. Clean. Prod. 2020, 245, 118857. [Google Scholar] [CrossRef]
- Carvalho, D.J.; Moretti, R.R.; Colodette, J.L.; Bizzo, W.A. Assessment of the self-sustained energy generation of an integrated first and second generation ethanol production from sugarcane through the characterization of the hydrolysis process residues. Energy Convers. Manag. 2020, 203, 112267. [Google Scholar] [CrossRef]
- Yuan, X.; Chen, X.; Shen, G.; Chen, S.; Yu, J.; Zhai, R.; Xu, Z.; Jin, M. Densifying lignocellulosic biomass with sulfuric acid provides a durable feedstock with high digestibility and high fermentability for cellulosic ethanol production. Renew. Energy 2022, 182, 377–389. [Google Scholar] [CrossRef]
- Devi, A.; Singh, A.; Bajar, S.; Pant, D.; Din, Z.U. Ethanol from lignocellulosic biomass: An in-depth analysis of pre-treatment methods, fermentation approaches and detoxification processes. J. Environ. Chem. Eng. 2021, 9, 105798. [Google Scholar] [CrossRef]
- Su, T.; Zhao, D.; Khodadadi, M.; Len, C. Lignocellulosic biomass for bioethanol: Recent advances, technology trends, and barriers to industrial development. Curr. Opin. Green Sustain. Chem. 2020, 24, 56–60. [Google Scholar] [CrossRef]
- Park, H.; Jeong, D.; Shin, M.; Kwak, S.; Oh, E.J.; Ko, J.K.; Kim, S.R. Xylose utilization in Saccharomyces cerevisiae during conversion of hydrothermally pretreated lignocellulosic biomass to ethanol. Appl. Microbiol. Biotechnol. 2020, 104, 3245–3252. [Google Scholar] [CrossRef]
- Priharto, N.; Ronsse, F.; Yildiz, G.; Heeres, H.J.; Deuss, P.J.; Prins, W. Fast pyrolysis with fractional condensation of lignin-rich digested stillage from second-generation bioethanol production. J. Anal. Appl. Pyrolsis 2020, 145, 104756. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, J.; Kou, L.; Zhang, X.; Tan, T. Bioethanol production from cellulose obtained from the catalytic hydro-deoxygenation (lignin-first refined to aviation fuel) of apple wood. Fuel 2019, 250, 245–253. [Google Scholar] [CrossRef]
- Rezania, S.; Oryani, B.; Cho, J.; Talaiekhozani, A.; Sabbagh, F.; Hashemi, B.; Rupani, P.F.; Mohammadi, A.A. Different pretreatment technologies of lignocellulosic biomass for bioethanol production: An overview. Energy 2020, 199, 117457. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, B.; Luo, L.; Zhang, F.; Yi, Y.; Shan, Y.; Liu, B.; Zhou, Y.; Wang, X.; Lü, X. A review on recycling techniques for bioethanol production from lignocellulosic biomass. Renew. Sustain. Energy Rev. 2021, 149, 111370. [Google Scholar] [CrossRef]
- Moysés, D.N.; Reis, V.C.B.; Almeida, J.R.M.d.; Moraes, L.M.P.d.; Torres, F.A.G. Xylose fermentation by Saccharomyces cerevisiae: Challenges and prospects. Int. J. Mol. Sci. 2016, 17, 207. [Google Scholar] [CrossRef] [PubMed]
- Cunha, J.T.; Soares, P.O.; Romaní, A.; Thevelein, J.M.; Domingues, L. Xylose fermentation efficiency of industrial Saccharomyces cerevisiae yeast with separate or combined xylose reductase/xylitol dehydrogenase and xylose isomerase pathways. Biotechnol. Biofuels 2019, 12, 20. [Google Scholar] [CrossRef] [PubMed]
- Martins, G.M.; Bocchini, D.A.; Bezzerra-Bussoli, C.; Pagnocca, F.C.; Boscolo, M.; Monteiro, D.A.; Da Silva, R.; Gomes, E. The isolation of pentose-assimilating yeasts and their xylose fermentation potential. Braz. J. Microbiol. 2018, 49, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Phaiboonsilpa, N.; Chysirichote, T.; Champreda, V.; Laosiripojana, N. Fermentation of xylose, arabinose, glucose, their mixtures and sugarcane bagasse hydrolyzate by yeast Pichia stipitis for ethanol production. Energy Rep. 2020, 6, 710–713. [Google Scholar] [CrossRef]
- Macwan, S.R.; Dabhi, B.K.; Parmar, S.; Aparnathi, K. Whey and its utilization. Int. J. Curr. Microbiol. 2016, 5, 134–155. [Google Scholar] [CrossRef]
- Kosaric, N.; Asher, Y. The utilization of cheese whey and its components. In Agricultural Feedstock and Waste Treatment and Engineering, 1st ed.; Scheper, T., Ulber, R., Eds.; Springer: Berlin/Heidelberg, Germany, 1985; Volume 32, pp. 25–60. [Google Scholar]
- Fox, P.F.; Guinee, T.P.; Cogan, T.M.; McSweeney, P.L. Whey and whey products. In Fundamentals of Cheese Science, 2nd ed.; Fox, P.F., Guinee, T.P., Cogan, T.M., McSweeney, P.L., Eds.; Springer: Boston, MA, USA, 2017; pp. 755–769. [Google Scholar]
- Eugeniya, A.; Alexandr, K.; Nikita, Z.; Nataliya, P.; Zinaida, B.; Tatyana, F. Processing cottage cheese whey components for functional food production. Food Raw Mater. 2020, 8, 1. [Google Scholar]
- Argenta, A.B.; Scheer, A.D.P. Membrane separation processes applied to whey: A review. Food Rev. Int. 2020, 36, 499–528. [Google Scholar] [CrossRef]
- Bosco, F.; Carletto, R.; Marmo, L. An integrated cheese whey valorization process. Chem. Eng. Trans. 2018, 64, 379–384. [Google Scholar]
- Menchik, P.; Zuber, T.; Zuber, A.; Moraru, C.I. Composition of coproduct streams from dairy processing: Acid whey and milk permeate. J. Dairy Sci. 2019, 102, 3978–3984. [Google Scholar] [CrossRef]
- Mansor, E.S.; Ali, E.A.; Shaban, A. Tight ultrafiltration polyethersulfone membrane for cheese whey wastewater treatment. Chem. Eng. J. 2021, 407, 127175. [Google Scholar] [CrossRef]
- Mabrouki, J.; Abbassi, M.A.; Khiari, B.; Jellali, S.; Zorpas, A.; Jeguirim, M. The dairy biorefinery: Integrating treatment process for Tunisian cheese whey valorization. Chemosphere 2022, 293, 133567. [Google Scholar] [CrossRef] [PubMed]
- Rao, R.; Basak, N. Fermentative molecular biohydrogen production from cheese whey: Present prospects and future strategy. Appl. Biochem. Biotechnol. 2021, 193, 2297–2330. [Google Scholar] [CrossRef] [PubMed]
- De Gioannis, G.; Friargiu, M.; Massi, E.; Muntoni, A.; Polettini, A.; Pomi, R.; Spiga, D. Biohydrogen production from dark fermentation of cheese whey: Influence of pH. Int. J. Hydrogen Energy 2014, 39, 20930–20941. [Google Scholar] [CrossRef]
- Pandey, A.; Srivastava, S.; Rai, P.; Duke, M. Cheese whey to biohydrogen and useful organic acids: A non-pathogenic microbial treatment by L. acidophilus. Sci. Rep. 2019, 9, 8320. [Google Scholar] [CrossRef]
- Rincón-Pérez, J.; Celis, L.B.; Morales, M.; Alatriste-Mondragón, F.; Tapia-Rodríguez, A.; Razo-Flores, E. Improvement of methane production at alkaline and neutral pH from anaerobic co-digestion of microalgal biomass and cheese whey. Biochem. Eng. J. 2021, 169, 107972. [Google Scholar] [CrossRef]
- Sousa, S.P.; Lovato, G.; Albanez, R.; Ratusznei, S.M.; Rodrigues, J.A. Improvement of sugarcane stillage (vinasse) anaerobic digestion with cheese whey as its co-substrate: Achieving high methane productivity and yield. Appl. Biochem. Biotechnol. 2019, 189, 987–1006. [Google Scholar] [CrossRef]
- Das, M.; Raychaudhuri, A.; Ghosh, S.K. Supply chain of bioethanol production from whey: A review. Procedia Environ. Sci. 2016, 35, 833–846. [Google Scholar] [CrossRef]
- Cunha, M.; Romaní, A.; Carvalho, M.; Domingues, L. Boosting bioethanol production from Eucalyptus wood by whey incorporation. Bioresour. Technol. 2018, 250, 256–264. [Google Scholar] [CrossRef]
- Becerra, M.; Cerdán, M.E.; González-Siso, M.I. Biobutanol from cheese whey. Microb. Cell Factories 2015, 14, 27. [Google Scholar] [CrossRef]
- Raganati, F.; Olivieri, G.; Procentese, A.; Russo, M.; Salatino, P.; Marzocchella, A. Butanol production by bioconversion of cheese whey in a continuous packed bed reactor. Bioresour. Technol. 2013, 138, 259–265. [Google Scholar] [CrossRef]
- Meng, W.; Zhang, Y.; Cao, M.; Zhang, W.; Lü, C.; Yang, C.; Gao, C.; Xu, P.; Ma, C. Efficient 2, 3-butanediol production from whey powder using metabolically engineered Klebsiella oxytoca. Microb. Cell Factories 2020, 19, 162. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Wang, Y.; Guo, J.; Wang, Q.; Zhang, Y.; Chen, Y.; Zhang, C.; Xiao, D. Efficient production of 2, 3-butanediol from cheese whey powder (CWP) solution by Klebsiella pneumoniae through integrating pulsed fed-batch fermentation with a two-stage pH control strategy. Fuel 2017, 203, 469–477. [Google Scholar] [CrossRef]
- Pais-Chanfrau, J.M.; Núñez-Pérez, J.; Espin-Valladares, R.d.C.; Lara-Fiallos, M.V.; Trujillo-Toledo, L.E. Bioconversion of Lactose from Cheese Whey to Organic Acids. In Lactose and Lactose Derivatives, 1st ed.; Gutiérrez-Méndez, N., Ed.; Intech Open: London, UK, 2020; pp. 53–74. [Google Scholar]
- Chwialkowska, J.; Duber, A.; Zagrodnik, R.; Walkiewicz, F.; Łężyk, M.; Oleskowicz-Popiel, P. Caproic acid production from acid whey via open culture fermentation–Evaluation of the role of electron donors and downstream processing. Bioresour. Technol. 2019, 279, 74–83. [Google Scholar] [CrossRef]
- Karim, A.; Aider, M. Bioconversion of electro-activated lactose, whey and whey permeate to produce single cell protein, ethanol, aroma volatiles, organic acids and fat by Kluyveromyces marxianus. Int. Dairy J. 2022, 129, 105334. [Google Scholar] [CrossRef]
- Szczerba, H.; Komoń-Janczara, E.; Dudziak, K.; Waśko, A.; Targoński, Z. A novel biocatalyst, Enterobacter aerogenes LU2, for efficient production of succinic acid using whey permeate as a cost-effective carbon source. Biotechnol. Biofuels 2020, 13, 96. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Guzmán, C.L.; Cisneros-de la Cueva, S.; Balderas-Hernández, V.E.; Smoliński, A.; De León-Rodríguez, A. Biohydrogen production from cheese whey powder by Enterobacter asburiae: Effect of operating conditions on hydrogen yield and chemometric study of the fermentative metabolites. Energy Rep. 2020, 6, 1170–1180. [Google Scholar] [CrossRef]
- Rao, R.; Basak, N. Optimization and modelling of dark fermentative hydrogen production from cheese whey by Enterobacter aerogenes 2822. Int. J. Hydrogen Energy 2021, 46, 1777–1800. [Google Scholar] [CrossRef]
- Lee, J.S.; Hyun, I.K.; Yoon, J.-W.; Seo, H.-J.; Kang, S.-S. Bioconversion products of whey by lactic acid bacteria exert anti-adipogenic effect. Food Sci. Anim. Resour. 2021, 41, 145. [Google Scholar] [CrossRef]
- Brown, K.; Harrison, J.; Bowers, K. Production of oxalic acid from Aspergillus niger and whey permeate. Water Air Soil Pollut. 2018, 229, 5. [Google Scholar] [CrossRef]
- Chroumpi, T.; Martínez-Reyes, N.; Kun, R.S.; Peng, M.; Lipzen, A.; Ng, V.; Tejomurthula, S.; Zhang, Y.; Grigoriev, I.V.; Mäkelä, M.R.; et al. Detailed analysis of the D-galactose catabolic pathways in Aspergillus niger reveals complexity at both metabolic and regulatory level. Fungal Genet. Biol. 2022, 159, 103670. [Google Scholar] [CrossRef]
- Fischer, C.; Kleinschmidt, T. Synthesis of galactooligosaccharides by Cryptococcus laurentii and Aspergillus oryzae using different kinds of acid whey. Int. Dairy J. 2021, 112, 104867. [Google Scholar] [CrossRef]
- Pirayre, A.; Duval, L.; Blugeon, C.; Firmo, C.; Perrin, S.; Jourdier, E.; Margeot, A.; Bidard, F. Glucose-lactose mixture feeds in industry-like conditions: A gene regulatory network analysis on the hyperproducing Trichoderma reesei strain Rut-C30. BMC Genom. 2020, 21, 885. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, Y.; Wu, C.; Liu, P.; Wang, W.; Wei, D. The transcription factor ACE3 controls cellulase activities and lactose metabolism via two additional regulators in the fungus Trichoderma reesei. J. Biol. Chem. 2019, 294, 18435–18450. [Google Scholar] [CrossRef] [PubMed]
- Sampaio, F.C.; de Faria, J.T.; da Silva, M.F.; de Souza Oliveira, R.P.; Converti, A. Cheese whey permeate fermentation by Kluyveromyces lactis: A combined approach to wastewater treatment and bioethanol production. Environ. Technol. 2020, 41, 3210–3218. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; He, J.; Xu, J.; Liu, X.; Qiu, Z.; Xu, N.; Su, L. Direct conversion of cheese whey to polymalic acid by mixed culture of Aureobasidium pullulans and permeabilized Kluyveromyces marxianus. Bioresour. Technol. 2021, 337, 125443. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, D.R.; Lopes, A.C.A.; Pereira, R.A.; Cardoso, P.G.; Duarte, W.F. Selection of potentially probiotic Kluyveromyces lactis for the fermentation of cheese whey–based beverage. Ann. Microbiol. 2019, 69, 1361–1372. [Google Scholar] [CrossRef]
- Zou, G.; Jiang, Y.P.; Liu, R.; Zhu, Z.H.; Zhou, Z.H. The putative beta-glucosidase BGL3I regulates cellulase induction in Trichoderma reesei. Biotechnol. Biofuels 2018, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.J.; Zhang, G.X.; Wang, W.; Wei, D.Z. Enhanced cellulase production in Trichoderma reesei RUT C30 via constitution of minimal transcriptional activators. Microb. Cell Factories 2018, 17, 14. [Google Scholar] [CrossRef] [PubMed]
- Harrison, M.-C.; LaBella, A.L.; Hittinger, C.T.; Rokas, A. The evolution of the GALactose utilization pathway in budding yeasts. Trends Genet. 2021, 38, 97–106. [Google Scholar] [CrossRef]
- Sunwoo, I.Y.; Sukwong, P.; Park, Y.R.; Jeong, D.Y.; Kim, S.R.; Jeong, G.-T.; Kim, S.-K. Enhancement of galactose uptake from Kappaphycus alvarezii hydrolysate using Saccharomyces cerevisiae through overexpression of Leloir pathway genes. Appl. Biochem. Biotechnol. 2021, 193, 335–348. [Google Scholar] [CrossRef]
- Wang, H.; Sun, T.; Zhao, Z.; Gu, S.; Liu, Q.; Wu, T.; Wang, D.; Tian, C.; Li, J. Transcriptional profiling of Myceliophthora thermophila on galactose and metabolic engineering for improved galactose utilization. Front. Microbiol. 2021, 12, 664011. [Google Scholar] [CrossRef] [PubMed]
- Varela, J.A.; Montini, N.; Scully, D.; Van der Ploeg, R.; Oreb, M.; Boles, E.; Hirota, J.; Akada, R.; Hoshida, H.; Morrissey, J.P. Polymorphisms in the LAC12 gene explain lactose utilisation variability in Kluyveromyces marxianus strains. FEMS Yeast Res. 2017, 17, 3. [Google Scholar] [CrossRef] [PubMed]
- Karim, A.; Gerliani, N.; Aïder, M. Kluyveromyces marxianus: An emerging yeast cell factory for applications in food and biotechnology. Int. J. Food Microbiol. 2020, 333, 108818. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhu, Y.; Dai, L.; Men, Y.; Wu, J.; Zhang, J.; Sun, Y. Efficiency analysis and mechanism Insight of that whole-cell biocatalytic production of melibiose from raffinose with Saccharomyces cerevisiae. Appl. Biochem. Biotechnol. 2017, 181, 407–423. [Google Scholar] [CrossRef]
- Álvarez Cao, M.E.; Cerdán, M.E.; González Siso, M.I.; Becerra, M. Optimization of Saccharomyces cerevisiae α-galactosidase production and application in the degradation of raffinose family oligosaccharides. Microb. Cell Factories 2019, 18, 172. [Google Scholar] [CrossRef] [PubMed]
- Sukwong, P.; Sunwoo, I.Y.; Jeong, D.Y.; Kim, S.R.; Jeong, G.-T.; Kim, S.-K. Improvement of bioethanol production by Saccharomyces cerevisiae through the deletion of GLK1, MIG1 and MIG2 and overexpression of PGM2 using the red seaweed Gracilaria verrucosa. Process. Biochem. 2020, 89, 134–145. [Google Scholar] [CrossRef]
- Oh, E.J.; Jin, Y.-S. Engineering of Saccharomyces cerevisiae for efficient fermentation of cellulose. FEMS Yeast Res. 2020, 20, foz089. [Google Scholar] [CrossRef] [PubMed]
- Sukwong, P.; Sunwoo, I.Y.; Jeong, D.Y.; Kim, S.R.; Jeong, G.-T.; Kim, S.-K. Enhancement of bioethanol production from Gracilaria verrucosa by Saccharomyces cerevisiae through the overexpression of SNR84 and PGM2. Bioprocess Biosyst. Eng. 2019, 42, 1421–1433. [Google Scholar] [CrossRef]
- Anders, A.; Breunig, K.D. Evolutionary aspects of a genetic network: Studying the lactose/galactose regulon of Kluyveromyces lactis. In Yeast Genetic Networks, 1st ed.; Becskei, A., Ed.; Humana Press: Clifton, NJ, USA, 2011; Volume 73, pp. 259–277. [Google Scholar]
- Pannala, V.R.; Ahammed Sherief, K.; Bhartiya, S.; Venkatesh, K. Dynamic analysis of the KlGAL regulatory system in Kluyveromyces lactis: A comparative study with Saccharomyces cerevisiae. Syst. Synth. Biol. 2011, 5, 69–85. [Google Scholar] [CrossRef][Green Version]
- Lavy, T.; Kumar, P.R.; He, H.; Joshua-Tor, L. The Gal3p transducer of the GAL regulon interacts with the Gal80p repressor in its ligand-induced closed conformation. Genes Dev. 2012, 26, 294–303. [Google Scholar] [CrossRef]
- da Silveira, F.A.; Diniz, R.H.S.; Sampaio, G.; Brandão, R.L.; da Silveira, W.B.; Castro, I.M. Sugar transport systems in Kluyveromyces marxianus CCT 7735. Antonie Leeuwenhoek 2019, 112, 211–223. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, B.I.; Whiteway, M. Evolutionary transition of GAL regulatory circuit from generalist to specialist function in ascomycetes. Trends Microbiol. 2018, 26, 692–702. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Fang, X.; Xu, N.; Yao, Z.; Xie, W.; Ye, L. Development of a Highly efficient copper-inducible GAL regulation system (CuIGR) in Saccharomyces cerevisiae. ACS Synth. Biol. 2021, 10, 3435–3444. [Google Scholar] [CrossRef]
- Zhou, P.; Xu, N.; Yang, Z.; Du, Y.; Yue, C.; Xu, N.; Ye, L. Directed evolution of the transcription factor Gal4 for development of an improved transcriptional regulation system in Saccharomyces cerevisiae. Enzym. Microb. Technol. 2020, 142, 109675. [Google Scholar] [CrossRef] [PubMed]
- Lavy, T.; Yanagida, H.; Tawfik, D.S. Gal3 binds Gal80 tighter than Gal1 indicating adaptive protein changes following duplication. Mol. Biol. Evol. 2016, 33, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Pannala, V.R.; Bhat, P.J.; Bhartiya, S.; Venkatesh, K. Systems biology of GAL regulon in Saccharomyces cerevisiae. Wiley Interdiscip. Rev. Syst. Biol. Med. 2010, 2, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Kayikci, Ö.; Nielsen, J. Glucose repression in Saccharomyces cerevisiae. FEMS Yeast Res. 2015, 15, fov068. [Google Scholar] [CrossRef]
- Alipourfard, I.; Datukishvili, N.; Bakhtiyari, S.; Haghani, K.; Di Renzo, L.; de Miranda, R.C.; Mikeladze, D. MIG1 glucose repression in metabolic processes of Saccharomyces cerevisiae: Genetics to metabolic engineering. Avicenna J. Med. Biotechnol. 2019, 11, 215–220. [Google Scholar]
- Lettow, J.; Aref, R.; Schüller, H.-J. Transcriptional repressor Gal80 recruits corepressor complex Cyc8–Tup1 to structural genes of the Saccharomyces cerevisiae GAL regulon. Curr. Genet. 2022, 68, 115–124. [Google Scholar] [CrossRef]
- Quarterman, J.; Skerker, J.M.; Feng, X.; Liu, I.Y.; Zhao, H.; Arkin, A.P.; Jin, Y.-S. Rapid and efficient galactose fermentation by engineered Saccharomyces cerevisiae. J. Biotechnol. 2016, 229, 13–21. [Google Scholar] [CrossRef]
- Lin, X. The regulation of Saccharomyces cerevisiae Snf1 protein kinase on glucose utilization is in a glucose-dependent manner. Curr. Genet. 2021, 67, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Shymansky, C.M.; Wang, G.; Baidoo, E.E.; Gin, J.; Apel, A.R.; Mukhopadhyay, A.; García Martín, H.; Keasling, J.D. Flux-enabled exploration of the role of Sip1 in galactose yeast metabolism. Front. Bioeng. Biotechnol. 2017, 5, 31. [Google Scholar] [CrossRef] [PubMed]
- Rogosa, M.; Browne, H.; Whittier, E. Ethyl alcohol from whey. J. Dairy Sci. 1947, 30, 263–269. [Google Scholar] [CrossRef]
- Browne, H. Ethyl alcohol from fermentation of lactose in whey. Chem. Eng. News 1941, 19, 1272–1273. [Google Scholar]
- Whittier, E.O. Lactose and its utilization: A review. J. Dairy Sci. 1944, 27, 505–537. [Google Scholar] [CrossRef]
- Lyons, T.; Cunningham, J. Fuel alcohol from whey. Am. Dairy Rev. 1980, 42, 42A. [Google Scholar]
- Maddox, I.S.; Gutierrez, N.A. Biotechnological developments in New Zealand. Crit. Rev. Biotechnol. 1996, 16, 119–143. [Google Scholar] [CrossRef]
- Ling, K.C. Whey to Ethanol: A Biofuel Role for Dairy Cooperatives? Rural Business and Cooperative Programs 2008; USDA Rural Development: Washington, DC, USA, 2008.
- Breunig, K.; Bolotin-Fukuhara, M.; Bianchi, M.; Bourgarel, D.; Falcone, C.; Ferrero, I.; Frontali, L.; Goffrini, P.; Krijger, J.; Mazzoni, C.; et al. Regulation of primary carbon metabolism in Kluyveromyces lactis. Enzym. Microb. Technol. 2000, 26, 771–780. [Google Scholar] [CrossRef]
- Schaffrath, R.; Breunig, K.D. Genetics and molecular physiology of the yeast Kluyveromyces lactis. Fungal Genet. Biol. 2000, 30, 173–190. [Google Scholar] [CrossRef][Green Version]
- Wellenbeck, W.; Mampel, J.; Naumer, C.; Knepper, A.; Neubauer, P. Fast-track development of a lactase production process with Kluyveromyces lactis by a progressive parameter-control workflow. Eng. Life Sci. 2017, 17, 1185–1194. [Google Scholar] [CrossRef]
- Rico-Díaz, A.; Álvarez-Cao, M.-E.; Escuder-Rodríguez, J.-J.; González-Siso, M.-I.; Cerdán, M.E.; Becerra, M. Rational mutagenesis by engineering disulphide bonds improves Kluyveromyces lactis beta-galactosidase for high-temperature industrial applications. Sci. Rep. 2017, 7, 45535. [Google Scholar] [CrossRef] [PubMed]
- De Freitas, M.d.F.M.; Hortêncio, L.C.; de Albuquerque, T.L.; Rocha, M.V.P.; Gonçalves, L.R.B. Simultaneous hydrolysis of cheese whey and lactulose production catalyzed by β-galactosidase from Kluyveromyces lactis NRRL Y1564. Bioprocess Biosyst. Eng. 2020, 43, 711–722. [Google Scholar] [CrossRef] [PubMed]
- Lima, P.C.; Gazoni, I.; de Carvalho, A.M.G.; Bresolin, D.; Cavalheiro, D.; de Oliveira, D.; Rigo, E. β-galactosidase from Kluyveromyces lactis in genipin-activated chitosan: An investigation on immobilization, stability, and application in diluted UHT milk. Food Chem. 2021, 349, 129050. [Google Scholar] [CrossRef] [PubMed]
- Pandey, R.; Veeranki, V.D. Optimizing secretory expression of recombinant human interferon gamma from Kluyveromyces lactis. Prep. Biochem. Biotechnol. 2018, 48, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Spohner, S.C.; Schaum, V.; Quitmann, H.; Czermak, P. Kluyveromyces lactis: An emerging tool in biotechnology. J. Biotechnol. 2016, 222, 104–116. [Google Scholar] [CrossRef]
- Madhavan, A.; Sukumaran, R.K. Signal peptides from filamentous fungi efficiently mediate the secretion of recombinant proteins in Kluyveromyces lactis. Biochem. Eng. J. 2015, 102, 31–37. [Google Scholar] [CrossRef]
- Madhavan, A.; Sukumaran, R.K. Secreted expression of an active human interferon-beta (HuIFNβ) in Kluyveromyces lactis. Eng. Life Sci. 2016, 16, 379–385. [Google Scholar] [CrossRef]
- Ceylan, H.K.; Tayhan, S.E.; Gökçe, İ. Secretory Expression of Human Vascular Endothelial Growth Factor (VEGF 165) in Kluyveromyces lactis and Characterization of Its Biological Activity. Int. J. Pept. Res. Ther. 2021, 27, 1989–2001. [Google Scholar] [CrossRef]
- Tamshybay, A. Features of Kluyveromyces lactis expression system. In Proceedings of the European Scientific Conference, Penza, Russia, 7 October 2020. [Google Scholar]
- Lane, M.M.; Burke, N.; Karreman, R.; Wolfe, K.H.; O’Byrne, C.P.; Morrissey, J.P. Physiological and metabolic diversity in the yeast Kluyveromyces marxianus. Antonie Leeuwenhoek 2011, 100, 507–519. [Google Scholar] [CrossRef]
- Pentjuss, A.; Stalidzans, E.; Liepins, J.; Kokina, A.; Martynova, J.; Zikmanis, P.; Mozga, I.; Scherbaka, R.; Hartman, H.; Poolman, M.G.; et al. Model-based biotechnological potential analysis of Kluyveromyces marxianus central metabolism. J. Ind. Microbiol. Biotechnol. 2017, 44, 1177–1190. [Google Scholar] [CrossRef]
- Silveira, W.; Passos, F.; Mantovani, H.; Passos, F. Ethanol production from cheese whey permeate by Kluyveromyces marxianus UFV-3: A flux analysis of oxido-reductive metabolism as a function of lactose concentration and oxygen levels. Enzym. Microb. Technol. 2005, 36, 930–936. [Google Scholar] [CrossRef]
- Das, B.; Sarkar, S.; Maiti, S.; Bhattacharjee, S. Studies on production of ethanol from cheese whey using Kluyveromyces marxianus. Mater. Today Proc. 2016, 3, 3253–3257. [Google Scholar] [CrossRef]
- Christensen, A.D.; Kádár, Z.; Oleskowicz-Popiel, P.; Thomsen, M.H. Production of bioethanol from organic whey using Kluyveromyces marxianus. Ind. Microbiol. Biotechnol. 2011, 38, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Murari, C.S.; Machado, W.R.C.; Schuina, G.L.; Del Bianchi, V.L. Optimization of bioethanol production from cheese whey using Kluyveromyces marxianus URM 7404. Biocatal. Agric. Biotechnol. 2019, 20, 101182. [Google Scholar] [CrossRef]
- Zoppellari, F.; Bardi, L. Production of bioethanol from effluents of the dairy industry by Kluyveromyces marxianus. New Biotechnol. 2013, 30, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Saini, P.; Beniwal, A.; Kokkiligadda, A.; Vij, S. Evolutionary adaptation of Kluyveromyces marxianus strain for efficient conversion of whey lactose to bioethanol. Process Biochem. 2017, 62, 69–79. [Google Scholar] [CrossRef]
- Tesfaw, A.; Oner, E.T.; Assefa, F. Evaluating crude whey for bioethanol production using non-Saccharomyces yeast, Kluyveromyces marxianus. SN Appl. Sci. 2021, 3, 1–8. [Google Scholar] [CrossRef]
- Diniz, R.H.S.; Villada, J.C.; Alvim, M.C.T.; Vidigal, P.M.P.; Vieira, N.M.; Lamas-Maceiras, M.; Cerdán, M.E.; González-Siso, M.-I.; Lahtvee, P.-J.; da Silveira, W.B. Transcriptome analysis of the thermotolerant yeast Kluyveromyces marxianus CCT 7735 under ethanol stress. Appl. Microbiol. Biotechnol. 2017, 101, 6969–6980. [Google Scholar] [CrossRef]
- Wang, D.; Wu, D.; Yang, X.; Hong, J. Transcriptomic analysis of thermotolerant yeast Kluyveromyces marxianus in multiple inhibitors tolerance. RSC Adv. 2018, 8, 14177–14192. [Google Scholar] [CrossRef]
- Sivarathnakumar, S.; Jayamuthunagai, J.; Baskar, G.; Praveenkumar, R.; Selvakumari, I.A.E.; Bharathiraja, B. Bioethanol production from woody stem Prosopis juliflora using thermo tolerant yeast Kluyveromyces marxianus and its kinetics studies. Bioresour. Technol. 2019, 293, 122060. [Google Scholar] [CrossRef]
- Ha-Tran, D.M.; Nguyen, T.T.M.; Huang, C.-C. Kluyveromyces marxianus: Current state of omics studies, strain improvement strategy and potential industrial implementation. Fermentation 2020, 6, 124. [Google Scholar] [CrossRef]
- Jhariya, U.; Dafale, N.A.; Srivastava, S.; Bhende, R.S.; Kapley, A.; Purohit, H.J. Understanding ethanol tolerance mechanism in Saccharomyces cerevisiae to enhance the bioethanol production: Current and future prospects. BioEnergy Res. 2021, 14, 670–688. [Google Scholar] [CrossRef]
- Lairón-Peris, M.; Routledge, S.; Linney, J.; Alonso-del-Real, J.; Spickett, C.; Pitt, A.; Guillamón, J.M.; Barrio, E.; Goddard, A.; Querol, A. Lipid composition analysis reveals mechanisms of ethanol tolerance in the model yeast Saccharomyces cerevisiae. Appl. Environ. Microb. 2021, 87, e00440-21. [Google Scholar] [CrossRef] [PubMed]
- Riles, L.; Fay, J.C. Genetic basis of variation in heat and ethanol tolerance in Saccharomyces cerevisiae. G3 Genes Genomes Genet. 2019, 9, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Da Costa, B.L.V.; Raghavendran, V.; Franco, L.F.M.; Chaves Filho, A.d.B.; Yoshinaga, M.Y.; Miyamoto, S.; Basso, T.O.; Gombert, A.K. Forever panting and forever growing: Physiology of Saccharomyces cerevisiae at extremely low oxygen availability in the absence of ergosterol and unsaturated fatty acids. FEMS Yeast Res. 2019, 19, foz054. [Google Scholar] [CrossRef] [PubMed]
- Da Costa, B.L.V.; Basso, T.O.; Raghavendran, V.; Gombert, A.K. Anaerobiosis revisited: Growth of Saccharomyces cerevisiae under extremely low oxygen availability. Appl. Microbiol. Biotechnol. 2018, 102, 2101–2116. [Google Scholar] [CrossRef]
- Parapouli, M.; Vasileiadis, A.; Afendra, A.-S.; Hatziloukas, E. Saccharomyces cerevisiae and its industrial applications. AIMS Microbiol. 2020, 6, 1–31. [Google Scholar] [CrossRef]
- Cripwell, R.A.; Favaro, L.; Viljoen-Bloom, M.; van Zyl, W.H. Consolidated bioprocessing of raw starch to ethanol by Saccharomyces cerevisiae: Achievements and challenges. Biotechnol. Adv. 2020, 42, 107579. [Google Scholar] [CrossRef]
- Boudjema, K.; Fazouane-Naimi, F.; Hellal, A. Optimization of the bioethanol production on sweet cheese whey by Saccharomyces cerevisiae DIV13-Z087C0VS using response surface methodology (RSM). Rom. Biotechnol. Lett. 2015, 20, 10814–10825. [Google Scholar]
- Kokkiligadda, A.; Beniwal, A.; Saini, P.; Vij, S. Utilization of cheese whey using synergistic immobilization of β-galactosidase and Saccharomyces cerevisiae cells in dual matrices. Appl. Biochem. Biotechnol. 2016, 179, 1469–1484. [Google Scholar] [CrossRef]
- Kisielewska, M. Ultrasonic stimulation of co-immobilized Saccharomyces cerevisiae cells and β-galactosidase enzyme for enhanced ethanol production from whey ultrafiltration permeate. Pol. J. Environ. Stud. 2012, 21, 387–393. [Google Scholar]
- Panagopoulos, V.; Dima, A.; Boura, K.; Bosnea, L.; Nigam, P.S.; Kanellaki, M.; Koutinas, A.A. Cell factory models of non-engineered S. cerevisiae containing lactase in a second layer for lactose fermentation in one batch. Enzym. Microb. Technol. 2021, 145, 109750. [Google Scholar] [CrossRef] [PubMed]
- Cunha, J.T.; Gomes, D.G.; Romaní, A.; Inokuma, K.; Hasunuma, T.; Kondo, A.; Domingues, L. Cell surface engineering of Saccharomyces cerevisiae for simultaneous valorization of corn cob and cheese whey via ethanol production. Energy Convers. Manag. 2021, 243, 114359. [Google Scholar] [CrossRef]
- Neri, D.F.; Balcão, V.M.; Carneiro-da-Cunha, M.G.; Carvalho Jr, L.B.; Teixeira, J.A. Immobilization of β-galactosidase from Kluyveromyces lactis onto a polysiloxane–polyvinyl alcohol magnetic (mPOS–PVA) composite for lactose hydrolysis. Catal. Commun. 2008, 9, 2334–2339. [Google Scholar] [CrossRef]
- Beniwal, A.; Saini, P.; Kokkiligadda, A.; Vij, S. Use of silicon dioxide nanoparticles for β-galactosidase immobilization and modulated ethanol production by co-immobilized K. marxianus and S. cerevisiae in deproteinized cheese whey. LWT 2018, 87, 553–561. [Google Scholar] [CrossRef]
- Soto, D.; Escobar, S.; Guzmán, F.; Cárdenas, C.; Bernal, C.; Mesa, M. Structure-activity relationships on the study of β-galactosidase folding/unfolding due to interactions with immobilization additives: Triton X-100 and ethanol. Int. J. Biol. Macromol. 2017, 96, 87–92. [Google Scholar] [CrossRef]
- Guo, X.; Zhou, J.; Xiao, D. Improved ethanol production by mixed immobilized cells of Kluyveromyces marxianus and Saccharomyces cerevisiae from cheese whey powder solution fermentation. Appl. Biochem. Biotechnol. 2010, 160, 532–538. [Google Scholar] [CrossRef]
- Eiadpum, A.; Limtong, S.; Phisalaphong, M. High-temperature ethanol fermentation by immobilized coculture of Kluyveromyces marxianus and Saccharomyces cerevisiae. J. Biosci. Bioeng. 2012, 114, 325–329. [Google Scholar] [CrossRef]
- Magalhatilde, K.T.; Rodrigues, A.K.; Gervasio, I.M.; Gervasio, I.; Schwan, R.F. Ethanol production from deproteinized cheese whey fermentations by co-cultures of Kluyveromyces marxianus and Saccharomyces cerevisiae. Afr. J. Microbiol. Res. 2013, 7, 1121–1127. [Google Scholar]
- Farkas, C.; Rezessy-Szabó, J.M.; Gupta, V.K.; Bujna, E.; Pham, T.M.; Pásztor-Huszár, K.; Friedrich, L.; Bhat, R.; Thakur, V.K.; Nguyen, Q.D. Batch and fed-batch ethanol fermentation of cheese-whey powder with mixed cultures of different yeasts. Energies 2019, 12, 4495. [Google Scholar] [CrossRef]
- Gabardo, S.; Pereira, G.F.; Klein, M.P.; Rech, R.; Hertz, P.F.; Ayub, M.A.Z. Dynamics of yeast immobilized-cell fluidized-bed bioreactors systems in ethanol fermentation from lactose-hydrolyzed whey and whey permeate. Bioprocess Biosyst. Eng. 2016, 39, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Díez-Antolínez, R.; Hijosa-Valsero, M.; Paniagua-García, A.I.; Garita-Cambronero, J.; Gómez, X. Yeast screening and cell immobilization on inert supports for ethanol production from cheese whey permeate with high lactose loads. PLoS ONE 2018, 13, e0210002. [Google Scholar] [CrossRef] [PubMed]
- Xin, Y.; Yang, M.; Yin, H.; Yang, J. Improvement of ethanol tolerance by inactive protoplast fusion in Saccharomyces cerevisiae. BioMed Res. Int. 2020, 2020, 1979318. [Google Scholar] [CrossRef] [PubMed]
- Lalithakumari, D. Fungal Protoplast: A Biotechnological Tool, 1st ed.; CRC Press: Boca Raton, FL, USA, 2019; pp. 127–154. [Google Scholar]
- Krishnamoorthy, R.N.; Vijila, K.; Kumutha, K. Intergeneric protoplast fusion of yeast for high ethanol production from cheese industry waste Whey. J. Yeast Fungal Res. 2010, 1, 81–87. [Google Scholar]
- Guo, X.; Wang, R.; Chen, Y.; Xiao, D. Intergeneric yeast fusants with efficient ethanol production from cheese whey powder solution: Construction of a Kluyveromyces marxianus and Saccharomyces cerevisiae AY-5 hybrid. Eng. Life Sci. 2012, 12, 656–661. [Google Scholar] [CrossRef]
- Sharma, S.; Arora, A.; Paul, D. Protoplast fusion of yeast strains for strain improvement to enhance mixed substrate utilization range. In Biotechnology and Biological Sciences, 1st ed.; Sen, S., Mukherjee, S., Paul, R., Narula, R., Eds.; CRC Press: London, UK, 2019; pp. 333–336. [Google Scholar]
- Ryu, Y.-W.; Jang, H.-W.; Lee, H.-S. Enhancement of ethanol tolerance of lactose assimilating yeast strain by protoplast fusion. J. Microbiol. Biotechnol. 1991, 1, 151–156. [Google Scholar]
- Ozbudak, E.M.; Thattai, M.; Lim, H.N.; Shraiman, B.I.; Van Oudenaarden, A. Multistability in the lactose utilization network of Escherichia coli. Nature 2004, 427, 737–740. [Google Scholar] [CrossRef]
- Beckwith, J.R. Regulation of the Lac Operon: Recent studies on the regulation of lactose metabolism in Escherichia coli support the operon model. Science 1967, 156, 597–604. [Google Scholar] [CrossRef]
- Ammar, E.M.; Wang, X.; Rao, C.V. Regulation of metabolism in Escherichia coli during growth on mixtures of the non-glucose sugars: Arabinose, lactose, and xylose. Sci. Rep. 2018, 8, 609. [Google Scholar] [CrossRef]
- Guarente, L.; Ptashne, M. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1981, 78, 2199–2203. [Google Scholar] [CrossRef]
- Porro, D.; Martegani, E.; Ranzi, B.M.; Alberghina, L. Lactose/whey utilization and ethanol production by transformed Saccharomyces cerevisiae cells. Biotechnol. Bioeng. 1992, 39, 799–805. [Google Scholar] [CrossRef] [PubMed]
- Martegani, E.; Brambilla, L.; Porro, D.; Ranzi, B.M.; Alberghina, L. Alteration of cell population structure due to cell lysis in Saccharomyces cerevisiae cells overexpressing the GAL4 gene. Yeast 1993, 9, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Show, P.L.; Oladele, K.O.; Siew, Q.Y.; Aziz Zakry, F.A.; Lan, J.C.-W.; Ling, T.C. Overview of citric acid production from Aspergillus niger. Front. Life Sci 2015, 8, 271–283. [Google Scholar] [CrossRef]
- Banjoko, I.O.; Adeyanju, M.M.; Ademuyiwa, O.; Adebawo, O.O. Hypolipidemic effects of lactic acid bacteria fermented cereal in rats. Lipids Health Dis. 2012, 11, 170. [Google Scholar] [CrossRef]
- Ramakrishnan, S.; Hartley, B.S. Fermentation of lactose by yeast cells secreting recombinant fungal lactase. Appl. Environ. Microbiol. 1993, 59, 4230–4235. [Google Scholar] [CrossRef]
- Domingues, L.; Onnela, M.-L.; Teixeira, J.; Lima, N.; Penttilä, M. Construction of a flocculent brewer’s yeast strain secreting Aspergillus niger β-galactosidase. Appl. Microbiol. Biotechnol. 2000, 54, 97–103. [Google Scholar] [CrossRef]
- Domingues, L.; Teixeira, J.; Penttilä, M.; Lima, N. Construction of a flocculent Saccharomyces cerevisiae strain secreting high levels of Aspergillus niger β-galactosidase. Appl. Microbiol. Biotechnol. 2002, 58, 645–650. [Google Scholar] [CrossRef]
- Sreekrishna, K.; Dickson, R.C. Construction of strains of Saccharomyces cerevisiae that grow on lactose. Proc. Natl. Acad. Sci. USA 1985, 82, 7909–7913. [Google Scholar] [CrossRef]
- Rubio-Texeira, M.; Castrillo, J.I.; Adam, A.C.; Ugalde, U.O.; Polaina, J. Highly efficient assimilation of lactose by a metabolically engineered strain of Saccharomyces cerevisiae. Yeast 1998, 14, 827–837. [Google Scholar] [CrossRef]
- Domingues, L.; Teixeira, J.; Lima, N. Construction of a flocculent Saccharomyces cerevisiae fermenting lactose. Appl. Microbiol. Biotechnol. 1999, 51, 621–626. [Google Scholar] [CrossRef]
- Divate, N.R.; Chen, G.-H.; Wang, P.-M.; Ou, B.-R.; Chung, Y.-C. Engineering Saccharomyces cerevisiae for improvement in ethanol tolerance by accumulation of trehalose. Bioengineered 2016, 7, 445–458. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, M. Diverse and common features of trehalases and their contributions to microbial trehalose metabolism. Appl. Microbiol. Biotechnol. 2020, 104, 1837–1847. [Google Scholar] [CrossRef] [PubMed]
- Maicas, S.; Guirao-Abad, J.P.; Argüelles, J.-C. Yeast trehalases: Two enzymes, one catalytic mission. Biochim. Biophys. Acta 2016, 1860, 2249–2254. [Google Scholar] [CrossRef]
- Zou, J.; Guo, X.; Shen, T.; Dong, J.; Zhang, C.; Xiao, D. Construction of lactose-consuming Saccharomyces cerevisiae for lactose fermentation into ethanol fuel. J. Ind. Microbiol. Biotechnol. 2013, 40, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Uncu, O.N.; Cekmecelioglu, D. Cost-effective approach to ethanol production and optimization by response surface methodology. Waste Manag. 2011, 31, 636–643. [Google Scholar] [CrossRef] [PubMed]
- Diniz, R.H.; Rodrigues, M.Q.; Fietto, L.G.; Passos, F.M.; Silveira, W.B. Optimizing and validating the production of ethanol from cheese whey permeate by Kluyveromyces marxianus UFV-3. Biocatal. Agric. Biotechnol. 2014, 3, 111–117. [Google Scholar] [CrossRef]
- Dasa, B.; Dasa, M.; Bhattacharjeeb, S.; Bhattacharjeea, C. Ethanol production from deproteinized cheese whey powder in a batch fermentation process: Optimization of process and kinetic modelling. Desalination Water Treat. 2017, 64, 198–206. [Google Scholar] [CrossRef]
- Wang, Y.; Li, K.; Huang, F.; Wang, J.; Zhao, J.; Zhao, X.; Garza, E.; Manow, R.; Grayburn, S.; Zhou, S. Engineering and adaptive evolution of Escherichia coli W for l-lactic acid fermentation from molasses and corn steep liquor without additional nutrients. Bioresour. Technol. 2013, 148, 394–400. [Google Scholar] [CrossRef]
- Martinez-Burgos, W.J.; Sydney, E.B.; de Paula, D.R.; Medeiros, A.B.P.; de Carvalho, J.C.; Molina, D.; Soccol, C.R. Hydrogen production by dark fermentation using a new low-cost culture medium composed of corn steep liquor and cassava processing water: Process optimization and scale-up. Bioresour. Technol. 2021, 320, 124370. [Google Scholar] [CrossRef]
- Agarwal, L.; Dutt, K.; Meghwanshi, G.K.; Saxena, R. Anaerobic fermentative production of lactic acid using cheese whey and corn steep liquor. Biotechnol. Lett. 2008, 30, 631–635. [Google Scholar] [CrossRef]
- Silva, A.C.; Guimarães, P.M.; Teixeira, J.A.; Domingues, L. Fermentation of deproteinized cheese whey powder solutions to ethanol by engineered Saccharomyces cerevisiae: Effect of supplementation with corn steep liquor and repeated-batch operation with biomass recycling by flocculation. J. Ind. Microbiol. Biotechnol. 2010, 37, 973–982. [Google Scholar] [CrossRef][Green Version]
- Bailey, J.E. Toward a science of metabolic engineering. Science 1991, 252, 1668–1675. [Google Scholar] [CrossRef] [PubMed]
- Ostergaard, S.; Olsson, L.; Johnston, M.; Nielsen, J. Increasing galactose consumption by Saccharomyces cerevisiae through metabolic engineering of the GAL gene regulatory network. Nat. Biotechnol. 2000, 18, 1283–1286. [Google Scholar] [CrossRef]
- Sunwoo, I.Y.; Sukwong, P.; Park, Y.R.; Jeong, D.Y.; Kim, S.R.; Jeong, G.-T.; Kim, S.-K. Enhancement of galactose uptake from Kappaphycus alvarezii using Saccharomyces cerevisiae through deletion of negative regulators of GAL genes. Appl. Biochem. Biotechnol. 2021, 193, 577–588. [Google Scholar] [CrossRef] [PubMed]
- Escalante-Chong, R.; Savir, Y.; Carroll, S.M.; Ingraham, J.B.; Wang, J.; Marx, C.J.; Springer, M. Galactose metabolic genes in yeast respond to a ratio of galactose and glucose. Proc. Natl. Acad. Sci. USA 2015, 112, 1636–1641. [Google Scholar] [CrossRef] [PubMed]
- Rønnow, B.; Olsson, L.; Nielsen, J.; Mikkelsen, J.D. Derepression of galactose metabolism in melibiase producing bakers’ and distillers’ yeast. J. Biotechnol. 1999, 72, 213–228. [Google Scholar] [CrossRef]
- Zou, J.; Chen, X.; Hu, Y.; Xiao, D.; Guo, X.; Chang, X.; Zhou, L. Uncoupling glucose sensing from GAL metabolism for heterologous lactose fermentation in Saccharomyces cerevisiae. Biotechnol. Lett. 2021, 43, 1607–1616. [Google Scholar] [CrossRef]
- Mans, R.; Daran, J.-M.G.; Pronk, J.T. Under pressure: Evolutionary engineering of yeast strains for improved performance in fuels and chemicals production. Curr. Opin. Biotechnol. 2018, 50, 47–56. [Google Scholar] [CrossRef]
- Çakar, Z.P.; Turanlı-Yıldız, B.; Alkım, C.; Yılmaz, Ü. Evolutionary engineering of Saccharomyces cerevisiae for improved industrially important properties. FEMS Yeast Res. 2012, 12, 171–182. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhang, J.; Ji, X.; Fang, Z.; Wu, Z.; Chen, J.; Du, G. Evolutionary engineering of industrial microorganisms-strategies and applications. Appl. Microbiol. Biotechnol. 2018, 102, 4615–4627. [Google Scholar] [CrossRef]
- Shepelin, D.; Hansen, A.S.L.; Lennen, R.; Luo, H.; Herrgård, M.J. Selecting the best: Evolutionary engineering of chemical production in microbes. Genes 2018, 9, 249. [Google Scholar] [CrossRef] [PubMed]
- Guimarães, P.M.; François, J.; Parrou, J.L.; Teixeira, J.A.; Domingues, L. Adaptive evolution of a lactose-consuming Saccharomyces cerevisiae recombinant. Appl. Environ. Microb. 2008, 74, 1748–1756. [Google Scholar] [CrossRef] [PubMed]
- Guimarães, P.M.; Le Berre, V.; Sokol, S.; François, J.; Teixeira, J.A.; Domingues, L. Comparative transcriptome analysis between original and evolved recombinant lactose-consuming Saccharomyces cerevisiae strains. Biotechnol. J. Healthc. Nutr. Technol. 2008, 3, 1591–1597. [Google Scholar]
- Królczyk, J.B.; Dawidziuk, T.; Janiszewska-Turak, E.; Sołowiej, B. Use of whey and whey preparations in the food industry—A review. Pol. J. Food Nutr. Sci. 2016, 66, 157. [Google Scholar] [CrossRef]
- Carvalho, F.; Prazeres, A.R.; Rivas, J. Cheese whey wastewater: Characterization and treatment. Sci. Total Environ. 2013, 445, 385–396. [Google Scholar] [CrossRef]
- Rogério, M.C.P.; Martins, E.C.; Shiotsuki, L.; Pompeu, R.C.F.F.; Muir, J.P.; Araújo, A.R.; de Sousa Oliveira, D.; Magalhães, J.L.L.; Campos, W.É.; Facó, O.; et al. Economic viability of finishing lambs in the feedlot using bovine cheese whey as a dietary ingredient. Small Rumin. Res. 2019, 170, 131–136. [Google Scholar] [CrossRef]
- Tsermoula, P.; Khakimov, B.; Nielsen, J.H.; Engelsen, S.B. Whey-The waste-stream that became more valuable than the food product. Trends Food Sci. Technol. 2021, 118, 230–241. [Google Scholar] [CrossRef]
- Kumar, R.; Chauhan, S.K.; Shinde, G.; Subramanian, V.; Nadanasabapathi, S. Whey proteins: A potential ingredient for food industry-a review. Asian J. Dairy Food Res. 2018, 37, 283–290. [Google Scholar]
- Tsiouris, V.; Kontominas, M.G.; Filioussis, G.; Chalvatzi, S.; Giannenas, I.; Papadopoulos, G.; Koutoulis, K.; Fortomaris, P.; Georgopoulou, I. The effect of whey on performance, gut health and bone morphology parameters in broiler chicks. Foods 2020, 9, 588. [Google Scholar] [CrossRef]
- Rtibi, K.; Marzouki, K.; Salhi, A.; Sebai, H. Dietary supplementation of carob and whey modulates gut morphology, hemato-biochemical indices, and antioxidant biomarkers in rabbits. J. Med. Food 2021, 24, 1124–1133. [Google Scholar] [CrossRef]
- Hausjell, J.; Miltner, M.; Herzig, C.; Limbeck, A.; Saracevic, Z.; Saracevic, E.; Weissensteiner, J.; Molitor, C.; Halbwirth, H.; Spadiut, O. Valorisation of cheese whey as substrate and inducer for recombinant protein production in E. coli HMS174(DE3). Bioresour. Technol. Rep. 2019, 8, 100340. [Google Scholar] [CrossRef]
- Panesar, P.S.; Kennedy, J.F.; Knill, C.J.; Kosseva, M. Production of L(+) lactic acid using Lactobacillus casei from whey. Braz. Arch. Biol. Technol. 2010, 53, 219–226. [Google Scholar] [CrossRef]
- Xu, J.; Hao, J.; Guzman, J.J.; Spirito, C.M.; Harroff, L.A.; Angenent, L.T. Temperature-phased conversion of acid whey waste into medium-chain carboxylic acids via lactic acid: No external e-donor. Joule 2018, 2, 280–295. [Google Scholar] [CrossRef]
- Rama, G.R.; Kuhn, D.; Beux, S.; Maciel, M.J.; de Souza, C.F.V. Potential applications of dairy whey for the production of lactic acid bacteria cultures. Int. Dairy J. 2019, 98, 25–37. [Google Scholar] [CrossRef]
- Singh, A.K.; Singh, K. Utilization of whey for the production of instant energy beverage by using response surface methodology. Adv. J. Food Sci. Technol. 2012, 4, 103–111. [Google Scholar]
- Luo, S.R.; DeMarsh, T.A.; DeRiancho, D.; Stelick, A.; Alcaine, S.D. Characterization of the fermentation and sensory profiles of novel yeast-fermented acid whey beverages. Foods 2021, 10, 1204. [Google Scholar] [CrossRef]
- Gantumur, M.-A.; Sukhbaatar, N.; Qayum, A.; Bilawal, A.; Tsembeltsogt, B.; Oh, K.-C.; Jiang, Z.; Hou, J. Characterization of major volatile compounds in whey spirits produced by different distillation stages of fermented lactose-supplemented whey. J. Dairy Sci. 2022, 105, 83–96. [Google Scholar] [CrossRef]
- Souza, F.P.; Balthazar, C.F.; Guimarães, J.T.; Pimentel, T.C.; Esmerino, E.A.; Freitas, M.Q.; Raices, R.S.; Silva, M.C.; Cruz, A.G. The addition of xyloligoosaccharide in strawberry-flavored whey beverage. LWT 2019, 109, 118–122. [Google Scholar] [CrossRef]
- AbdulAlim, T.; Zayan, A.; Campelo, P.; Bakry, A. Development of new functional fermented product: Mulberry-whey beverage. J. Nutr. Food Technol. 2018, 1, 64–69. [Google Scholar] [CrossRef]
- Yadav, R.B.; Yadav, B.S.; Kalia, N. Development and storage studies on whey-based banana herbal (Mentha arvensis) beverage. Am. J. Food Technol. 2010, 5, 121–129. [Google Scholar] [CrossRef]
- Soccol, C.R.; Faraco, V.; Karp, S.G.; Vandenberghe, L.P.; Thomaz-Soccol, V.; Woiciechowski, A.L.; Pandey, A. Lignocellulosic bioethanol: Current status and future perspectives. In Biofuels: Alternative Feedstocks and Conversion Processes for the Production of Liquid and Gaseous Biofuels, 2nd ed.; Pandey, A., Larroche, C., Dussap, C.G., Gnansounou, E., Khanal, S.K., Ricke, S., Eds.; Elsevier: London, UK, 2019; pp. 331–354. [Google Scholar]
- Gomes, D.G.; Teixeira, J.A.; Domingues, L. Economic determinants on the implementation of a Eucalyptus wood biorefinery producing biofuels, energy and high added-value compounds. Appl. Energy 2021, 303, 117662. [Google Scholar] [CrossRef]
- Gomes, D.G.; Michelin, M.; Romaní, A.; Domingues, L.; Teixeira, J.A. Co-production of biofuels and value-added compounds from industrial Eucalyptus globulus bark residues using hydrothermal treatment. Fuel 2021, 285, 119265. [Google Scholar] [CrossRef]
- Ferreira, P.G.; da Silveira, F.A.; dos Santos, R.C.V.; Genier, H.L.A.; Diniz, R.H.S.; Ribeiro, J.I.; Fietto, L.G.; Passos, F.M.L.; da Silveira, W.B. Optimizing ethanol production by thermotolerant Kluyveromyces marxianus CCT 7735 in a mixture of sugarcane bagasse and ricotta whey. Food Sci. Biotechnol. 2015, 24, 1421–1427. [Google Scholar] [CrossRef]
- Fischer, J.; Lopes, V.; Galvão, C.; Teodoro, J.; Coutinho Filho, U.; Cardoso, V.L. Utilization of cheese whey and cellulosic biomass for production of ethanol by selected fungi strain from brazilian savannas. Chem. Eng. Trans. 2013, 32, 1075–1080. [Google Scholar]
- Coman, G.; Andreea, N.; Constanta, S.; Bahrim, G. Bioethanol production by solid state fermentation from cheese whey mixed with brewer’s spent grains. Ann. Univ. Dunarea Jos Galati. Fascicle VI Food Technol. 2015, 39, 49. [Google Scholar]
- Lee, K.-S.; Hong, M.-E.; Jung, S.-C.; Ha, S.-J.; Yu, B.J.; Koo, H.M.; Park, S.M.; Seo, J.-H.; Kweon, D.-H.; Park, J.C.; et al. Improved galactose fermentation of Saccharomyces cerevisiae through inverse metabolic engineering. Biotechnol. Bioeng. 2011, 108, 621–631. [Google Scholar] [CrossRef]
- Garcia Sanchez, R.; Hahn-Hägerdal, B.; Gorwa-Grauslund, M.F. PGM2 overexpression improves anaerobic galactose fermentation in Saccharomyces cerevisiae. Microb. Cell Factories 2010, 9, 40. [Google Scholar] [CrossRef]
- Hong, K.-K.; Vongsangnak, W.; Vemuri, G.N.; Nielsen, J. Unravelling evolutionary strategies of yeast for improving galactose utilization through integrated systems level analysis. Proc. Natl. Acad. Sci. USA 2011, 108, 12179–12184. [Google Scholar] [CrossRef]

| Parameter | Sweet Whey | Acid Whey |
|---|---|---|
| Total solid (%) | 6.21 | 5.70 |
| Lactose (%) | 4.82 | 4.60 |
| Protein (%) | 0.75 | 0.30 |
| Fat (%) | 0.05 | <0.01 |
| Ash (%) | 0.60 | 0.80 |
| pH | 5.80–6.10 | 4.0–5.0 |
| Category | S. cerevisiae | K. lactis | ||
|---|---|---|---|---|
| Gene Name | Function | Gene Name | Function | |
| Catabolic genes | MEL11 | α-galactosidase | LAC4 | β-galactosidase |
| GAL2 | Galactose permease | LAC12 | Lactose/galactose permease | |
| GAL1 | Bifunctional galactokinase/sensor | KlGAL1 | Bifunctional galactokinase/sensor inducer [74,75] | |
| GAL7 | Galactose-1-phosphate uridylyltransferase | KlGAL7 | Galactose-1-phosphate uridylyltransferase | |
| GAL10 | Uridine diphoshpoglucose 4-epimerase | KlGAL10 | Uridine diphoshpoglucose 4-epimerase | |
| GAL5(PGM2) | Phosphoglucomutase | KlGAL5 | Phosphoglucomutase | |
| Regulatory genes | GAL4 | Transcriptional activator [75] | KlGAL4(LAC9) | Transcriptional activator [74,75] |
| GAL80 | Gal4p repressor [74,75] | KlGAL80 | Gal4p repressor | |
| GAL3 | Gal80 repressor (sensor/inducer) [76] | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, J.; Chang, X. Past, Present, and Future Perspectives on Whey as a Promising Feedstock for Bioethanol Production by Yeast. J. Fungi 2022, 8, 395. https://doi.org/10.3390/jof8040395
Zou J, Chang X. Past, Present, and Future Perspectives on Whey as a Promising Feedstock for Bioethanol Production by Yeast. Journal of Fungi. 2022; 8(4):395. https://doi.org/10.3390/jof8040395
Chicago/Turabian StyleZou, Jing, and Xuedong Chang. 2022. "Past, Present, and Future Perspectives on Whey as a Promising Feedstock for Bioethanol Production by Yeast" Journal of Fungi 8, no. 4: 395. https://doi.org/10.3390/jof8040395
APA StyleZou, J., & Chang, X. (2022). Past, Present, and Future Perspectives on Whey as a Promising Feedstock for Bioethanol Production by Yeast. Journal of Fungi, 8(4), 395. https://doi.org/10.3390/jof8040395







