Assessment of Yeasts as Potential Probiotics: A Review of Gastrointestinal Tract Conditions and Investigation Methods
Abstract
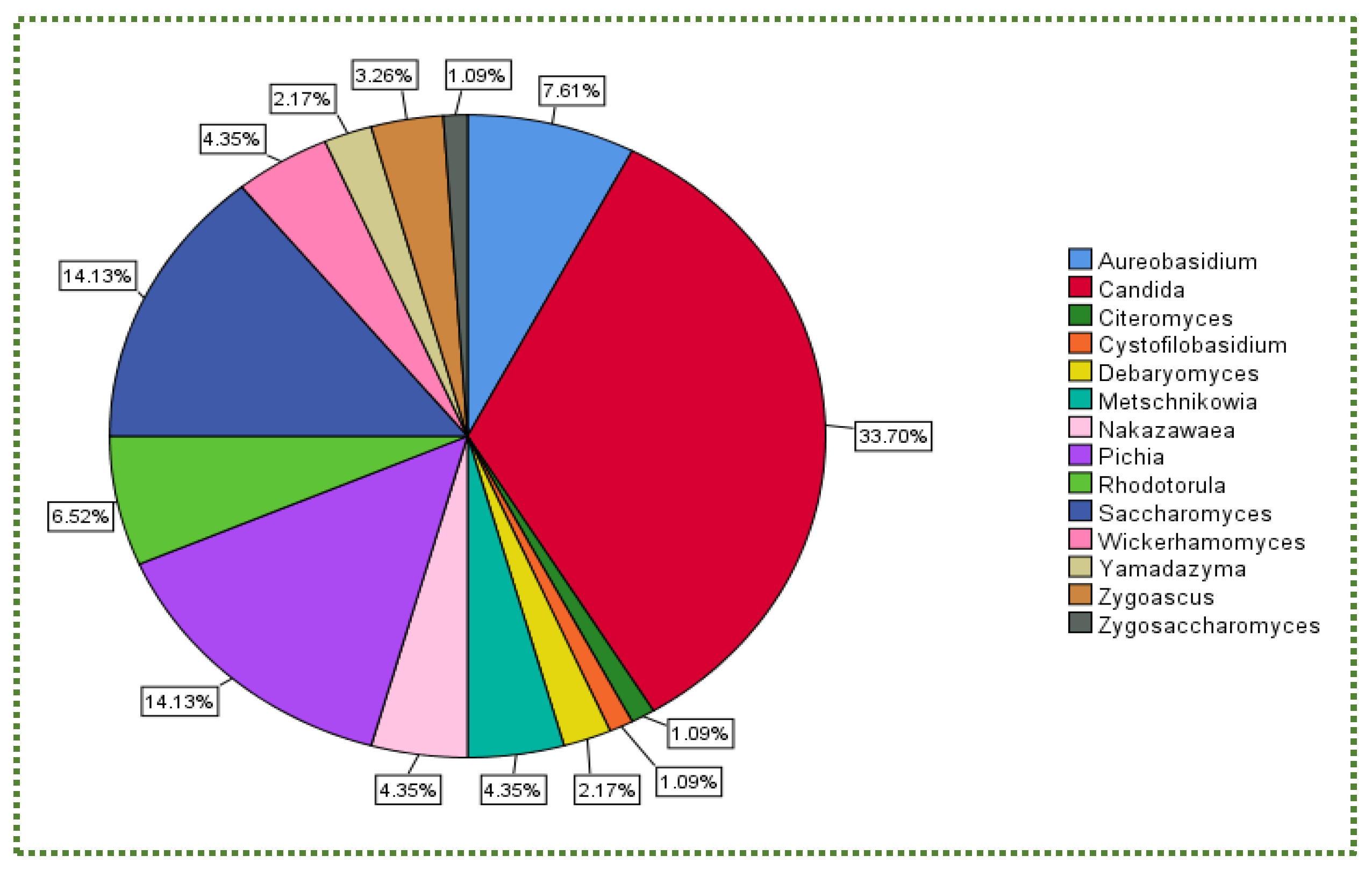
:1. Introduction
2. The Gastric Environment
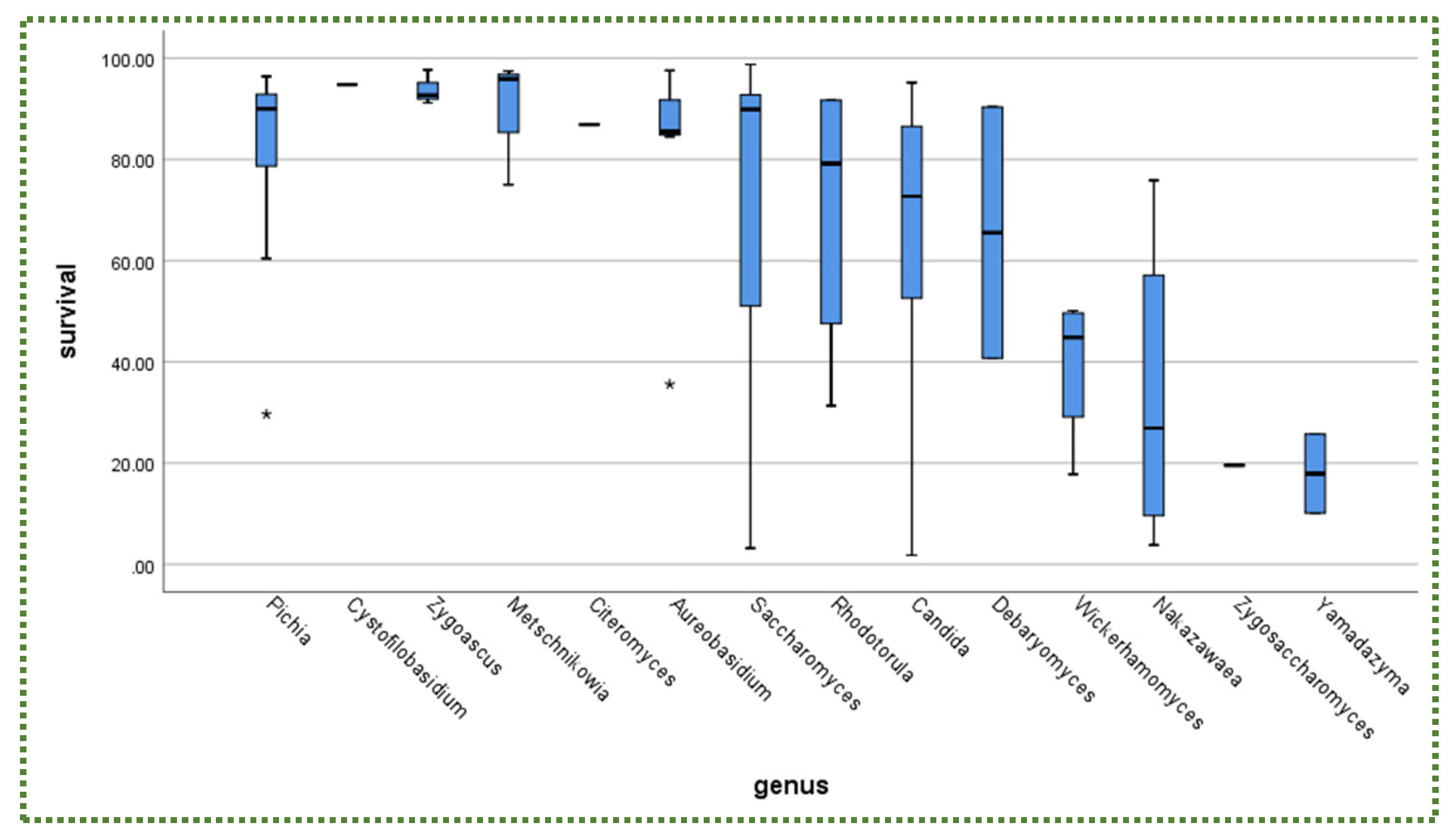
2.1. Tolerance to Gastric Conditions
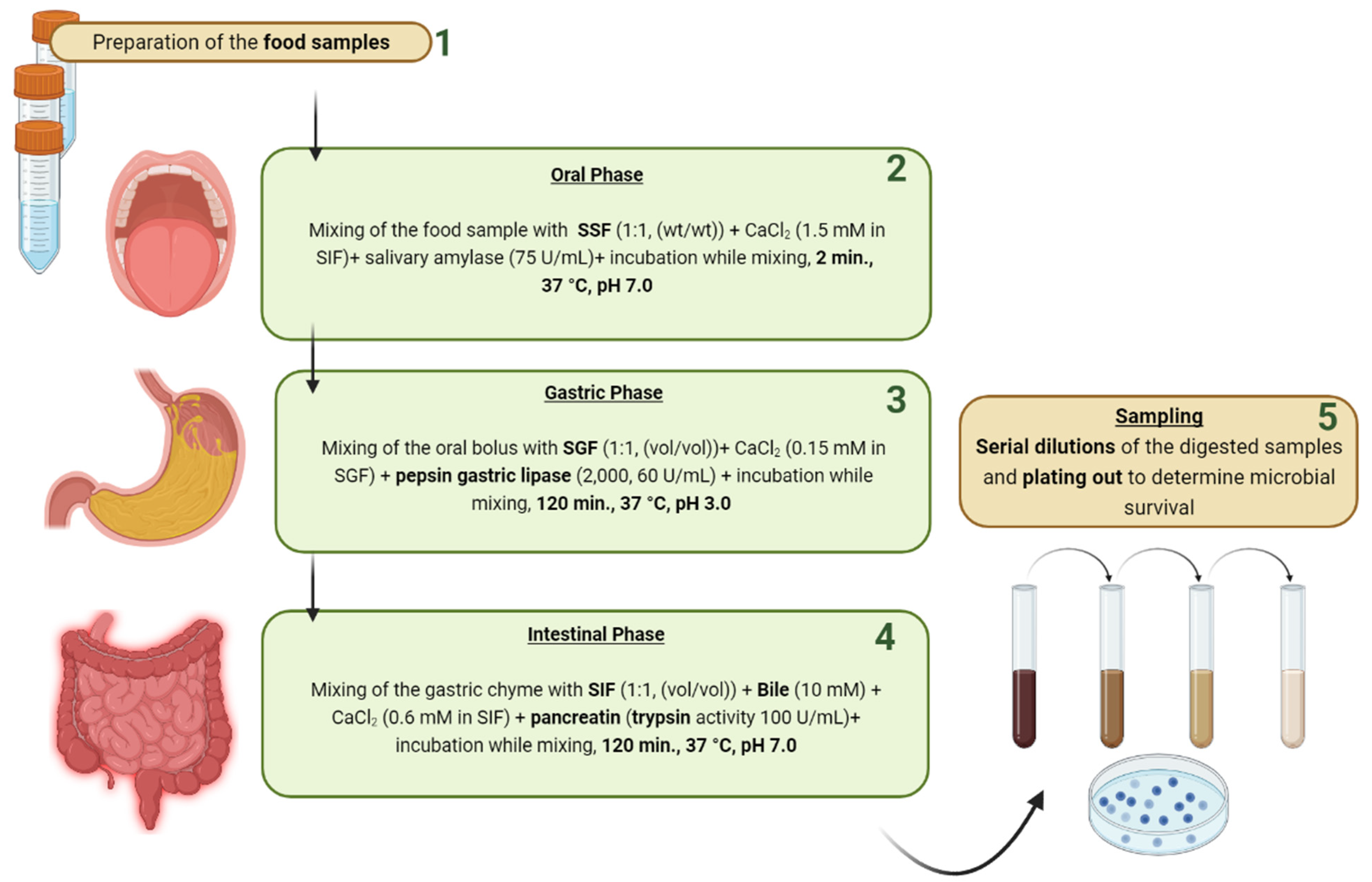
2.2. Assessment Methods of Gastric Tolerance
2.3. Mechanisms of Gastric Tolerance
3. Intestinal Conditions
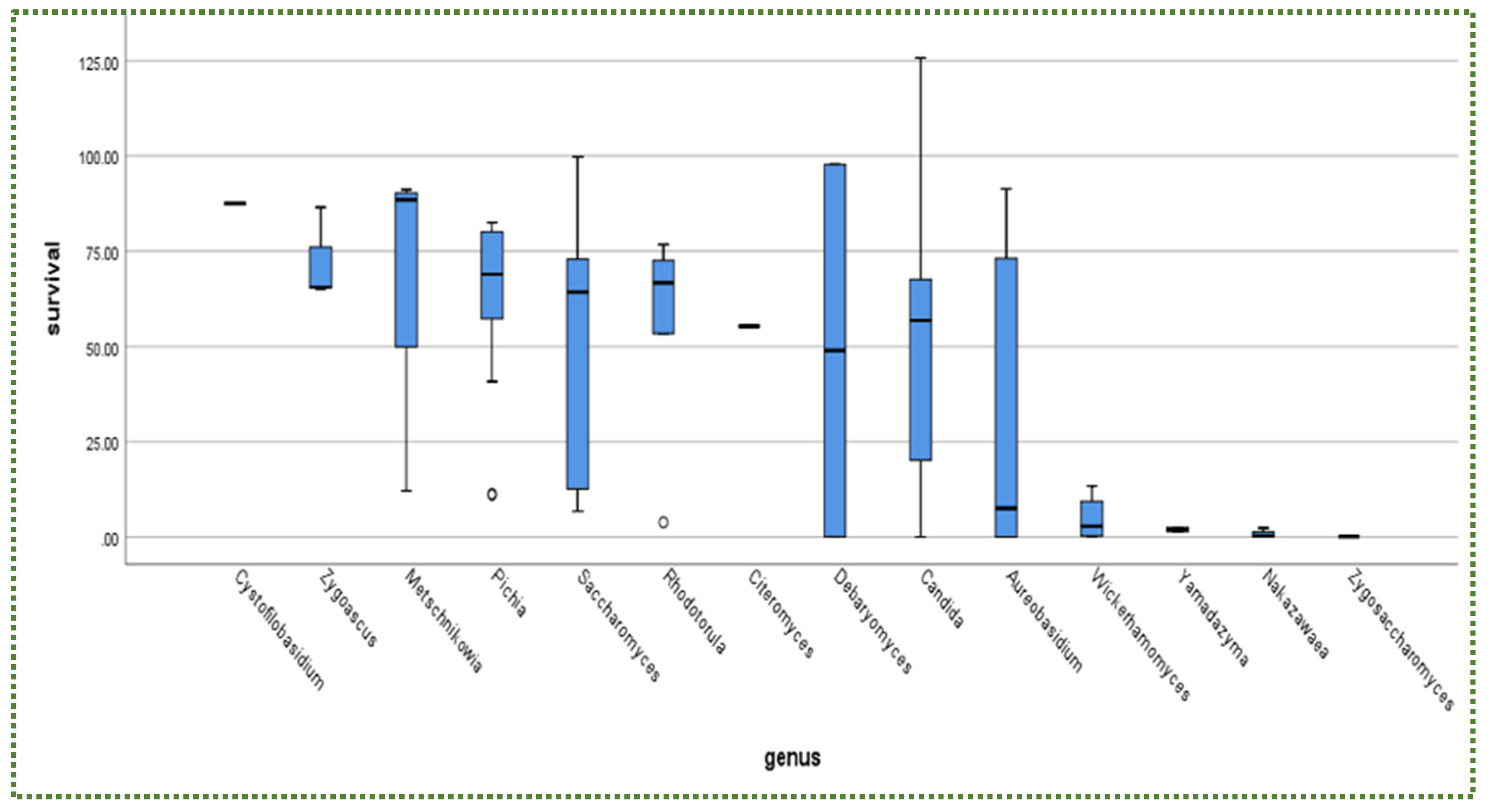
3.1. Tolerance to Intestinal Conditions
3.2. Assessment Methods of Intestinal Tolerance
3.3. Mechanisms of Intestinal Tolerance
4. Salt Conditions
4.1. Salt Stress
4.2. Assessment Methods of Salt Tolerance
4.3. Mechanisms of Salt Tolerance
5. Autoaggregation
5.1. Assessment Methods of Autoaggregation
5.2. Mode of Action of Autoaggregation
6. Hydrophobicity
6.1. Assessment Methods of Hydrophobicity
6.2. Mechanisms of Hydrophobicity
7. Insights and Future Recommendations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document: The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashaolu, T.J. Immune boosting functional foods and their mechanisms: A critical evaluation of probiotics and prebiotics. Biomed. Pharmacother. 2020, 130, 110625. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Al-Shabib, N.A. Functional Food Products and Sustainable Health; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar]
- Wang, X.; Han, M.; Zhang, M.; Wang, Y.; Ren, Y.; Yue, T.; Gao, Z. In vitro evaluation of the hypoglycemic properties of lactic acid bacteria and its fermentation adaptability in apple juice. LWT 2021, 136, 110363. [Google Scholar] [CrossRef]
- Dargahi, N.; Johnson, J.; Donkor, O.; Vasiljevic, T.; Apostolopoulos, V. Immunomodulatory effects of Streptococcus thermophilus on U937 monocyte cell cultures. J. Funct. Foods 2018, 49, 241–249. [Google Scholar] [CrossRef]
- Dargahi, N.; Johnson, J.; Apostolopoulos, V. Streptococcus thermophilus alters the expression of genes associated with innate and adaptive immunity in human peripheral blood mononuclear cells. PLoS ONE 2020, 15, e0228531. [Google Scholar] [CrossRef] [Green Version]
- Dargahi, N.; Matsoukas, J.; Apostolopoulos, V. Streptococcus thermophilus ST285 alters pro-inflammatory to anti-inflammatory cytokine secretion against multiple sclerosis peptide in mice. Brain Sci. 2020, 10, 126. [Google Scholar] [CrossRef] [Green Version]
- Dargahi, N.; Johnson, J.C.; Apostolopoulos, V. Immune Modulatory Effects of Probiotic Streptococcus thermophilus on Human Monocytes. Biologics 2021, 1, 396–415. [Google Scholar] [CrossRef]
- Dargahi, N.; Johnson, J.; Donkor, O.; Vasiljevic, T.; Apostolopoulos, V. Immunomodulatory effects of probiotics: Can they be used to treat allergies and autoimmune diseases? Maturitas 2019, 119, 25–38. [Google Scholar] [CrossRef]
- Lee, S.J.; Bose, S.; Seo, J.-G.; Chung, W.-S.; Lim, C.-Y.; Kim, H. The effects of co-administration of probiotics with herbal medicine on obesity, metabolic endotoxemia and dysbiosis: A randomized double-blind controlled clinical trial. Clin. Nutr. 2014, 33, 973–981. [Google Scholar] [CrossRef]
- Wong, R.K.; Yang, C.; Song, G.-H.; Wong, J.; Ho, K.-Y. Melatonin regulation as a possible mechanism for probiotic (VSL# 3) in irritable bowel syndrome: A randomized double-blinded placebo study. Dig. Dis. 2015, 60, 186–194. [Google Scholar]
- Gu, J.; Ahn-Jarvis, J.H.; Riedl, K.M.; Schwartz, S.J.; Clinton, S.K.; Vodovotz, Y. Characterization of black raspberry functional food products for cancer prevention human clinical trials. J. Agric. Food Chem. 2014, 62, 3997–4006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daba, G.M.; Elnahas, M.O.; Elkhateeb, W.A. Contributions of exopolysaccharides from lactic acid bacteria as biotechnological tools in food, pharmaceutical, and medical applications. Int. J. Biol. 2021, 173, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Vera-Pingitore, E.; Jimenez, M.E.; Dallagnol, A.; Belfiore, C.; Fontana, C.; Fontana, P.; von Wright, A.; Vignolo, G.; Plumed-Ferrer, C. Screening and characterization of potential probiotic and starter bacteria for plant fermentations. LWT-Food Sci. Technol. 2016, 71, 288–294. [Google Scholar] [CrossRef]
- FAO; WHO. Report of a Joint FAO/WHO Working Group on Drafting Guidelines for the Evaluation of Probiotics in Food; FAO: Rome, Lazio, Italy; WHO: Geneva, Switzerland, 2002. [Google Scholar]
- Czerucka, D.; Piche, T.; Rampal, P. Review article: Yeast as probiotics—Saccharomyces boulardii. Aliment. Pharmacol. Ther. 2007, 26, 767–778. [Google Scholar] [CrossRef]
- Silva, T.; Reto, M.; Sol, M.; Peito, A.; Peres, C.; Peres, C.; Malcata, F.X. Characterization of yeasts from Portuguese brined olives, with a focus on their potentially probiotic behavior. LWT-Food Sci. Technol. 2011, 44, 1349–1354. [Google Scholar] [CrossRef]
- Zahoor, F.; Sooklim, C.; Songdech, P.; Duangpakdee, O.; Soontorngun, N. Selection of potential yeast probiotics and a cell factory for xylitol or acid production from honeybee samples. Metabolites 2021, 11, 312. [Google Scholar] [CrossRef]
- Perricone, M.; Bevilacqua, A.; Corbo, M.R.; Sinigaglia, M. Technological characterization and probiotic traits of yeasts isolated from Altamura sourdough to select promising microorganisms as functional starter cultures for cereal-based products. Food Microbiol. 2014, 38, 26–35. [Google Scholar] [CrossRef]
- Greppi, A.; Saubade, F.; Botta, C.; Humblot, C.; Guyot, J.-P.; Cocolin, L. Potential probiotic Pichia kudriavzevii strains and their ability to enhance folate content of traditional cereal-based African fermented food. Food Microbiol. 2017, 62, 169–177. [Google Scholar] [CrossRef]
- Rai, A.K.; Appaiah, K.A. Application of native yeast from Garcinia (Garcinia xanthochumus) for the preparation of fermented beverage: Changes in biochemical and antioxidant properties. Food Biosci. 2014, 5, 101–107. [Google Scholar] [CrossRef]
- Amorim, J.C.; Piccoli, R.H.; Duarte, W.F. Probiotic potential of yeasts isolated from pineapple and their use in the elaboration of potentially functional fermented beverages. Food Res. Int. 2018, 107, 518–527. [Google Scholar] [CrossRef]
- Di Cagno, R.; Filannino, P.; Cantatore, V.; Polo, A.; Celano, G.; Martinovic, A.; Cavoski, I.; Gobbetti, M. Design of potential probiotic yeast starters tailored for making a cornelian cherry (Cornus mas L.) functional beverage. Int. J. Food Microbiol. 2020, 323, 108591. [Google Scholar] [CrossRef] [PubMed]
- Arévalo-Villena, M.; Fernandez-Pacheco, P.; Castillo, N.; Bevilacqua, A.; Pérez, A.B. Probiotic capability in yeasts: Set-up of a screening method. LWT 2018, 89, 657–665. [Google Scholar] [CrossRef]
- Gut, A.M.; Vasiljevic, T.; Yeager, T.; Donkor, O.N. Characterization of yeasts isolated from traditional kefir grains for potential probiotic properties. J. Funct. Foods 2019, 58, 56–66. [Google Scholar] [CrossRef]
- Ayyash, M.; Liu, S.-Q.; Al Mheiri, A.; Aldhaheri, M.; Raeisi, B.; Al-Nabulsi, A.; Osaili, T.; Olaimat, A. In vitro investigation of health-promoting benefits of fermented camel sausage by novel probiotic Lactobacillus plantarum: A comparative study with beef sausages. LWT 2019, 99, 346–354. [Google Scholar] [CrossRef]
- Ogunremi, O.R.; Agrawal, R.; Sanni, A.I. Development of cereal-based functional food using cereal-mix substrate fermented with probiotic strain—Pichia kudriavzevii OG32. Food Sci. Nutr. 2015, 3, 486–494. [Google Scholar] [CrossRef] [Green Version]
- Sarwar, A.; Aziz, T.; Al-Dalali, S.; Zhao, X.; Zhang, J.; Ud Din, J.; Chen, C.; Cao, Y.; Yang, Z. Physicochemical and microbiological properties of synbiotic yogurt made with probiotic yeast Saccharomyces boulardii in combination with inulin. Foods 2019, 8, 468. [Google Scholar] [CrossRef] [Green Version]
- Senkarcinova, B.; Graça Dias, I.A.; Nespor, J.; Branyik, T. Probiotic alcohol-free beer made with Saccharomyces cerevisiae var. boulardii. LWT 2019, 100, 362–367. [Google Scholar] [CrossRef]
- Rosa, C.; Peter, G. Biodiversity and Ecophysiology of Yeasts; Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Roto, S.M.; Rubinelli, P.M.; Ricke, S.C. An introduction to the avian gut microbiota and the effects of yeast-based prebiotic-type compounds as potential feed additives. Front. Vet. Sci. 2015, 2, 28. [Google Scholar] [CrossRef] [Green Version]
- Hassanein, S.M.; Soliman, N.K. Effect of Probiotic (Saccharomyces cerevisiae) Adding to Diets on Intestinal Microflora and Performance of Hy-Line Layers Hens. J. Am. Sci. 2010, 6, 159–169. [Google Scholar]
- Vohra, A.; Syal, P.; Madan, A. Probiotic yeasts in livestock sector. Anim. Feed Sci. Technol. 2016, 219, 31–47. [Google Scholar] [CrossRef]
- Marden, J.P.; Julien, C.; Monteils, V.; Auclair, E.; Moncoulon, R.; Bayourthe, C. How does live yeast differ from sodium bicarbonate to stabilize ruminal pH in high-yielding dairy cows? J. Dairy Sci. 2008, 91, 3528–3535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reyes-Becerril, M.; Tovar-Ramírez, D.; Ascencio-Valle, F.; Civera-Cerecedo, R.; Gracia-López, V.; Barbosa-Solomieu, V. Effects of dietary live yeast Debaryomyces hansenii on the immune and antioxidant system in juvenile leopard grouper Mycteroperca rosacea exposed to stress. Aquaculture 2008, 280, 39–44. [Google Scholar] [CrossRef]
- Andrighetto, C.; Psomas, E.; Tzanetakis, N.; Suzzi, G.; Lombardi, A. Randomly amplified polymorphic DNA (RAPD) PCR for the identification of yeasts isolated from dairy products. Lett. Appl. Microbiol. 2000, 30, 5–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Querol, A.; Ramon, D. The application of molecular techniques in wine microbiology. Trends Food Sci. Technol. 1996, 7, 73–78. [Google Scholar] [CrossRef]
- Liu, T.; Li, Y.; Sadiq, F.A.; Yang, H.; Gu, J.; Yuan, L.; Lee, Y.K.; He, G. Predominant yeasts in Chinese traditional sourdough and their influence on aroma formation in Chinese steamed bread. Food Chem. 2018, 242, 404–411. [Google Scholar] [CrossRef]
- Soll, D.R. The ins and outs of DNA fingerprinting the infectious fungi. Clin. Microbiol. Rev. 2000, 13, 332–370. [Google Scholar] [CrossRef]
- Chen, K.W.; Chen, Y.C.; Lin, Y.H.; Chou, H.H.; Li, S.Y. The molecular epidemiology of serial Candida tropicalis isolates from ICU patients as revealed by multilocus sequence typing and pulsed-field gel electrophoresis. Infect. Genet. Evol. 2009, 9, 912–920. [Google Scholar] [CrossRef]
- Arastehfar, A.; Boekhout, T.; Butler, G.; Buda De Cesare, G.; Dolk, E.; Gabaldón, T.; Hafez, A.; Hube, B.; Hagen, F.; Hovhannisyan, H.; et al. Recent trends in molecular diagnostics of yeast infections: From PCR to NGS. FEMS Microbiol. Rev. 2019, 43, 517–547. [Google Scholar] [CrossRef] [Green Version]
- Prillinger, H.; Molnár, O.; Eliskases-Lechner, F.; Lopandic, K. Phenotypic and genotypic identification of yeasts from cheese. Antonie van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 1999, 75, 267–283. [Google Scholar] [CrossRef]
- Kunyeit, L.; Anu-Appaiah, K.A.; Rao, R.P. Application of probiotic yeasts on candida species associated infection. J. Fungi 2020, 6, 189. [Google Scholar] [CrossRef]
- Saber, A.; Alipour, B.; Faghfoori, Z.; Yari Khosroushahi, A. Cellular and molecular effects of yeast probiotics on cancer. Crit. Rev. Microbiol. 2017, 43, 96–115. [Google Scholar] [CrossRef] [PubMed]
- Shurson, G.C. Yeast and yeast derivatives in feed additives and ingredients: Sources, characteristics, animal responses, and quantification methods. Anim. Feed Sci. Technol. 2018, 235, 60–76. [Google Scholar] [CrossRef]
- Liu, D.; Dhital, S.; Wu, P.; Chen, X.D.; Gidley, M.J. In Vitro Digestion of Apple Tissue Using a Dynamic Stomach Model: Grinding and Crushing Effects on Polyphenol Bioaccessibility. J. Agric. Food Chem. 2020, 68, 574–583. [Google Scholar] [CrossRef] [PubMed]
- Camilleri, M. Integrated Upper Gastrointestinal Response to Food Intake. Gastroenterology 2006, 131, 640–658. [Google Scholar] [CrossRef] [PubMed]
- Welcome, M.O. Chemical Digestion, Absorption, and Transport. In Gastrointestinal Physiology; Springer: Berlin/Heidelberg, Germany, 2018; pp. 871–972. [Google Scholar]
- Meyer, J.H.; Ohashi, H.; Jehn, D.; Thomson, J.B. Size of liver particles emptied from the human stomach. Gastroenterology 1981, 80, 1489–1496. [Google Scholar] [CrossRef]
- Carlson, B.M. Chapter 12—The Digestive System. In The Human Body; Carlson, B.M., Ed.; Academic Press: Cambridge, MA, USA, 2019; pp. 321–355. [Google Scholar]
- Herdt, T.H. 29—Secretions of the Gastrointestinal Tract. In Cunningham’s Textbook of Veterinary Physiology, 6th ed.; Klein, B.G., Ed.; W.B. Saunders: St. Louis, MO, USA, 2020; pp. 307–315. [Google Scholar]
- Schubert, M.L. Gastric secretion. Curr. Opin. Gastroenterol. 2003, 19, 519–525. [Google Scholar] [CrossRef]
- Schubert, M.L. Physiologic, pathophysiologic, and pharmacologic regulation of gastric acid secretion. Curr. Opin. Gastroenterol. 2017, 33, 430–438. [Google Scholar] [CrossRef]
- Berin, M.C.; Furuta, G.T.; Aceves, S.S. 67—Gastrointestinal Mucosal Immunology. In Middleton’s Allergy, 8th ed.; Adkinson, N.F., Bochner, B.S., Burks, A.W., Busse, W.W., Holgate, S.T., Lemanske, R.F., O’Hehir, R.E., Eds.; W.B. Saunders: London, UK, 2014; pp. 1084–1094. [Google Scholar]
- da Silva Guedes, J.; Pimentel, T.C.; Diniz-Silva, H.T.; Tayse da Cruz Almeida, E.; Tavares, J.F.; Leite de Souza, E.; Garcia, E.F.; Magnani, M. Protective effects of β-glucan extracted from spent brewer yeast during freeze-drying, storage and exposure to simulated gastrointestinal conditions of probiotic lactobacilli. LWT 2019, 116, 108496. [Google Scholar] [CrossRef]
- Rui, Y.; Wan, P.; Chen, G.; Xie, M.; Sun, Y.; Zeng, X.; Liu, Z. Simulated digestion and fermentation in vitro by human gut microbiota of intra- and extra-cellular polysaccharides from Aspergillus cristatus. LWT 2019, 116, 108508. [Google Scholar] [CrossRef]
- Sekova, V.Y.; Dergacheva, D.I.; Tereshina, V.M.; Isakova, E.P.; Deryabina, Y.I. Carbohydrate Spectrum of Extremophilic Yeasts Yarrowia lipolytica under pH Stress. Microbiology 2018, 87, 173–182. [Google Scholar] [CrossRef]
- Guerzoni, M.E.; Serrazanetti, D.I.; Vernocchi, P.; Gianotti, A. Physiology and biochemistry of sourdough yeasts. In Handbook on Sourdough Biotechnology; Springer: Boston, MA, USA, 2013; pp. 155–181. [Google Scholar]
- Orij, R.; Brul, S.; Smits, G.J. Intracellular pH is a tightly controlled signal in yeast. Biochim. Biophys. Acta Gen. Subj. 2011, 1810, 933–944. [Google Scholar] [CrossRef] [PubMed]
- Horák, J. Yeast nutrient transporters. Biochim. Biophys. Acta Int. J. Biochem. Biophys. 1997, 1331, 41–79. [Google Scholar] [CrossRef]
- Guan, N.; Liu, L. Microbial response to acid stress: Mechanisms and applications. Appl. Microbiol. Biotechnol. 2020, 104, 51–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaichumjai, P.; Valyasevi, R.; Assavanig, A.; Kurdi, P. Isolation and characterization of acid-sensitive Lactobacillus plantarum with application as starter culture for Nham production. Food Microbiol. 2010, 27, 741–748. [Google Scholar] [CrossRef] [PubMed]
- Estruch, F. Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol. Rev. 2000, 24, 469–486. [Google Scholar] [CrossRef]
- Viljoen, B.C. Yeast Ecological Interactions. Yeast’Yeast, Yeast’Bacteria, Yeast’Fungi Interactions and Yeasts as Biocontrol Agents. In Yeasts in Food and Beverages; Querol, A., Fleet, G., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 83–110. [Google Scholar]
- Bonatsou, S.; Karamouza, M.; Zoumpopoulou, G.; Mavrogonatou, E.; Kletsas, D.; Papadimitriou, K.; Tsakalidou, E.; Nychas, G.J.E.; Panagou, E.Ζ. Evaluating the probiotic potential and technological characteristics of yeasts implicated in cv. Kalamata natural black olive fermentation. Int. J. Food Microbiol. 2018, 271, 48–59. [Google Scholar] [CrossRef]
- Zullo, B.A.; Ciafardini, G. Evaluation of physiological properties of yeast strains isolated from olive oil and their in vitro probiotic trait. Food Microbiol. 2019, 78, 179–187. [Google Scholar] [CrossRef]
- Palla, M.; Agnolucci, M.; Calzone, A.; Giovannetti, M.; Di Cagno, R.; Gobbetti, M.; Rizzello, C.G.; Pontonio, E. Exploitation of autochthonous Tuscan sourdough yeasts as potential starters. Int. J. Food Microbiol. 2019, 302, 59–68. [Google Scholar] [CrossRef]
- Puppala, K.R.; Ravi Kumar, V.; Khire, J.; Dharne, M. Dephytinizing and Probiotic Potentials of Saccharomyces cerevisiae (NCIM 3662) Strain for Amelioration of Nutritional Quality of Functional Foods. Probiotics Antimicrob. Proteins 2019, 11, 604–617. [Google Scholar] [CrossRef]
- Ragavan, M.L.; Das, N. In Vitro Studies on Therapeutic Potential of Probiotic Yeasts Isolated from Various Sources. Curr. Microbiol. 2020, 77, 2821–2830. [Google Scholar] [CrossRef]
- Porru, C.; Rodríguez-Gómez, F.; Benítez-Cabello, A.; Jiménez-Díaz, R.; Zara, G.; Budroni, M.; Mannazzu, I.; Arroyo-López, F.N. Genotyping, identification and multifunctional features of yeasts associated to Bosana naturally black table olive fermentations. Food Microbiol. 2018, 69, 33–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du Toit, M.; Franz, C.M.A.P.; Dicks, L.M.T.; Schillinger, U.; Haberer, P.; Warlies, B.; Ahrens, F.; Holzapfel, W.H. Characterisation and selection of probiotic lactobacilli for a preliminary minipig feeding trial and their effect on serum cholesterol levels, faeces pH and faeces moisture content. Int. J. Food Microbiol. 1998, 40, 93–104. [Google Scholar] [CrossRef]
- Kenfack, C.H.M.; Kaktcham, P.M.; Ngoufack, F.Z.; Wang, Y.R.; Yin, L.; Zhu, T. Screening and Characterization of Putative Probiotic Lactobacillus Strains from Honey Bee Gut (Apis mellifera). J. Adv. Microbiol. 2018, 10, 1–18. [Google Scholar] [CrossRef]
- Oliveira, T.; Ramalhosa, E.; Nunes, L.; Pereira, J.A.; Colla, E.; Pereira, E.L. Probiotic potential of indigenous yeasts isolated during the fermentation of table olives from Northeast of Portugal. Innov. Food Sci. Emerg. Technol. 2017, 44, 167–172. [Google Scholar] [CrossRef] [Green Version]
- Bonatsou, S.; Benitez, A.; Rodriguez-Gomez, F.; Panagou, E.Z.; Arroyo-Lopez, F.N. Selection of yeasts with multifunctional features for application as starters in natural black table olive processing. Food Microbiol. 2015, 46, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Kapteyn, J.; Ter Riet, B.; Vink, E.; Blad, S.; De Nobel, H.; Van Den Ende, H.; Klis, F.M. Low external pH induces HOG1-dependent changes in the organization of the Saccharomyces cerevisiae cell wall. Mol. Microbiol. 2001, 39, 469–480. [Google Scholar] [CrossRef] [Green Version]
- Lucena, R.M.; Dolz-Edo, L.; Brul, S.; de Morais, M.A.; Smits, G. Extreme Low Cytosolic pH Is a Signal for Cell Survival in Acid Stressed Yeast. Genes 2020, 11, 656. [Google Scholar] [CrossRef]
- Chen, A.K.-L.; Gelling, C.; Rogers, P.L.; Dawes, I.W.; Rosche, B. Response of Saccharomyces cerevisiae to stress-free acidification. J. Microbiol. 2009, 47, 1–8. [Google Scholar] [CrossRef]
- de Lucena, R.M.; Elsztein, C.; de Barros Pita, W.; de Souza, R.B.; Júnior, S.d.S.L.P.; de Morais Junior, M.A. Transcriptomic response of Saccharomyces cerevisiae for its adaptation to sulphuric acid-induced stress. Antonie Leeuwenhoek 2015, 108, 1147–1160. [Google Scholar] [CrossRef]
- Levin, D.E. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: The cell wall integrity signaling pathway. Genetics 2011, 189, 1145–1175. [Google Scholar] [CrossRef] [Green Version]
- De Melo, H.; Bonini, B.; Thevelein, J.; Simões, D.; Morais, M.A., Jr. Physiological and molecular analysis of the stress response of Saccharomyces cerevisiae imposed by strong inorganic acid with implication to industrial fermentations. J. Appl. Microbiol. 2010, 109, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Brandão, R.L.; Rosa, J.C.C.; Nicoli, J.R.; Almeida, M.V.S.; do Carmo, A.P.; Queiros, H.T.; Castro, I.M. Investigating acid stress response in different Saccharomyces strains. J. Mycol. 2014, 2014, 178274. [Google Scholar]
- Claret, S.; Gatti, X.; Doignon, F.; Thoraval, D.; Crouzet, M. The Rgd1p Rho GTPase-activating protein and the Mid2p cell wall sensor are required at low pH for protein kinase C pathway activation and cell survival in Saccharomyces cerevisiae. Eukaryot. Cell 2005, 4, 1375–1386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Lucena, R.; Elsztein, C.; Simoes, D.; de Morais, M.A., Jr. Participation of CWI, HOG and Calcineurin pathways in the tolerance of Saccharomyces cerevisiae to low pH by inorganic acid. J. Appl. Microbiol. 2012, 113, 629–640. [Google Scholar] [CrossRef]
- Lin, X.; Qi, Y.; Yan, D.; Liu, H.; Chen, X.; Liu, L. CgMED3 changes membrane sterol composition to help Candida glabrata tolerate low-pH stress. Appl. Environ. Microbiol. 2017, 83, e00972-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fletcher, E.; Feizi, A.; Bisschops, M.M.M.; Hallström, B.M.; Khoomrung, S.; Siewers, V.; Nielsen, J. Evolutionary engineering reveals divergent paths when yeast is adapted to different acidic environments. Metab. Eng. 2017, 39, 19–28. [Google Scholar] [CrossRef]
- Abbott, A.M.; Armstrong, L.; Jensen, E.H. Chapter 68—Small Intestine. In Shackelford’s Surgery of the Alimentary Tract, 7th ed.; Yeo, C.J., Ed.; W.B. Saunders: Philadelphia, PA, USA, 2013; pp. 839–863. [Google Scholar]
- Kiela, P.R.; Ghishan, F.K. Physiology of intestinal absorption and secretion. Best Pract. Res. Clin. Gastroenterol. 2016, 30, 145–159. [Google Scholar] [CrossRef] [Green Version]
- Silk, D.B. Digestion and absorption of carbohydrate protein and fat. Contemp. Issues Clin. Biochem. 1986, 4, 7–40. [Google Scholar]
- Alexander, S.P.H. Diacylglycerol Lipase (DAG Lipase). In xPharm: The Comprehensive Pharmacology Reference; Enna, S.J., Bylund, D.B., Eds.; Elsevier: New York, NY, USA, 2009; pp. 1–6. [Google Scholar]
- Chen, J.-M.; Radisky, E.S.; Férec, C. Chapter 576—Human Trypsins. In Handbook of Proteolytic Enzymes, 3rd ed.; Rawlings, N.D., Salvesen, G., Eds.; Academic Press: Cambridge, MA, USA, 2013; pp. 2600–2609. [Google Scholar]
- Semrin, M.G.; Russo, M.A. Anatomy, histology, embryology, and developmental anomalies of the stomach and duodenum. In Sleisenger and Fordtran’s Gastrointestinal and Liver Disease; Elsevier: Amsterdam, The Netherlands, 2010; pp. 773–788.e2. [Google Scholar]
- Hofmann, A.F. Bile acid secretion, bile flow and biliary lipid secretion in humans. Hepatology 1990, 12, 17S–25S. [Google Scholar]
- Drury, D.R.; McMaster, P.D.; Rous, P. Observations on some causes of gall stone formation. III. The relation of the reaction of the bile to experimental cholelitiasis. J. Exp. Med. 1924, 39, 403–424. [Google Scholar] [CrossRef]
- Maldonado-Valderrama, J.; Wilde, P.; MacIerzanka, A.; MacKie, A. The role of bile salts in digestion. Adv. Colloid Interface Sci. 2011, 165, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Pais, P.; Almeida, V.; Yılmaz, M.; Teixeira, M.C. Saccharomyces boulardii: What makes it tick as successful probiotic? J. Fungi 2020, 6, 78. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Ballester, M.; Herrero-Cervera, A.; Vinué, Á.; Martínez-Hervás, S.; González-Navarro, H. Impact of cholesterol metabolism in immune cell function and atherosclerosis. Nutrients 2020, 12, 2021. [Google Scholar] [CrossRef]
- Urdaneta, V.; Casadesús, J. Interactions between bacteria and bile salts in the gastrointestinal and hepatobiliary tracts. Front. Med. 2017, 4, 163. [Google Scholar] [CrossRef] [PubMed]
- Merchán, A.V.; Benito, M.J.; Galván, A.I.; Ruiz-Moyano Seco de Herrera, S. Identification and selection of yeast with functional properties for future application in soft paste cheese. LWT 2020, 124, 109173. [Google Scholar] [CrossRef]
- Canadell, D.; García-Martínez, J.; Alepuz, P.; Pérez-Ortín, J.E.; Ariño, J. Impact of high pH stress on yeast gene expression: A comprehensive analysis of mRNA turnover during stress responses. Biochim. Biophys. Acta Gene Regul. Mech. 2015, 1849, 653–664. [Google Scholar] [CrossRef]
- Tominaga, A.; Higuchi, Y.; Mori, H.; Akai, M.; Suyama, A.; Yamada, N.; Takegawa, K. Catechol O-methyltransferase homologs in Schizosaccharomyces pombe are response factors to alkaline and salt stress. Appl. Microbiol. Biotechnol. 2019, 103, 4881–4887. [Google Scholar] [CrossRef]
- Zvyagilskaya, R.; Parchomenko, O.; Abramova, N.; Allard, P.; Panaretakis, T.; Pattison-Granberg, J.; Persson, B.L. Proton-and sodium-coupled phosphate transport systems and energy status of Yarrowia lipolytica cells grown in acidic and alkaline conditions. J. Membr. Biol. 2001, 183, 39–50. [Google Scholar] [CrossRef]
- Peñalva, M.A.; Arst, H.N., Jr. Recent advances in the characterization of ambient pH regulation of gene expression in filamentous fungi and yeasts. Annu. Rev. Microbiol. 2004, 58, 425–451. [Google Scholar] [CrossRef]
- Causton, H.C.; Ren, B.; Sang Seok, K.; Harbison, C.T.; Kanin, E.; Jennings, E.G.; Tong Ihn, L.; True, H.L.; Lander, E.S.; Young, R.A. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 2001, 12, 323–337. [Google Scholar] [CrossRef] [Green Version]
- Serrano, R.; Ruiz, A.; Bernal, D.; Chambers, J.R.; Ariño, J. The transcriptional response to alkaline pH in Saccharomyces cerevisiae: Evidence for calcium-mediated signalling. Mol. Microbiol. 2002, 46, 1319–1333. [Google Scholar] [CrossRef] [PubMed]
- Serrano, R.; Martín, H.; Casamayor, A.; Ariño, J. Signaling alkaline pH stress in the yeast Saccharomyces cerevisiae through the Wsc1 cell surface sensor and the Slt2 MAPK pathway. J. Biol. Chem. 2006, 281, 39785–39795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skoneczny, M.; Skoneczna, A. Response mechanisms to chemical and physical stresses in yeast and filamentous fungi. In Stress Response Mechanisms in Fungi: Theoretical and Practical Aspects; Springer: Cham, Switzerland, 2018; pp. 35–85. [Google Scholar]
- Casamayor, A.; Serrano, R.; Platara, M.; Casado, C.; Ruiz, A.; Ariño, J. The role of the Snf1 kinase in the adaptive response of Saccharomyces cerevisiae to alkaline pH stress. Biochem. J. 2012, 444, 39–49. [Google Scholar] [CrossRef] [Green Version]
- Serra-Cardona, A.; Petrezsélyová, S.; Canadell, D.; Ramos, J.; Ariño, J. Coregulated expression of the Na+/phosphate pho89 transporter and ena1 Na+-ATPase allows their functional coupling under high-pH stress. Mol. Cell. Biol. 2014, 34, 4420–4435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Higuchi, Y.; Mori, H.; Kubota, T.; Takegawa, K. Analysis of ambient pH stress response mediated by iron and copper intake in Schizosaccharomyces pombe. J. Biosci. Bioeng. 2018, 125, 92–96. [Google Scholar] [CrossRef]
- Sen, S.; Mansell, T.J. Yeasts as probiotics: Mechanisms, outcomes, and future potential. Fungal Genet. Biol. 2020, 137, 103333. [Google Scholar] [CrossRef]
- Helmy, E.; Soliman, S.; Abdel-Ghany, T.M.; Ganash, M. Evaluation of potentially probiotic attributes of certain dairy yeast isolated from buffalo sweetened Karish cheese. Heliyon 2019, 5, e01649. [Google Scholar] [CrossRef] [Green Version]
- Brodkorb, A.; Egger, L.; Alminger, M.; Alvito, P.; Assunção, R.; Ballance, S.; Bohn, T.; Bourlieu-Lacanal, C.; Boutrou, R.; Carrière, F.; et al. INFOGEST static in vitro simulation of gastrointestinal food digestion. Nat. Protoc. 2019, 14, 991–1014. [Google Scholar] [CrossRef]
- Ayyash, M.; Abdalla, A.; Alhammadi, A.; Senaka Ranadheera, C.; Affan Baig, M.; Al-Ramadi, B.; Chen, G.; Kamal-Eldin, A.; Huppertz, T. Probiotic survival, biological functionality and untargeted metabolomics of the bioaccessible compounds in fermented camel and bovine milk after in vitro digestion. Food Chem. 2021, 363, 130243. [Google Scholar] [CrossRef]
- Ta, L.P.; Bujna, E.; Antal, O.; Ladányi, M.; Juhász, R.; Szécsi, A.; Kun, S.; Sudheer, S.; Gupta, V.K.; Nguyen, Q.D. Effects of various polysaccharides (alginate, carrageenan, gums, chitosan) and their combination with prebiotic saccharides (resistant starch, lactosucrose, lactulose) on the encapsulation of probiotic bacteria Lactobacillus casei 01 strain. Int. J. Biol. Macromol. 2021, 183, 1136–1144. [Google Scholar] [CrossRef]
- Gomez-Mascaraque, L.G.; Morfin, R.C.; Pérez-Masiá, R.; Sanchez, G.; Lopez-Rubio, A. Optimization of electrospraying conditions for the microencapsulation of probiotics and evaluation of their resistance during storage and in-vitro digestion. LWT-Food Sci. Technol. 2016, 69, 438–446. [Google Scholar] [CrossRef] [Green Version]
- FAO; WHO. Report of a Joint FAO/WHO Expert Consultation on Guidelines for the Evaluation of Probiotics in Food; FAO: Rome, Lazio, Italy; WHO: Geneva, Switzerland, 2002. [Google Scholar]
- Begley, M.; Hill, C.; Gahan, C.G.M. Bile salt hydrolase activity in probiotics. Appl. Environ. Microbiol. 2006, 72, 1729–1738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Begley, M.; Gahan, C.G.M.; Hill, C. The interaction between bacteria and bile. FEMS Microbiol. Rev. 2005, 29, 625–651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elkins, C.A.; Moser, S.A.; Savage, D.C. Genes encoding bile salt hydrolases and conjugated bile salt transporters in Lactobacillus johnsonii 100-100 and other Lactobacillus species. Microbiology 2001, 147, 3403–3412. [Google Scholar] [CrossRef] [Green Version]
- Martins, F.S.; Miranda, I.C.; Rosa, C.A.; Nicoli, J.R.; Neves, M.J. Effect of the trehalose levels on the screening of yeast as probiotic by in vivo and in vitro assays. Braz. J. Microbiol. 2008, 39, 50–55. [Google Scholar] [CrossRef] [Green Version]
- Hossain, M.N.; Afrin, S.; Humayun, S.; Ahmed, M.M.; Saha, B.K. Identification and Growth Characterization of a Novel Strain of Saccharomyces boulardii Isolated from Soya Paste. Front. Nutr. 2020, 7, 27. [Google Scholar] [CrossRef]
- Hernández-Gómez, J.G.; López-Bonilla, A.; Trejo-Tapia, G.; Ávila-Reyes, S.V.; Jiménez-Aparicio, A.R.; Hernández-Sánchez, H. In vitro bile salt hydrolase (BSH) Activity screening of different probiotic microorganisms. Foods 2021, 10, 674. [Google Scholar] [CrossRef]
- Nakayama, H.; Yoshida, K.; Shinmyo, A. Yeast plasma membrane Ena1p ATPase alters alkali-cation homeostasis and confers increased salt tolerance in tobacco cultured cells. Biotechnol. Bioeng. 2004, 85, 776–789. [Google Scholar] [CrossRef]
- Casado, C.; González, A.; Platara, M.; Ruiz, A.; Ariño, J. The role of the protein kinase A pathway in the response to alkaline pH stress in yeast. Biochem. J. 2011, 438, 523–533. [Google Scholar] [CrossRef] [Green Version]
- Wilms, T.; Swinnen, E.; Eskes, E.; Dolz-Edo, L.; Uwineza, A.; Van Essche, R.; Rosseels, J.; Zabrocki, P.; Cameroni, E.; Franssens, V.; et al. The yeast protein kinase Sch9 adjusts V-ATPase assembly/disassembly to control pH homeostasis and longevity in response to glucose availability. PLoS Genet. 2017, 13, e1006835. [Google Scholar] [CrossRef]
- Diab, H.I.; Kane, P.M. Loss of vacuolar H+-ATPase (V-ATPase) activity in yeast generates an iron deprivation signal that is moderated by induction of the peroxiredoxin TSA2. J. Biol. Chem. 2013, 288, 11366–11377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.Y.; Huang, T.K.; Kuo, H.F.; Chiou, T.J. Role of vacuoles in phosphorus storage and remobilization. J. Exp. Bot. 2017, 68, 3045–3055. [Google Scholar] [CrossRef] [PubMed]
- Capusoni, C.; Arioli, S.; Donzella, S.; Guidi, B.; Serra, I.; Compagno, C. Hyper-Osmotic Stress Elicits Membrane Depolarization and Decreased Permeability in Halotolerant Marine Debaryomyces hansenii Strains and in Saccharomyces cerevisiae. Front. Microbiol. 2019, 10, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deparis, Q.; Claes, A.; Foulquié-Moreno, M.R.; Thevelein, J.M. Engineering tolerance to industrially relevant stress factors in yeast cell factories. FEMS Yeast Res. 2017, 17, fox036. [Google Scholar] [CrossRef]
- Gandhi, A.; Shah, N.P. Effect of salt stress on morphology and membrane composition of Lactobacillus acidophilus, Lactobacillus casei, and Bifidobacterium bifidum, and their adhesion to human intestinal epithelial-like Caco-2 cells. J. Dairy Sci. 2016, 99, 2594–2605. [Google Scholar] [CrossRef] [Green Version]
- Romero-Gil, V.; Bautista-Gallego, J.; Rodríguez-Gómez, F.; García-García, P.; Jiménez-Díaz, R.; Garrido-Fernández, A.; Arroyo-López, F.N. Evaluating the individual effects of temperature and salt on table olive related microorganisms. Food Microbiol. 2013, 33, 178–184. [Google Scholar] [CrossRef]
- Hou, L.; Wang, M.; Wang, C.; Wang, C.; Wang, H. Analysis of salt-tolerance genes in Zygosaccharomyces rouxii. Appl. Biochem. Biotechnol. 2013, 170, 1417–1425. [Google Scholar] [CrossRef]
- Song, Y.R.; Jeong, D.Y.; Baik, S.H. Effects of indigenous yeasts on physicochemical and microbial properties of Korean soy sauce prepared by low-salt fermentation. Food Microbiol. 2015, 51, 171–178. [Google Scholar] [CrossRef]
- Yao, S.J.; Zhou, R.Q.; Jin, Y.; Zhang, L.Q.; Huang, J.; Wu, C.D. Co-culture with Tetragenococcus halophilus changed the response of Zygosaccharomyces rouxii to salt stress. Process Biochem. 2020, 95, 279–287. [Google Scholar] [CrossRef]
- Fleet, G.H. Yeast spoilage of foods and beverages. In The Yeasts: A Taxonomic Study; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier Science: San Diego, CA, USA, 2011; Volume 1, pp. 53–63. [Google Scholar]
- Hong, J.; Li, W.; Lin, B.; Zhan, M.; Liu, C.; Chen, B.Y. Deciphering the effect of salinity on the performance of submerged membrane bioreactor for aquaculture of bacterial community. Desalination 2013, 316, 23–30. [Google Scholar] [CrossRef]
- Heinisch, J.J.; Rodicio, R. Stress responses in wine yeast. In Biology of Microorganisms on Grapes, in Must and in Wine; Springer: Berlin/Heidelberg, Germany, 2017; pp. 377–395. [Google Scholar]
- Hohmann, S. An integrated view on a eukaryotic osmoregulation system. Curr. Genet. 2015, 61, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Sharma, S.C. Physiological Basis for the Tolerance of Yeast Zygosaccharomyces bisporus to Salt Stress. HAYATI J. Biosci. 2017, 24, 176–181. [Google Scholar] [CrossRef]
- Pascual-Ahuir, A.; Manzanares-Estreder, S.; Timón-Gómez, A.; Proft, M. Ask yeast how to burn your fats: Lessons learned from the metabolic adaptation to salt stress. Curr. Genet. 2018, 64, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Dakal, T.C.; Solieri, L.; Giudici, P. Adaptive response and tolerance to sugar and salt stress in the food yeast Zygosaccharomyces rouxii. Int. J. Food Microbiol. 2014, 185, 140–157. [Google Scholar] [CrossRef] [PubMed]
- Hounsa, C.G.; Brandt, E.V.; Thevelein, J.; Hohmann, S.; Prior, B.A. Role of trehalose in survival of Saccharomyces cerevisiae under osmotic stress. Microbiology 1998, 144, 671–680. [Google Scholar] [CrossRef] [PubMed]
- Brewster, J.L.; Gustin, M.C. Hog1: 20 years of discovery and impact. Sci. Signal. 2014, 7, re7. [Google Scholar] [CrossRef]
- Rodrigues-Pousada, C.A.; Nevitt, T.; Menezes, R.; Azevedo, D.; Pereira, J.; Amaral, C. Yeast activator proteins and stress response: An overview. FEBS Lett. 2004, 567, 80–85. [Google Scholar] [CrossRef]
- Dhar, R.; Sägesser, R.; Weikert, C.; Yuan, J.; Wagner, A. Adaptation of Saccharomyces cerevisiae to saline stress through laboratory evolution. J. Evol. Biol. 2011, 24, 1135–1153. [Google Scholar] [CrossRef] [Green Version]
- Isenring, J.; Geirnaert, A.; Lacroix, C.; Stevens, M.J.A. Bistable auto-aggregation phenotype in Lactiplantibacillus plantarum emerges after cultivation in in vitro colonic microbiota. BMC Microbiol. 2021, 21, 268. [Google Scholar] [CrossRef]
- Galdiero, F.; Carratelli, C.R.; Nuzzo, I.; Bentivoglio, C.; Galdiero, M. Phagocytosis of bacterial aggregates by granulocytes. Eur. J. Epidemiol. 1988, 4, 456–460. [Google Scholar] [CrossRef]
- Monteagudo-Mera, A.; Rastall, R.A.; Gibson, G.R.; Charalampopoulos, D.; Chatzifragkou, A. Adhesion mechanisms mediated by probiotics and prebiotics and their potential impact on human health. Appl. Microbiol. Biotechnol. 2019, 103, 6463–6472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verstrepen, K.J.; Klis, F.M. Flocculation, adhesion and biofilm formation in yeasts. Mol. Microbiol. 2006, 60, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Pacheco Rodríguez, P.; Arévalo-Villena, M.; Zaparoli Rosa, I.; Briones Pérez, A. Selection of potential non-Sacharomyces probiotic yeasts from food origin by a step-by-step approach. Food Res. Int. 2018, 112, 143–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suvarna, S.; Dsouza, J.; Ragavan, M.L.; Das, N. Potential probiotic characterization and effect of encapsulation of probiotic yeast strains on survival in simulated gastrointestinal tract condition. Food Sci. Biotechnol. 2018, 27, 745–753. [Google Scholar] [CrossRef]
- Lima, M.D.S.F.D.; Souza, K.M.S.D.; Albuquerque, W.W.C.; Teixeira, J.A.C.; Cavalcanti, M.T.H.; Porto, A.L.F. Saccharomyces cerevisiae from Brazilian kefir-fermented milk: An in vitro evaluation of probiotic properties. Microb. Pathog. 2017, 110, 670–677. [Google Scholar] [CrossRef] [Green Version]
- García-Cayuela, T.; Korany, A.M.; Bustos, I.; de Cadiñanos, L.P.G.; Requena, T.; Peláez, C.; Martínez-Cuesta, M.C. Adhesion abilities of dairy Lactobacillus plantarum strains showing an aggregation phenotype. Food Res. Int. 2014, 57, 44–50. [Google Scholar] [CrossRef]
- Nandhra, G.K.; Mark, E.B.; Di Tanna, G.L.; Haase, A.M.; Poulsen, J.; Christodoulides, S.; Kung, V.; Klinge, M.W.; Knudsen, K.; Borghammer, P.; et al. Normative values for region-specific colonic and gastrointestinal transit times in 111 healthy volunteers using the 3D-Transit electromagnet tracking system: Influence of age, gender, and body mass index. Neurogastroenterol. Motil. 2020, 32, e13734. [Google Scholar] [CrossRef]
- Asnicar, F.; Leeming, E.R.; Dimidi, E.; Mazidi, M.; Franks, P.W.; Al Khatib, H.; Valdes, A.M.; Davies, R.; Bakker, E.; Francis, L.; et al. Blue poo: Impact of gut transit time on the gut microbiome using a novel marker. Gut 2021, 70, 1665–1674. [Google Scholar] [CrossRef]
- Roland, B.C.; Ciarleglio, M.M.; Clarke, J.O.; Semler, J.R.; Tomakin, E.; Mullin, G.E.; Pasricha, P.J. Small Intestinal Transit Time Is Delayed in Small Intestinal Bacterial Overgrowth. J. Clin. Gastroenterol. 2015, 49, 571–576. [Google Scholar] [CrossRef]
- Gil-Rodríguez, A.M.; Carrascosa, A.V.; Requena, T. Yeasts in foods and beverages: In vitro characterisation of probiotic traits. LWT-Food Sci. Technol. 2015, 64, 1156–1162. [Google Scholar] [CrossRef] [Green Version]
- Stewart, G.G. Yeast flocculation—sedimentation and flotation. Fermentation 2018, 4, 28. [Google Scholar] [CrossRef] [Green Version]
- Di Gianvito, P.; Tesnière, C.; Suzzi, G.; Blondin, B.; Tofalo, R. FLO5 gene controls flocculation phenotype and adhesive properties in a Saccharomyces cerevisiae sparkling wine strain. Sci. Rep. 2017, 7, 10786. [Google Scholar] [CrossRef] [PubMed]
- Miki, B.L.A.; Poon, N.H.; James, A.P.; Seligy, V.L. Possible mechanism for flocculation interactions governed by gene FLO1 in Saccharomyces cerevisiae. J. Bacteriol. 1982, 150, 878–889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Touhami, A.; Hoffmann, B.; Vasella, A.; Denis, F.A.; Dufrêne, Y.F. Aggregation of yeast cells: Direct measurement of discrete lectin-carbohydrate interactions. Microbiology 2003, 149, 2873–2878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goossens, K.V.Y.; Ielasi, F.S.; Nookaew, I.; Stals, I.; Alonso-Sarduy, L.; Daenen, L.; Van Mulders, S.E.; Stassen, C.; Van Eijsden, R.G.E.; Siewers, V.; et al. Molecular mechanism of flocculation self-recognition in yeast and its role in mating and survival. mBio 2015, 6, e00427-15. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Yu, Z.; Tian, F.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. Surface components and metabolites of probiotics for regulation of intestinal epithelial barrier. Microb. Cell Fact. 2020, 19, 23. [Google Scholar] [CrossRef] [PubMed]
- Somashekaraiah, R.; Shruthi, B.; Deepthi, B.V.; Sreenivasa, M.Y. Probiotic properties of lactic acid bacteria isolated from neera: A naturally fermenting coconut palm nectar. Front. Microbiol. 2019, 10, 1382. [Google Scholar] [CrossRef]
- Krasowska, A.; Sigler, K. How microorganisms use hydrophobicity and what does this mean for human needs? Front. Cell. Infect. Microbiol. 2014, 4, 112. [Google Scholar] [CrossRef] [Green Version]
- Kochkodan, V.; Tsarenko, S.; Potapchenko, N.; Kosinova, V.; Goncharuk, V. Adhesion of microorganisms to polymer membranes: A photobactericidal effect of surface treatment with TiO2. Desalination 2008, 220, 380–385. [Google Scholar] [CrossRef]
- Lugea, A.; Salas, A.; Casalot, J.; Guarner, F.; Malagelada, J.R. Surface hydrophobicity of the rat colonic mucosa is a defensive barrier against macromolecules and toxins. Gut 2000, 46, 515–521. [Google Scholar] [CrossRef] [Green Version]
- Farhadi, A.; Banan, A.; Fields, J.; Keshavarzian, A. Intestinal barrier: An interface between health and disease. J. Gastroenterol. Hepatol. 2003, 18, 479–497. [Google Scholar] [CrossRef] [PubMed]
- Van Tassell, M.L.; Miller, M.J. Lactobacillus adhesion to mucus. Nutrients 2011, 3, 613–636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGinnis, M.R. Pathogenesis of indoor fungal diseases. Med. Mycol. 2004, 42, 107–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ehrmann, M.A.; Kurzak, P.; Bauer, J.; Vogel, R.F. Characterization of lactobacilli towards their use as probiotic adjuncts in poultry. J. Appl. Microbiol. 2002, 92, 966–975. [Google Scholar] [CrossRef]
- França, R.C.; Conceição, F.R.; Mendonça, M.; Haubert, L.; Sabadin, G.; de Oliveira, P.D.; Amaral, M.G.; Silva, W.P.; Moreira, Â.N. Pichia pastoris X-33 has probiotic properties with remarkable antibacterial activity against Salmonella Typhimurium. Appl. Microbiol. Biotechnol. 2015, 99, 7953–7961. [Google Scholar] [CrossRef]
- Menezes, A.G.T.; Ramos, C.L.; Cenzi, G.; Melo, D.S.; Dias, D.R.; Schwan, R.F. Probiotic Potential, Antioxidant Activity, and Phytase Production of Indigenous Yeasts Isolated from Indigenous Fermented Foods. Probiotics Antimicrob Proteins 2020, 12, 280–288. [Google Scholar] [CrossRef]
- Rezac, S.; Kok, C.R.; Heermann, M.; Hutkins, R. Fermented foods as a dietary source of live organisms. Front. Microbiol. 2018, 9, 1785. [Google Scholar] [CrossRef]
- Goktas, H.; Dikmen, H.; Demirbas, F.; Sagdic, O.; Dertli, E. Characterisation of probiotic properties of yeast strains isolated from kefir samples. Int. J. Dairy Technol. 2021, 74, 715–722. [Google Scholar] [CrossRef]
- Romero-Luna, H.E.; Hernández-Sánchez, H.; Ribas-Aparicio, R.M.; Cauich-Sánchez, P.I.; Dávila-Ortiz, G. Evaluation of the probiotic potential of Saccharomyces cerevisiae Strain (C41) isolated from Tibicos by in vitro studies. Probiotics Antimicrob. Proteins 2019, 11, 794–800. [Google Scholar] [CrossRef]
- Servin, A.L. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol. Rev. 2004, 28, 405–440. [Google Scholar] [CrossRef] [Green Version]
- Linder, M.B. Hydrophobins: Proteins that self assemble at interfaces. Curr. Opin. Colloid Interface Sci. 2009, 14, 356–363. [Google Scholar] [CrossRef]
- Hazen, K.C.; Hazen, B.W. Surface hydrophobic and hydrophilic protein alterations in Candida albicans. FEMS Microbiol. Lett. 1993, 107, 83–87. [Google Scholar] [CrossRef] [PubMed]
- Willaert, R.G. Adhesins of yeasts: Protein structure and interactions. J. Fungi 2018, 4, 119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirayama, S.; Furukawa, S.; Ogihara, H.; Morinaga, Y. Yeast mannan structure necessary for co-aggregation with Lactobacillus plantarum ML11-11. Biochem. Biophys. Res. Commun. 2012, 419, 652–655. [Google Scholar] [CrossRef]
- Lara-Hidalgo, C.; Dorantes-Álvarez, L.; Hernández-Sánchez, H.; Santoyo-Tepole, F.; Martínez-Torres, A.; Villa-Tanaca, L.; Hernández-Rodríguez, C. Isolation of yeasts from guajillo pepper (Capsicum annuum L.) fermentation and study of some probiotic characteristics. Probiotics Antimicrob. Proteins 2019, 11, 748–764. [Google Scholar] [CrossRef]
- Kaczorek, E.; Chrzanowski, Ł.; Pijanowska, A.; Olszanowski, A. Yeast and bacteria cell hydrophobicity and hydrocarbon biodegradation in the presence of natural surfactants: Rhamnolipides and saponins. Bioresour. Technol. 2008, 99, 4285–4291. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alkalbani, N.S.; Osaili, T.M.; Al-Nabulsi, A.A.; Olaimat, A.N.; Liu, S.-Q.; Shah, N.P.; Apostolopoulos, V.; Ayyash, M.M. Assessment of Yeasts as Potential Probiotics: A Review of Gastrointestinal Tract Conditions and Investigation Methods. J. Fungi 2022, 8, 365. https://doi.org/10.3390/jof8040365
Alkalbani NS, Osaili TM, Al-Nabulsi AA, Olaimat AN, Liu S-Q, Shah NP, Apostolopoulos V, Ayyash MM. Assessment of Yeasts as Potential Probiotics: A Review of Gastrointestinal Tract Conditions and Investigation Methods. Journal of Fungi. 2022; 8(4):365. https://doi.org/10.3390/jof8040365
Chicago/Turabian StyleAlkalbani, Nadia S., Tareq M. Osaili, Anas A. Al-Nabulsi, Amin N. Olaimat, Shao-Quan Liu, Nagendra P. Shah, Vasso Apostolopoulos, and Mutamed M. Ayyash. 2022. "Assessment of Yeasts as Potential Probiotics: A Review of Gastrointestinal Tract Conditions and Investigation Methods" Journal of Fungi 8, no. 4: 365. https://doi.org/10.3390/jof8040365
APA StyleAlkalbani, N. S., Osaili, T. M., Al-Nabulsi, A. A., Olaimat, A. N., Liu, S.-Q., Shah, N. P., Apostolopoulos, V., & Ayyash, M. M. (2022). Assessment of Yeasts as Potential Probiotics: A Review of Gastrointestinal Tract Conditions and Investigation Methods. Journal of Fungi, 8(4), 365. https://doi.org/10.3390/jof8040365








