Selection by UV Mutagenesis and Physiological Characterization of Mutant Strains of the Yeast Saprochaete suaveolens (Former Geotrichum fragrans) with Higher Capacity to Produce Flavor Compounds
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Strains, UV Mutagenesis and Screening
2.2. Volatile Organic Compound (VOCs) Semi-Quantitative Analysis
2.3. Preparation of Crude Extracts and Assays of Enzymatic Activities
2.4. Determination of Strain Growth Rates
2.5. Statistical Analysis
3. Results and Discussion
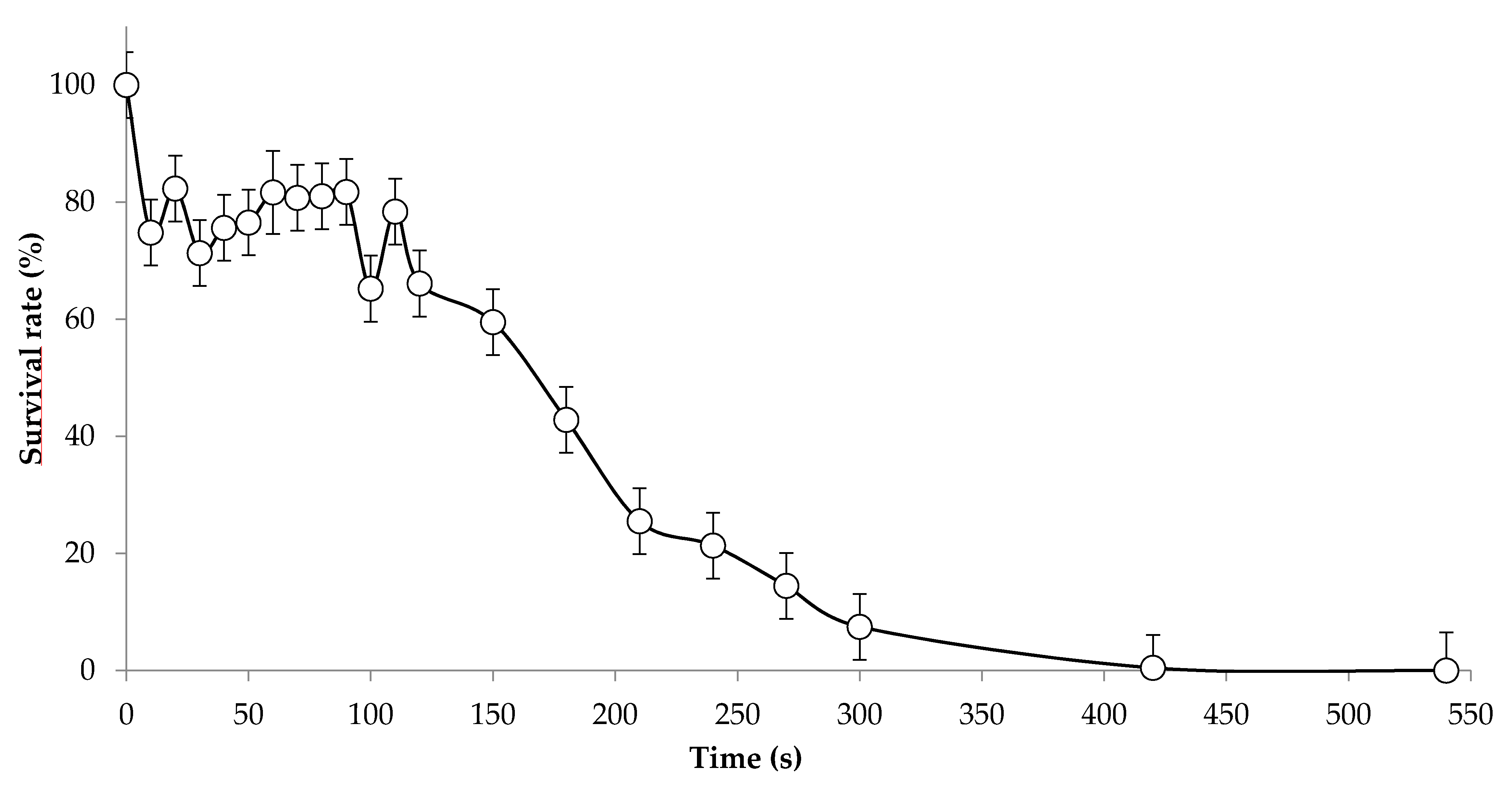
3.1. Condition of UV Mutagenesis and Screening Strategy
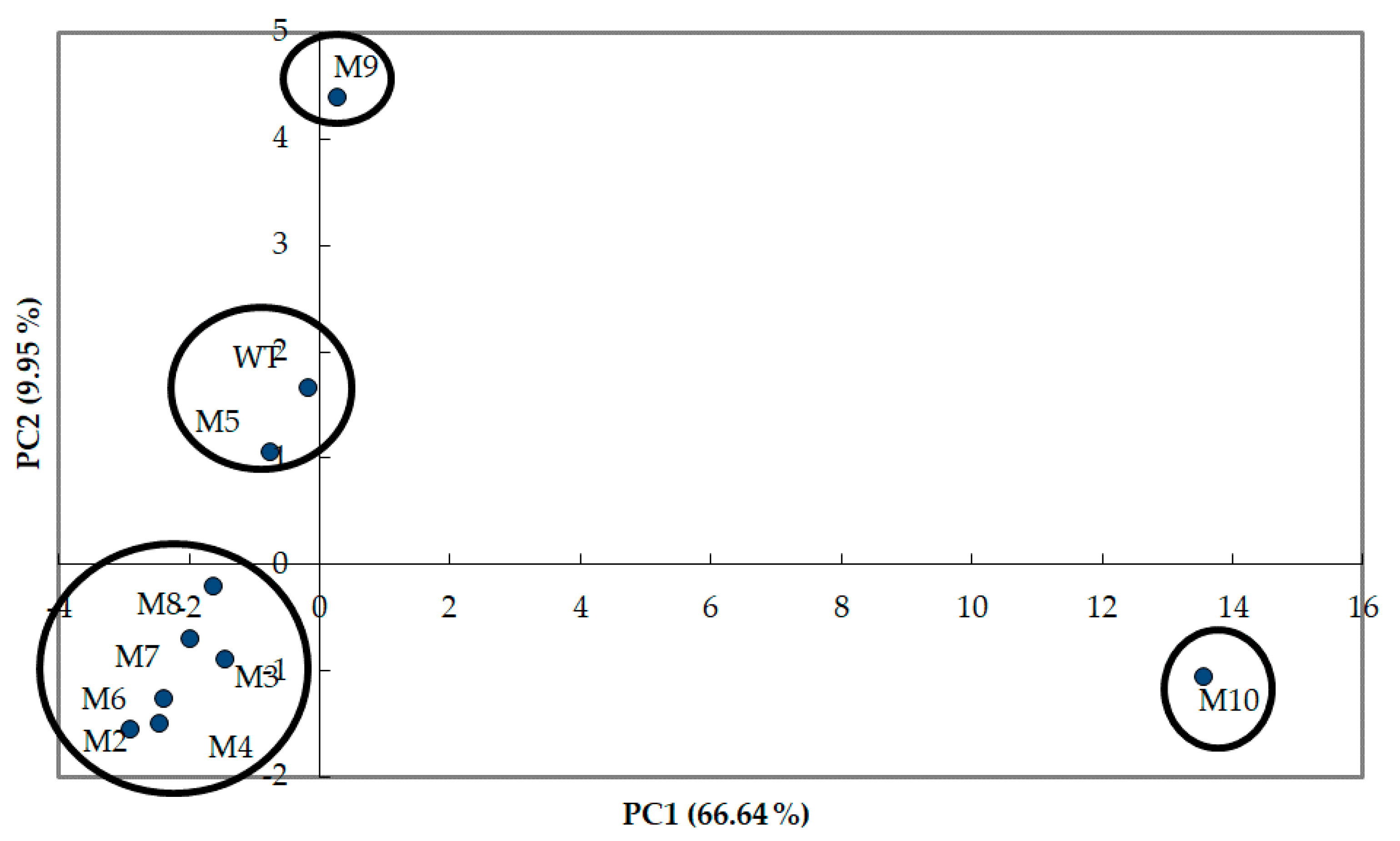
3.2. Multivariate Analysis of The Wild-Type of S. suaveolens and Its 9 ILV-Mutants Based on Their VOCs Production
3.3. Comparative Analysis VOCs Production between S. suaveolens and the 9 ILV-Mutants
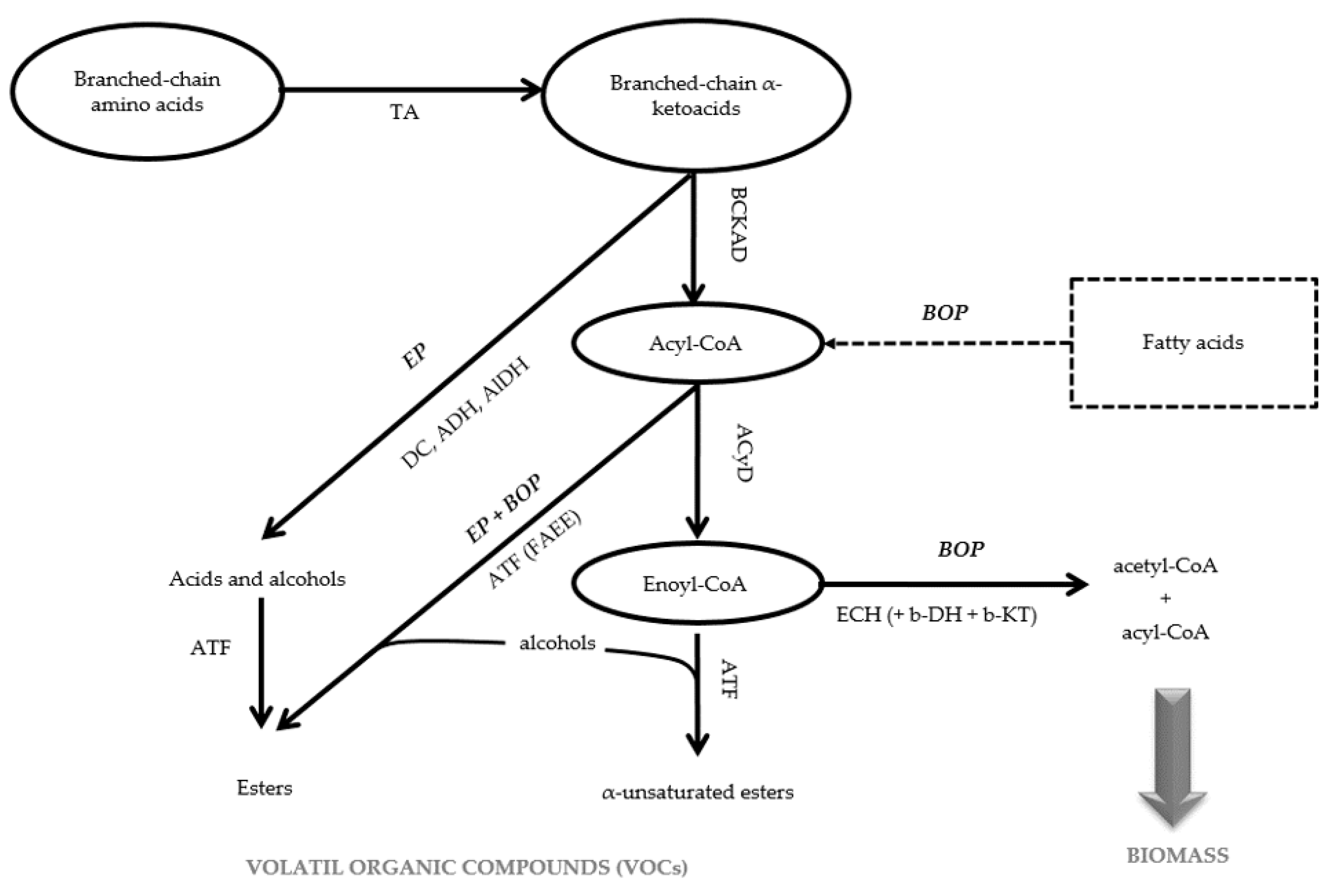
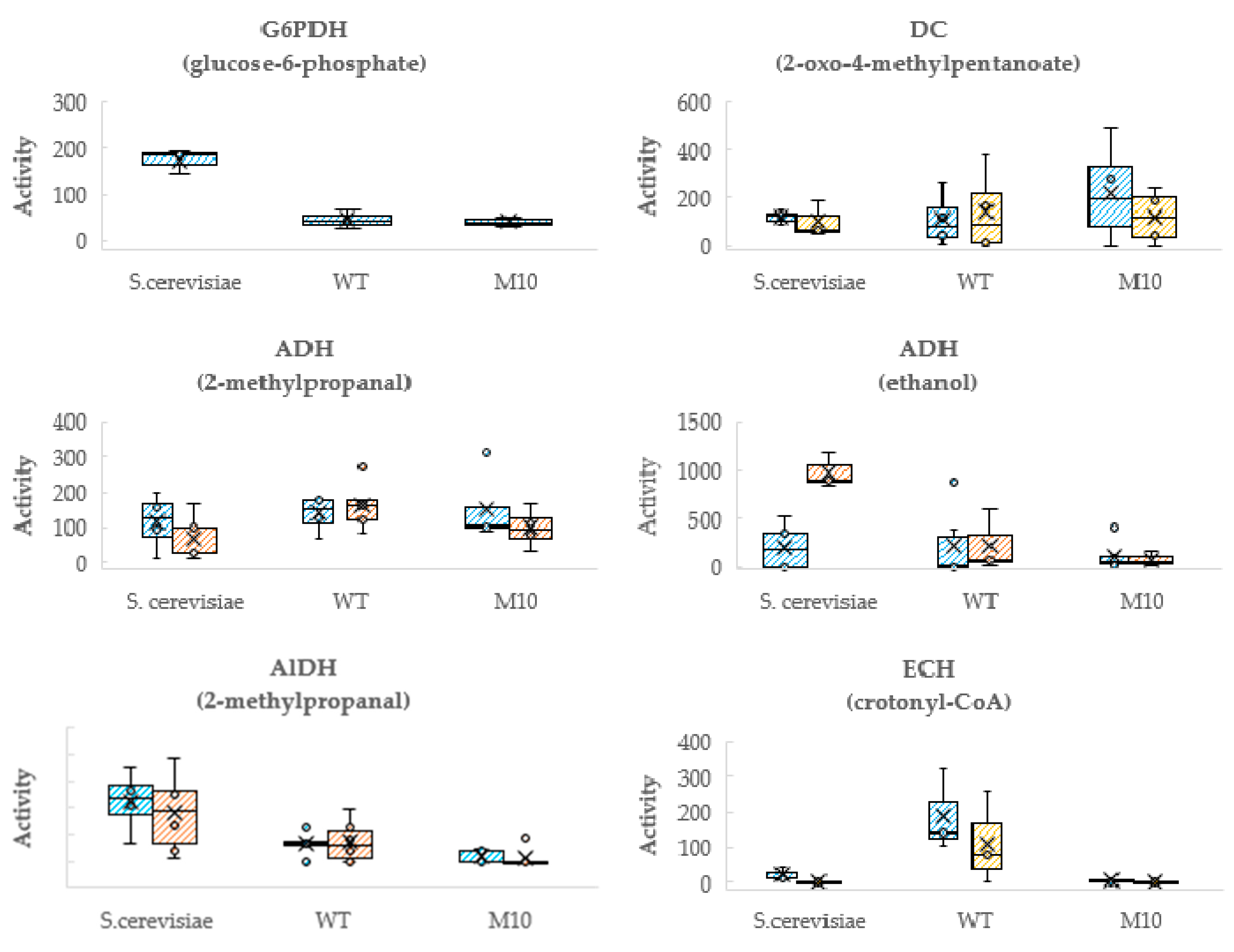
3.4. Loss of Enoyl-CoA Hydratase Activity in the β-Oxidation Pathway May Account for Higher VOCs Production in Mutant M10
4. Conclusions
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bogacz-Radomska, L.; Pietkiewicz, J.J.; Górska, K. Aroma Production and Application in Food Products. In Proceedings of the 35th International Conference of Slovak Society of Chemical Engineering, Vysoké Tatry, Slovakia, 26–30 May 2008. [Google Scholar]
- Hua, D.; Xu, P. Recent advances in biotechnological production of 2-phenylethanol. Biotechnol. Adv. 2011, 29, 654–660. [Google Scholar] [CrossRef]
- Saraiva, A.; Carrascosa, C.; Raheem, D.; Ramos, F.; Raposo, A. Natural Sweeteners: The Relevance of Food Naturalness for Consumers, Food Security Aspects, Sustainability and Health Impacts. IJERPH 2020, 17, 6285. [Google Scholar] [CrossRef]
- Chemat, F.; Abert Vian, M.; Fabiano-Tixier, A.-S.; Nutrizio, M.; Režek Jambrak, A.; Munekata, P.E.S.; Lorenzo, J.M.; Barba, F.J.; Binello, A.; Cravotto, G. A review of sustainable and intensified techniques for extraction of food and natural products. Green Chem. 2020, 22, 2325–2353. [Google Scholar] [CrossRef]
- Buzzini, P.; Martini, A.; Cappelli, F.; Pagnoni, U.M.; Davoli, P. A study on volatile organic compounds (VOCs) produced by tropical ascomycetous yeasts. Antonie Van Leeuwenhoek 2003, 84, 301–311. [Google Scholar] [CrossRef]
- Buzzini, P.; Gasparetti, C.; Turchetti, B.; Cramarossa, M.R.; Vaughan-Martini, A.; Martini, A.; Pagnoni, U.M.; Forti, L. Production of volatile organic compounds (VOCs) by yeasts isolated from the ascocarps of black (Tuber melanosporum Vitt.) and white (Tuber magnatum Pico) truffles. Arch. Microbiol. 2005, 184, 187–193. [Google Scholar] [CrossRef]
- Mdaini, N.; Gargouri, M.; Hammami, M.; Monser, L.; Hamdi, M. Production of natural fruity aroma by Geotrichum candidum. Appl. Biochem. Biotechnol. 2006, 128, 227–235. [Google Scholar] [CrossRef]
- Bhari, R.; Singh, R.S. Microbial Production of Natural Flavours. In Technology of Handling, Packaging, Processing, Preservation of Fruits and Vegetables: Theory and Practicals; New India Publishing Agency: New Dehli, India, 2019; pp. 767–813. [Google Scholar]
- Holt, S.; Mukherjee, V.; Lievens, B.; Verstrepen, K.J.; Thevelein, J.M. Bioflavoring by non-conventional yeasts in sequential beer fermentations. Food Microbiol. 2018, 72, 55–66. [Google Scholar] [CrossRef]
- Tufariello, M.; Fragasso, M.; Pico, J.; Panighel, A.; Castellarin, S.D.; Flamini, R.; Grieco, F. Influence of Non-Saccharomyces on Wine Chemistry: A Focus on Aroma-Related Compounds. Molecules 2021, 26, 644. [Google Scholar] [CrossRef] [PubMed]
- Grondin, E.; Shum Cheong Sing, A.; Caro, Y.; Raherimandimby, M.; Randrianierenana, A.L.; James, S.; Nueno-Palop, C.; François, J.M.; Petit, T. A comparative study on the potential of epiphytic yeasts isolated from tropical fruits to produce flavoring compounds. Int. J. Food Microbiol. 2015, 203, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C. Recognition of yeast speciesgene sequence comparisons. Open Appl. Inf. J. 2011, 5, 20–29. [Google Scholar] [CrossRef]
- Grondin, E.; Shum Cheong Sing, A.; James, S.; Nueno-Palop, C.; François, J.M.; Petit, T. Flavour production by Saprochaete and Geotrichum yeasts and their close relatives. Food Chem. 2017, 237, 677–684. [Google Scholar] [CrossRef]
- Grondin, E.; Shum Cheong Sing, A.; Caro, Y.; Billerbeck, G.M.; François, J.M.; Petit, T. Physiological and biochemical characteristics of the ethyl tiglate production pathway in the yeast Saprochaete suaveolens. Yeast 2015, 32, 57–66. [Google Scholar] [PubMed]
- Hazelwood, L.A.; Daran, J.-M.; van Maris, A.J.A.; Pronk, J.T.; Dickinson, J.R. The Ehrlich Pathway for Fusel Alcohol Production: A Century of Research on Saccharomyces cerevisiae Metabolism. Appl. Environ. Microbiol. 2008, 74, 3920. [Google Scholar] [CrossRef]
- Verstrepen, K.J.; Derdelinckx, G.; Dufour, J.-P.; Winderickx, J.; Thevelein, J.M.; Pretorius, I.S.; Delvaux, F.R. Flavor-active esters: Adding fruitiness to beer. J. Biosci. Bioeng. 2003, 96, 110–118. [Google Scholar] [CrossRef]
- Maggio-Hall, L.A.; Lyne, P.; Wolff, J.A.; Keller, N.P. A single acyl-CoA dehydrogenase is required for catabolism of isoleucine, valine and short-chain fatty acids in Aspergillus nidulans. Fungal Genet. Biol. 2008, 45, 180–189. [Google Scholar] [CrossRef]
- Liu, S.Q.; Holland, R.; Crow, V.L. Esters and their biosynthesis in fermented dairy products: A review. Int. Dairy J. 2004, 14, 923. [Google Scholar] [CrossRef]
- Saerens, S.M.G.; Verstrepen, K.J.; Van Laere, S.D.M.; Voet, A.R.D.; Van Dijck, P.; Delvaux, F.R.; Thevelein, J.M. The Saccharomyces cerevisiae EHT1 and EEB1 Genes Encode Novel Enzymes with Medium-chain Fatty Acid Ethyl Ester Synthesis and Hydrolysis Capacity. J. Biol. Chem. 2006, 281, 4446–4456. [Google Scholar] [CrossRef]
- Sun, W.; Wang, H. Recent advances of genome editing and related technologies in China. Gene Ther. 2020, 27, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Lichancová, H.; Hodorová, V.; Sienkiewicz, K.; Penir, S.M.U.; Afanasyev, P.; Boceck, D.; Bonnin, S.; Hakobyan, S.; Krawczyk, P.S.; Smyczynska, U.; et al. Genome Sequence of Flavor-Producing Yeast Saprochaete suaveolens NRRL Y-17571. Microbiol. Resour. Announc. 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Winston, F. EMS and UV Mutagenesis in Yeast. In Current Protocols in Molecular Biology; Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., Struhl, K., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2008; ISBN 978-0-471-14272-0. [Google Scholar]
- Conseil de l’Union Européenne. Directive 88/388/CEE du Conseil du 22 Juin 1988 Relative au Rapprochement des Législations des États Membres dans le Domaine des Arômes Destinés à être Employés dans les Denrées Alimentaires et des Matériaux de Base Pour Leur Production. Available online: http://op.europa.eu/en/publication-detail/-/publication/4f729209-04b9-42ac-a202-4487fad12882/language-fr/format-PDFA1B (accessed on 22 September 2020).
- Spicer, A.; Molnar, A. Gene Editing of Microalgae: Scientific Progress and Regulatory Challenges in Europe. Biology 2018, 7, 21. [Google Scholar] [CrossRef]
- Patston, P.A.; Espinal, J.; Beggs, M.; Randle, P.J. Assay of total complex and activity state of branched-chain alpha-keto acid dehydrogenase complex and of activator protein in mitochondria, cells, and tissues. In Methods in Enzymoloy, Branched-Chain Amino Acids; Elsevier Inc.: Amsterdam, The Netherlands, 1988; Volume 166, pp. 175–189. [Google Scholar]
- Park, S.J.; Lee, S.Y. Identification and Characterization of a New Enoyl Coenzyme A Hydratase Involved in Biosynthesis of Medium-Chain-Length Polyhydroxyalkanoates in Recombinant Escherichia coli. J. Bacteriol. 2003, 185, 5391–5397. [Google Scholar] [CrossRef]
- Gilham, D.; Lehner, R. Techniques to measure lipase and esterase activity in vitro. Methods 2005, 36, 139–147. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Capozzi, V.; Yener, S.; Khomenko, I.; Farneti, B.; Cappellin, L.; Gasperi, F.; Scampicchio, M.; Biasioli, F. PTR-ToF-MS Coupled with an Automated Sampling System and Tailored Data Analysis for Food Studies: Bioprocess Monitoring, Screening and Nose-space Analysis. J. Vis. Exp. 2017, 54075. [Google Scholar] [CrossRef] [PubMed]
- Fujii, T.; Nagasawa, N.; Iwamatsu, A.; Bogaki, T.; Tamai, Y.; Hamachi, M. Molecular cloning, sequence analysis, and expression of the yeast alcohol acetyltransferase gene. Appl. Environ. Microbiol. 1994, 60, 2786–2792. [Google Scholar] [CrossRef]
- Nagasawa, N.; Bogaki, T.; Iwamatsu, A.; Hamachi, M.; Kumagai, C. Cloning and nucleotide sequence of the alcohol acetyltransferase II gene (AFT2) from Saccharomyces cerevisiae kyokai no. 7. Biosci. Biotechnol. Biochem. 1998, 62, 1852–1857. [Google Scholar] [CrossRef]
- Knight, M.J.; Bull, I.D.; Curnow, P. The yeast enzyme Eht1 is an octanoyl-CoA: Ethanol acyltransferase that also functions as a thioesterase. Yeast 2014, 31, 463–474. [Google Scholar] [CrossRef]
- Saerens, S.M.; Delvaux, F.R.; Verstrepen, K.J.; Thevelein, J.M. Production and biological function of volatile esters in Saccharomyces cerevisiae. Microb. Biotechnol. 2010, 3, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-S.; Chuang, D.T. Regulation of Branched-Chain α-Keto Acid Dehydrogenase Kinase Gene Expression by Glucocorticoids in Hepatoma Cells and Rat Liver. Methods Enzymol. 2000, 324, 498–511. [Google Scholar]
- Gethins, L.; Guneser, O.; Demirkol, A.; Rea, M.C.; Stanton, C.; Ross, R.P.; Karagul Yuceer, Y.; Morrissey, J.P. Influence of Carbon and Nitrogen source on production of volatile fragrance and flavour metabolites by the yeast Kluyveromyces marxianus: Nutrient effects on volatiles in K. marxianus. Yeast 2014, 32, 67–76. [Google Scholar] [CrossRef] [PubMed]
- İşleten Hoşoğlu, M. Study of increase the production of flavor compounds by the yeast Kluyveromyces marxianus through optimization of carbon and nitrogen sources. Food Health 2018, 4, 112–123. [Google Scholar] [CrossRef]
- Carroll, A.L.; Desai, S.H.; Atsumi, S. Microbial production of scent and flavor compounds. Curr. Opin. Biotechnol. 2016, 37, 8–15. [Google Scholar] [CrossRef]
- Pinotti, T.; Carvalho, P.M.B.; Garcia, K.M.G.; Silva, T.R.; Hagler, A.N.; Leite, S.G.F. Media components and amino acid supplements influencing the production of fruity aroma by Geotrichum candidum. Braz. J. Microbiol. 2006, 37, 494–498. [Google Scholar] [CrossRef][Green Version]
- van Wyk, N.; Kroukamp, H.; Pretorius, I. The Smell of Synthetic Biology: Engineering Strategies for Aroma Compound Production in Yeast. Fermentation 2018, 4, 54. [Google Scholar] [CrossRef]




| WT | M2 | M3 | M4 | M5 | M6 | M7 | M8 | M9 | M10 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Total VOCs (mg.L−1) | 231 ± 14 | 66. ± 14 | 251 ± 38 | 42 ± 16 | 278 ± 40 | 126 ± 17 | 143 ± 104 | 204 ± 125 | 424 ± 171 | 1862 ± 247 |
| Number of total VOCs | 34 | 25 | 32 | 29 | 33 | 24 | 31 | 29 | 26 | 31 |
| WT | M2 | M3 | M4 | M5 | M6 | M7 | M8 | M9 | M10 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Ester from the EHRLICH PATHWAY | ||||||||||
| 2-methylbutyl 2-methylbutanoate | 25.8 ± 3.1 a | 13.2± 2.9 a | 3.4 ± 1.4 a | 4.6 ±3.6 a | 48.7 ± 25.5 a | 0.2 ± 0.4 a | 1.6 ± 1.3 a | 27.2 ± 46.0 a | 74.4 ± 32.5 a | 533.2 ± 113.0 a |
| 2-methylbutyl 3-methylbutanoate | 5.0 ± 0.3 a | 1,5±1,1 a | 2.0 ± 3.6 a | 0.4 ± 0.3 a | 23.7 ± 18.1 a,b | n.da | n.da | 4.0 ± 6.9 a | 7.0 ± 3.5 a | 55.0 ± 30.0 b |
| 2-methylbutyl acetate | 4.0 ± 0.8 a | 1.8±1.1 a | 3.3 ± 2.7 a | 0.4 ± 0.2 a | 7.8 ± 5.5 a | 6.6 ± 2.9 a | n.da | 2.3 ± 0.9 a | n.da | 42.1 ± 34.0 b |
| 2-methylbutyl butanoate | n.da | n.da | n.da | n.da | 0.3 ± 0.1 a | n.da | n.da | n.da | n.da | 0.9 ± 0.5 b |
| 2-methylbutyl propanoate | 1.0 ± 0.9 a | 0.1 ± 0.1 a | 0.2 ± 0.4 a | 0.1 ± 0.1 a | 0.4 ± 0.4 a | n.da | 0.1 ± 0.1 a | n.da | 4.9 ± 3.0 a,b | 6.2 ± 3.5 b |
| 2-methylpropyl 2-methylbutanoate | 20.2 ± 4.0 a | 5.3 ± 4.9 a | 16.7 ± 2.7 a | 4.4 ± 1.3 a | 10.6 ± 8.4 a | 3.6 ± 2.8 a | 48.8 ± 69.5 a,b | 32.7 ± 21.5 a | 37.0 ± 19.0 a | 140.5 ± 57.5 c |
| 2-methylpropyl 3-methylbutanoate | 2.30 ± 2.6 a | 1.0 ± 3.0 a | 2.2 ± 1.3 a | 0.2 ± 0.3 a | 1.0 ± 1.7 a | 0.1 ± 0.1 a | 5.0 ± 8.4 a | 2.6± 1.8 a | 1.2 ± 1.0 a | 17.1 ± 15.7 a |
| 2-methylpropyl acetate | 1.7 ± 0.6 a | 0.7 ± 0.9 a | 0.3 ± 0.1 a | 0.4 ±0.4 a | 4.1 ± 2.2 a,b | 0.7 ± 0.3 a | 0.2 ± 0.1 a | 0.6 ± 0.7 a | 0.7± 0.3 a | 12.0 ± 5.5 c |
| 2-methylpropyl butanoate | 0.1 ± 0.1 a | n.d a | n.d a | n.d a | n.d a | 0.1 ± 0.0 a | n.d a | 0.1 ± 0.0 a | n.d a | n.d a |
| 2-methylpropyl propanoate | 1.1 ± 0.6 a,b,c | 0.6 ± 0.4 a,b,c | 0.4 ± 0.1 a,b | 0.3 ± 0.2 a,b | 0.3 ± 0.0 a | n.d a | n.d a | 7.0 ±2.5 d | n.d a | 4.9 ± 2.5 c,d |
| 2-phenylethanol | 0.1 ± 0.1 a,b | n.d a | n.d a | n.d a | 0.1 ± 0.1 a,b | n.d a | 0.1 ± 0.1 a,b | n.d a | n.d a | n.d a |
| 2-phenylethyl acetate | 0.2 ± 0.1 a | 0.1 ± 0.1 a | 0.6 ± 0.8 a | 0.1 ± 0.1 a | 0.6 ± 0.2 a | 0.1 ± 0.2 a | 0.7 ± 1.1 a | 0.1 ± 0.2 a | 0.1 ± 0.1 a | 0.6 ± 0.7 a |
| 3-methylbutyl 2-methylbutanoate | 6.0 ± 3.0 a | 3.8 ± 2.2 a | 28.1 ± 25.1 a | 4.1 ± 4.0 a | 2.3 ± 1.9 a | 5.3 ± 5.3 a | 18.0 ± 3.6 a | 7.4 ± 9.4 a | 37.2 ± 40.3 a,b | 67.4 ± 34.0 a,b |
| 3-methylbutyl 2-methylpropanotae | 2.4 ± 1.2 a | 0.8 ± 0.7 a | 1.1 ± 1.5 a | 0.3 ± 0.2 a | 2.4 ± 0.3 a | 0.2 ± 0.0 a | 1.0 ± 0.9 a | 2.0 ± 1.9 a | 3.0 ± 1.4 a | 12.5 ± 9.3 b |
| 3-methylbutyl acetate | 1.2 ± 0.9 a | 0.5 ± 0.5 a | 0 ± 1.0 a | 1.2 ± 1.9 a | 2.0 ± 2.2 a | 0.3 ± 0.4 a | 1.4 ± 0.4 a | 0.8 ± 0.8 a | 5.3 ± 2.2 a,b | 17.4 ± 2.7 b |
| 3-methylbutyl propanoate | 0.4 ± 0.4 a | 0.1 ± 0.0 a | 0.1 ± 0.1 a | 0.1 ± 0.0 a | 0.1 ± 0.0 a | n.d a | 0.1 ± 0.0 a | 0.3 ± 0.2 a | 0.8 ± 0.9 a | n.d a |
| ethyl 2-methylbutanoate | 28.8 ± 10.6 a | 15.60 ± 6.4 a | 32.60 ± 5.7 a | 9.2 ± 3.5 a | 41.0 ± 5.7 a | 52.3 ± 10.5 a | 17.7 ± 7.0 a | 42.8 ± 19.9 a | 66.9 ± 31.4 a | 171.7± 21.7 b |
| ethyl 2-methylpropanoate | 2.4 ± 0.6 a | 0.4 ± 0.3 a | 0.4 ± 1.0 a | 0.4 ± 0.4 a | 0.9 ± 0.7 a | 0.6 ± 0.4 a | 1.5 ± 0.9 a | 1.0 ± 0.9 a | 1.7 ± 0.9 a | 17.6 ± 3.2 b |
| ethyl 3-methylbutanoate | 9.5 ± 0.7 a | 4.3 ± 1.9 a | 5.1 ± 0.1 a | 1.5 ± 0.9 a | 3.9 ± 3.8 a | 1.2 ± 0.1 a | 3.3 ± 3.0 a | 6.3 ± 4.1 a | 11.5 ± 6.4 a | 56.7 ± 26.0 b |
| ethyl 3-methylpentanoate | 0.1 ± 0.0 a | 0.1 ± 0.0 a | 0.1 ± 0.1 a | 0.1 ± 0.1 a | 0.3 ± 0.1 a | n.d a | 0.1 ± 0.1 a | 0.1 ± 0.2 a | n.d a | n.d a |
| phenethyl 2-methylbutanoate | 1.7 ± 2.0 a,b | 0.1 ± 0.1 a | 1.0 ± 1.3 a | 0.1 ± 0.1 a | 0.8 ± 0.5 a | 0.1 ± 0.0 a | 0.3 ± 0.2 a | 0.4 ± 0.5 a | 0.7 ± 0.3 a | 2.4± 2.4 a,b |
| propyl 2-methylbutanoate | 0.2 ± 0.1 a | n.d a | 0.4 ± 0.2 a | n.d a | 0.3 ± 0.0 a | 0.3 ± 0.0 a | 0.2 ± 0.1 a | 0.7 ± 0.2 a | 0.6 ± 1.0 a | 3.8 ± 3.1 a,b |
| Esters from the Acyl-CoA of BOP | ||||||||||
| ethyl octanoate | 16.9 ± 12.6 a | 3.1 ± 4.2 a | 9.0 ± 9.5 a | n.d a | 9.2 ± 9.8 a | n.d a | 3.9 ± 6.4 a | 5.6 ± 9.7 a | 11.4 ± 19.0 a | 54.5 ± 94.5 a |
| octyl 2-methylpropanoate | 0.7 ± 0.7 a,b | 0.1 ± 0.1 a,b | 0.5 ± 0.5 a,b | 0.1 ± 0.1 a | 0.7 ± 0.3 a,b | 0.2± 0.0 a,b | 0.3 ± 0.1 a,b | 0.5 ± 0.0 a,b | 1.7 ± 0.6 a,b | 1.2 ± 1.0 a,b |
| octyl acetate | 8.60 ± 8.9 a | 1.1 ± 0.5 a | 2.9 ± 2.4 a | 0.5 ± 0.5 a | 11.3 ± 6.4 a | 5.7 ± 5.4 a | 2.0 ± 1.6 a | 4.2 ± 4.6 a | 7.4 ± 2.4 a | 13.9 ± 10.6 a |
| ethyl butanoate | 0.4 ± 0.1 a | 0.2 ± 0.1 a | 44.9 ± 12.1 a | 0.11 ± 0.1 a | 0.6 ± 0.4 a | 0.3± 0.2 a | 0.7 ± 0.7 a | 2.2 ± 3.1 a | 0.4 ± 0.0 a | 4.2 ± 2.8 a |
| octyl butanoate | 9.2 ± 9.5 a,b | 2.2 ± 1.9 a | 9.4 ± 12.1 a,b | 1.2 ± 1.6 a | 15.7 ± 10.8 a,b | 2.5 ± 0.4 a | 6.2 ± 1.9 a | 6.2 ± 5.4 a | 31 ± 15.4 b | 5.4 ± 2.3 a |
| octyl propanoate | 0.1 ± 0.1 a | n.d a | n.d a | 0.1 ± 0.1 a | n.d a | n.d a | 0.1 ± 0.0 a | n.d a | 0.2 ± 0.3 a | 0.8 ± 0.7 a |
| Ester from the Enoyl-CoA of BOP | ||||||||||
| 2-methylbutyl 2-methylbut-2Z-enoate | n.d a | n.d a | 0.3 ± 0.20 a,b | 0.05 ± 0.06 a,b | 0.15 ± 0.01 a,b | n.d a | 0.1 ± 0.1 a,b | 0.3 ± 0.4 a,b | n.d a | n.d a |
| 2-methylpropyl 2-methylbut-2E-enoate | n.d a | 3.0 ± 4.9 a | 15.9 ± 2.9 a,b | 1.9 ± 2.4 a | 0.3 ± 0.5 a | 0.1 ± 0.1 a | 9.4 ± 4.8 a,b | 4.1 ± 5.1 a | 8.8 ± 5.4 a,b | 37.4 ± 20.0 c |
| 2-methylpropyl 2-methylbut-2Z-enoate | 1.1 ± 0.0 a | n.da | 0.2 ± 0.2 a | 0.1 ± 0.0 a | 0.3 ± 0.1 a,b | n.d a | 0.1 ± 0.1 a | 0.1 ± 0.0 a | n.d a | 1.1 ± 0.7 a,b |
| 3-methylbutyl 2-methylbut-2E-enoate | 0.2 ± 0.2 a | n.d b | n.d b | n.d b | 0.1 ± 0.1 a,b | n.d b | n.d b | n.d b | n.d b | n.d b |
| 3-methylbutyl 2-methylbut-2Z-enoate | 0.4 ± 0.5 a | n.d a | 0.1 ± 0.1 a | n.d a | 0.1 ± 0.1 a | n.d a | 0.1 ± 0.0 a | n.d a | 0. 2 ± 0.2 a | 0.6 ± 1.0 a |
| butyl 2-methylbut-2E-enoate | 28.5 ± 2.2 a | n.d b | 1.7 ± 1.4 b | 0.5 ± 0.7 b | 7.7 ± 8.5 a,b | 0.1 ± 0.1 b | 3.3 ± 4.8 b | 7.9 ± 9 a,b | 4.9 ± 3.1 a | 90.2 ± 10.5 c |
| ethyl 2-methylbut-2 E-enoate | 48.4 ± 3.0 a,b | n.d a | 36.0 ± 7.0 a,b | 8.3 ± 10.2 a | 5.5 ± 2.0 a | 1.5 ± 1.4 a | 3.5 ± 1.7 a | 34.1± 20.4 a,b | 32.0 ± 38.5 a,b | 115.9± 30.7 a,b |
| ethyl but-2E-enoate | 0.1 ± 0.1 a | 1.4 ± 1.4 a | 1.7 ± 1.9 a | 0.5 ± 0.7 a | n.d a | 36.6 ± 0.7 a,b | 0.1 ± 0.1 a | n.d a | n.d a | 155.8 ± 136.0 b |
| propyl 2-methylbut-2E-enoate | 0.1 ± 0.0 a | n.d a | 0.1 ± 0.1 a | n.d a | n.d a | n.d a | 0.1 ± 0.1 a | n.d a | n.d a | 1.0 ± 1.2 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, M.; Caro, Y.; Sing, A.S.C.; Reiss, H.; Francois, J.-M.; Petit, T. Selection by UV Mutagenesis and Physiological Characterization of Mutant Strains of the Yeast Saprochaete suaveolens (Former Geotrichum fragrans) with Higher Capacity to Produce Flavor Compounds. J. Fungi 2021, 7, 1031. https://doi.org/10.3390/jof7121031
Tan M, Caro Y, Sing ASC, Reiss H, Francois J-M, Petit T. Selection by UV Mutagenesis and Physiological Characterization of Mutant Strains of the Yeast Saprochaete suaveolens (Former Geotrichum fragrans) with Higher Capacity to Produce Flavor Compounds. Journal of Fungi. 2021; 7(12):1031. https://doi.org/10.3390/jof7121031
Chicago/Turabian StyleTan, Melissa, Yanis Caro, Alain Shum Cheong Sing, Héloïse Reiss, Jean-Marie Francois, and Thomas Petit. 2021. "Selection by UV Mutagenesis and Physiological Characterization of Mutant Strains of the Yeast Saprochaete suaveolens (Former Geotrichum fragrans) with Higher Capacity to Produce Flavor Compounds" Journal of Fungi 7, no. 12: 1031. https://doi.org/10.3390/jof7121031
APA StyleTan, M., Caro, Y., Sing, A. S. C., Reiss, H., Francois, J.-M., & Petit, T. (2021). Selection by UV Mutagenesis and Physiological Characterization of Mutant Strains of the Yeast Saprochaete suaveolens (Former Geotrichum fragrans) with Higher Capacity to Produce Flavor Compounds. Journal of Fungi, 7(12), 1031. https://doi.org/10.3390/jof7121031








