Novel Insights into Aspergillus fumigatus Pathogenesis and Host Response from State-of-the-Art Imaging of Host–Pathogen Interactions during Infection
Abstract
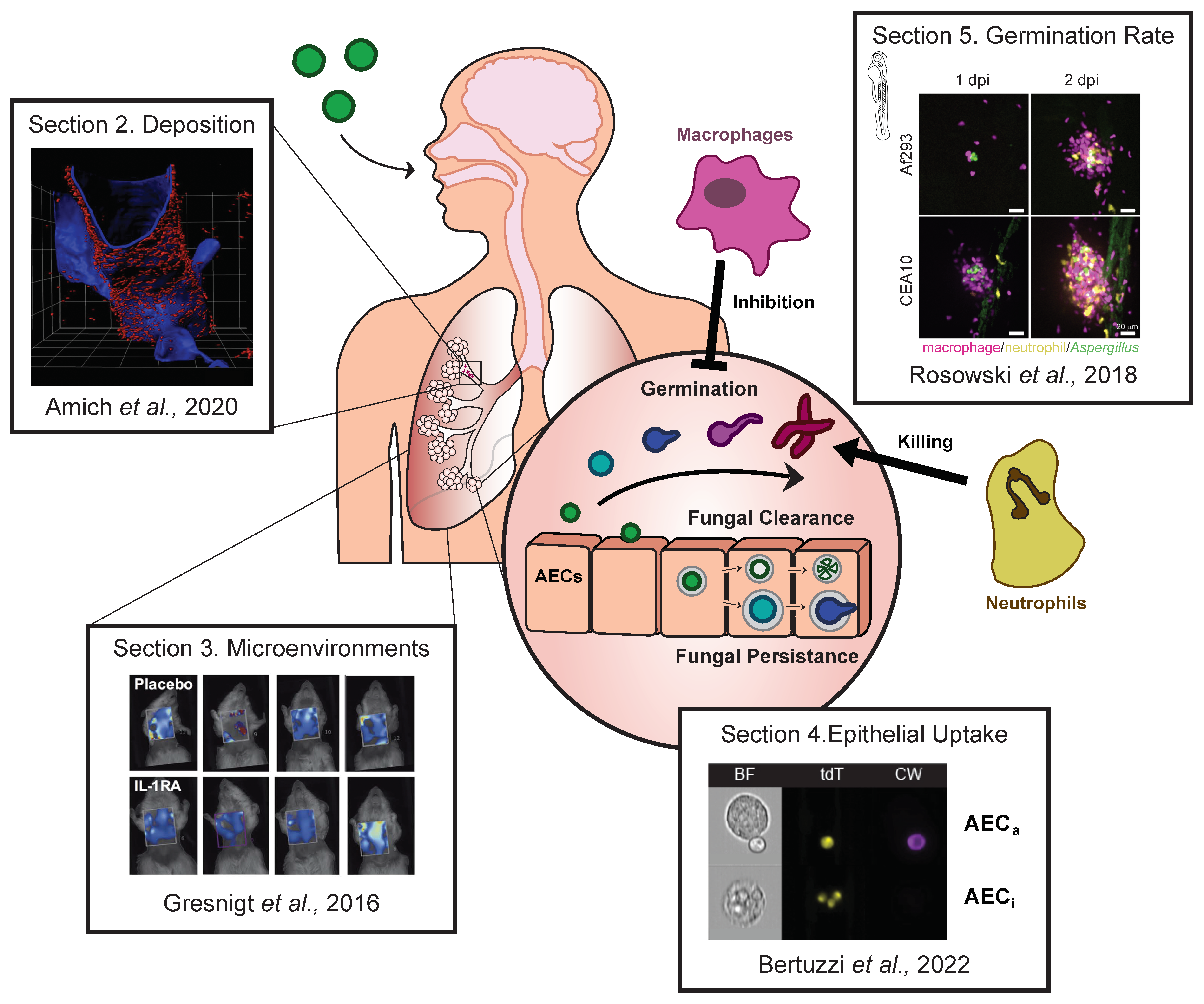
1. Introduction
2. Are A. fumigatus Spores Deposited in the Alveoli?
3. What Microenvironments Does A. fumigatus Encounter upon Lung Deposition?
4. Are Airway Respiratory Cells Internalising A. fumigatus during Infection?
5. How Does Germination Alter the Interactions of A. fumigatus with Immune Cells?
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- O’Gorman, C. Airborne Aspergillus fumigatus conidia: A risk factor for aspergillosis. Fungal Biol. Rev. 2021, 25, 151–157. [Google Scholar] [CrossRef]
- Brown, G.D.; Denning, D.W.; Gow, N.A.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012, 4, 165rv13. [Google Scholar] [CrossRef]
- Bongomin, F.; Gago, S.; Oladele, R.O.; Denning, D.W. Global and multi-national prevalence of fungal diseases-estimate precision. J. Fungi 2017, 3, 57. [Google Scholar] [CrossRef] [PubMed]
- Guinea, J.; Torres-Narbona, M.; Gijón, P.; Muñoz, P.; Pozo, F.; Peláez, T.; de Miguel, J.; Bouza, E. Pulmonary aspergillosis in patients with chronic obstructive pulmonary disease: Incidence, risk factors, and outcome. Clin. Microbiol. Infect. 2010, 16, 870–877. [Google Scholar] [CrossRef] [PubMed]
- Hammond, E.E.; McDonald, C.S.; Vestbo, J.; Denning, D.W. The global impact of Aspergillus infection on COPD. BMC Pulm. Med. 2020, 20, 241. [Google Scholar] [CrossRef] [PubMed]
- Knutsen, A.P.; Slavin, R.G. Allergic bronchopulmonary aspergillosis in asthma and cystic fibrosis. Clin. Dev. Immunol. 2011, 2021, 843763. [Google Scholar] [CrossRef]
- Denning, D.W.; Bromley, M.J. Infectious Disease. How to bolster the antifungal pipeline. Science 2015, 347, 1414–1416. [Google Scholar] [CrossRef]
- Toda, M.; Beer, K.D.; Kuivila, K.M.; Chiller, T.M.; Jackson, B.R. Trends in agricultural triazole fungicide use in the United States, 1992-2016 and possible implications for antifungal-resistant fungi in human disease. Environ. Health Perspect. 2021, 129, 55001. [Google Scholar] [CrossRef]
- Verweij, P.E.; Lucas, J.A.; Arendrup, M.C.; Bowyer, P.; Brinkmann, A.J.; Denning, D.W.; Dyer, P.S.; Fisher, M.C.; Geenen, P.L.; Gisi, U. The one health problem of azole resistance in Aspergillus fumigatus: Current insights and future research agenda. Fungal Biol. Rev. 2020, 34, 202–214. [Google Scholar] [CrossRef]
- Gunzer, M.; Thornton, C.R.; Beziere, N. Advances in the in vivo molecular imaging of invasive aspergillosis. J. Fungi 2020, 6, 338. [Google Scholar] [CrossRef]
- Latgé, J.P.; Chamilos, G. Aspergillus fumigatus and Aspergillosis in 2019. Clin. Microbiol. Rev. 2019, 33, e00140-18. [Google Scholar] [CrossRef] [PubMed]
- Mackel, J.J.; Steele, C. Host defense mechanisms against Aspergillus fumigatus lung colonization and invasion. Curr. Opin. Microbiol. 2019, 52, 14–19. [Google Scholar] [CrossRef] [PubMed]
- van de Veerdonk, F.L.; Gresnigt, M.S.; Romani, L.; Netea, M.G.; Latgé, J.P. Aspergillus fumigatus morphology and dynamic host interactions. Nat. Rev. Microbiol. 2017, 15, 661–674. [Google Scholar] [CrossRef] [PubMed]
- Latgé, J.P.; Beauvais, A.; Chamilos, G. The cell wall of the human fungal pathogen Aspergillus fumigatus: Biosynthesis, organization, immune response, and virulence. Annu. Rev. Microbiol. 2017, 71, 99–116. [Google Scholar] [CrossRef]
- Amich, J.; Mokhtari, Z.; Strobel, M.; Vialetto, E.; Sheta, D.; Yu, Y.; Hartweg, J.; Kalleda, N.; Jarick, K.J.; Brede, C.; et al. Three-dimensional light sheet fluorescence microscopy of lungs to dissect local host immune-Aspergillus fumigatus interactions. mBio 2020, 11, e02752-19. [Google Scholar] [CrossRef]
- Gresnigt, M.S.; Rekiki, A.; Rasid, O.; Savers, A.; Jouvion, G.; Dannaoui, E.; Parlato, M.; Fitting, C.; Brock, M.; Cavaillon, J.M.; et al. Reducing hypoxia and inflammation during invasive pulmonary aspergillosis by targeting the Interleukin-1 receptor. Sci. Rep. 2016, 6, 26490. [Google Scholar] [CrossRef]
- Bertuzzi, M.; Howell, G.J.; Thomson, D.D.; Fortune-Grant, R.; Moeslinger, A.; Dancer, P.; Van Rhijn, N.; Motsi, N.; Du, X.; Codling, A.; et al. Epithelial uptake of Aspergillus fumigatus drives efficient fungal clearance in vivo and is aberrant in Chronic Obstructive Pulmonary Disease (COPD). bioRxiv 2022. [CrossRef]
- Rosowski, E.E.; Raffa, N.; Knox, B.P.; Golenberg, N.; Keller, N.P.; Huttenlocher, A. Macrophages inhibit Aspergillus fumigatus germination and neutrophil-mediated fungal killing. PLoS Pathog. 2018, 14, e1007229. [Google Scholar] [CrossRef]
- American Society for Microbiology. One Health: Fungal Pathogens of Humans, Animals, and Plants: Report on an American Academy of Microbiology Colloquium Held in Washington, DC, USA on 18 October 2017; American Society for Microbiology: Washington, DC, USA, 2019. Available online: https://www.ncbi.nlm.nih.gov/books/NBK549988/ (accessed on 2 February 2022). [CrossRef]
- Nicod, L.P. Lung defences: An overview. Eur. Respir. Rev. 2005, 14, 45–50. [Google Scholar] [CrossRef]
- Bertuzzi, M.; Hayes, G.E.; Icheoku, U.J.; van Rhijn, N.; Denning, D.W.; Osherov, N.; Bignell, E.M. Anti-Aspergillus activities of the respiratory epithelium in health and disease. J. Fungi 2018, 4, 8. [Google Scholar] [CrossRef]
- McCormick, A.; Loeffler, J.; Ebel, F. Aspergillus fumigatus: Contours of an opportunistic human pathogen. Cell. Microbiol. 2010, 12, 1535–1543. [Google Scholar] [CrossRef] [PubMed]
- Latgé, J.P. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 1999, 12, 310–350. [Google Scholar] [CrossRef] [PubMed]
- Brakhage, A.A.; Langfelder, K. Menacing mold: The molecular biology of Aspergillus fumigatus. Annu. Rev. Microbiol. 2002, 56, 433–455. [Google Scholar] [CrossRef] [PubMed]
- Dagenais, T.R.; Keller, N.P. Pathogenesis of Aspergillus fumigatus in invasive aspergillosis. Clin. Microbiol. Rev. 2009, 22, 447–465. [Google Scholar] [CrossRef] [PubMed]
- Brede, C.; Friedrich, M.; Jordán-Garrote, A.L.; Riedel, S.S.; Bäuerlein, C.A.; Heinze, K.G.; Bopp, T.; Schulz, S.; Mottok, A.; Kiesel, C.; et al. Mapping immune processes in intact tissues at cellular resolution. J. Clin. Investig. 2012, 122, 4439–4446. [Google Scholar] [CrossRef]
- Rolle, A.M.; Hasenberg, M.; Thornton, C.R.; Solouk-Saran, D.; Männ, L.; Weski, J.; Maurer, A.; Fischer, E.; Spycher, P.R.; Schibli, R.; et al. ImmunoPET/MR imaging allows specific detection of Aspergillus fumigatus lung infection in vivo. Proc. Natl. Acad. Sci. USA 2016, 113, E1026–E1033. [Google Scholar] [CrossRef]
- Soutiere, S.E.; Tankersley, C.G.; Mitzner, W. Differences in alveolar size in inbred mouse strains. Respir. Physiol. Neurobiol. 2004, 140, 283–291. [Google Scholar] [CrossRef]
- Knust, J.; Ochs, M.; Gundersen, H.J.; Nyengaard, J.R. Stereological estimates of alveolar number and size and capillary length and surface area in mice lungs. Anat. Rec. 2009, 292, 113–122. [Google Scholar] [CrossRef]
- Mercer, R.R.; Russell, M.L.; Crapo, J.D. Alveolar septal structure in different species. J. Appl. Physiol. 1994, 77, 1060–1066. [Google Scholar] [CrossRef]
- Bertuzzi, M.; Schrettl, M.; Alcazar-Fuoli, L.; Cairns, T.C.; Muñoz, A.; Walker, L.A.; Herbst, S.; Safari, M.; Cheverton, A.M.; Chen, D.; et al. The pH-responsive PacC transcription factor of Aspergillus fumigatus governs epithelial entry and tissue invasion during pulmonary aspergillosis. PLoS Pathog. 2014, 10, e1004413. [Google Scholar] [CrossRef]
- Croft, C.A.; Culibrk, L.; Moore, M.M.; Tebbutt, S.J. Interactions of Aspergillus fumigatus conidia with airway epithelial cells: A Critical Review. Front. Microbiol. 2016, 7, 472. [Google Scholar] [CrossRef] [PubMed]
- Geiser, M.; Leupin, N.; Maye, I.; Hof, V.I.; Gehr, P. Interaction of fungal spores with the lungs: Distribution and retention of inhaled puffball (Calvatia excipuliformis) spores. J. Allergy Clin. Immunol. 2000, 106 Pt 1, 92–100. [Google Scholar] [CrossRef]
- Rammaert, B.; Jouvion, G.; de Chaumont, F.; Garcia-Hermoso, D.; Szczepaniak, C.; Renaudat, C.; Olivo-Marin, J.C.; Chrétien, F.; Dromer, F.; Bretagne, S. Absence of fungal spore internalization by bronchial epithelium in mouse models evidenced by a new bioimaging approach and transmission electronic microscopy. Am. J. Pathol. 2015, 185, 2421–2430. [Google Scholar] [CrossRef] [PubMed]
- Secondo, L.E.; Sagona, J.A.; Calderón, L.; Wang, Z.; Plotnik, D.; Senick, J.; Sorensen-Allacci, M.; Wener, R.; Andrews, C.J.; Mainelis, G. Estimating lung deposition of fungal spores using actual airborne spore concentrations and physiological data. Environ. Sci. Technol. 2021, 55, 1852–1863. [Google Scholar] [CrossRef]
- Sagona, J.; Secondo, L.; Mainelis, G. Comparison of two models to estimate deposition of fungi and bacteria in the human respiratory tract. Atmosphere 2020, 11, 561. [Google Scholar] [CrossRef]
- Cho, S.; Seo, S.-C.; Schmechel, D.; Grinshpun, S.; Reponen, T. Aerodynamic characteristic and respiratory deposition of fungal particles. Atmos. Environ. 2005, 39, 5454–5465. [Google Scholar] [CrossRef]
- Madhwal, S.; Prabhu, V.; Sundriyal, S.; Shridhar, V. Distribution, characterization and health risk assessment of size fractionated bioaerosols at an open landfill site in Dehradun, India. Atmos. Pollut. Res. 2019, 11, 156–169. [Google Scholar] [CrossRef]
- Reponen, T. Aerodynamic diameters and respiratory deposition estimates of viable fungal particles in mold problem dwellings. Aerosol Sci. Technol. 1995, 22, 11–23. [Google Scholar] [CrossRef]
- Bogorodskiy, A.O.; Bolkhovitina, E.L.; Gensch, T.; Troyanova, N.I.; Mishin, A.V.; Okhrimenko, I.S.; Braun, A.; Spies, E.; Gordeliy, V.I.; Sapozhnikov, A.M.; et al. Murine intraepithelial dendritic cells interact with phagocytic cells during Aspergillus fumigatus-induced inflammation. Front. Immunol. 2020, 11, 298. [Google Scholar] [CrossRef]
- Maslov, I.V.; Bogorodskiy, A.O.; Pavelchenko, M.V.; Zykov, I.O.; Troyanova, N.I.; Borshchevskiy, V.I.; Shevchenko, M.A. Confocal laser scanning microscopy-based quantitative analysis of Aspergillus fumigatus conidia distribution in whole-mount optically cleared mouse lung. J. Vis. Exp. JoVE 2021, 175, e62436. [Google Scholar] [CrossRef]
- King, J.; Brunel, S.F.; Warris, A. Aspergillus infections in cystic fibrosis. J. Infect. 2016, 72, S50–S55. [Google Scholar] [CrossRef] [PubMed]
- Belchi, F.; Pirashvili, M.; Conway, J.; Bennett, M.; Djukanovic, R.; Brodzki, J. Lung topology characteristics in patients with chronic obstructive pulmonary disease. Sci. Rep. 2018, 8, 5341. [Google Scholar] [CrossRef] [PubMed]
- Meyerholz, D.K.; Stoltz, D.A.; Namati, E.; Ramachandran, S.; Pezzulo, A.A.; Smith, A.R.; Rector, M.V.; Suter, M.J.; Kao, S.; McLennan, G.; et al. Loss of cystic fibrosis transmembrane conductance regulator function produces abnormalities in tracheal development in neonatal pigs and young children. Am. J. Respir. Crit. Care Med. 2010, 182, 1251–1261. [Google Scholar] [CrossRef]
- Semaniakou, A.; Croll, R.P.; Chappe, V. Animal models in the pathophysiology of cystic fibrosis. Front. Pharmacol. 2019, 9, 1475. [Google Scholar] [CrossRef]
- Sugui, J.A.; Kwon-Chung, K.J.; Juvvadi, P.R.; Latgé, J.P.; Steinbach, W.J. Aspergillus fumigatus and related species. Cold Spring Harb. Perspect. Med. 2014, 5, a019786. [Google Scholar] [CrossRef] [PubMed]
- Grahl, N.; Puttikamonkul, S.; Macdonald, J.M.; Gamcsik, M.P.; Ngo, L.Y.; Hohl, T.M.; Cramer, R.A. In vivo hypoxia and a fungal alcohol dehydrogenase influence the pathogenesis of invasive pulmonary aspergillosis. PLoS Pathog. 2011, 7, e1002145. [Google Scholar] [CrossRef] [PubMed]
- Warn, P.A.; Sharp, A.; Guinea, J.; Denning, D.W. Effect of hypoxic conditions on in vitro susceptibility testing of amphotericin B, itraconazole and micafungin against Aspergillus and Candida. J. Antimicrob. Chemother. 2004, 53, 743–749. [Google Scholar] [CrossRef] [PubMed]
- West, J.B. Respiratory Physiology—The Essentials, 3rd ed.; Williams & Wilkins: Baltimore, MD, USA, 1985. [Google Scholar]
- Simmen, H.P.; Battaglia, H.; Giovanoli, P.; Blaser, J. Analysis of pH, pO2 and pCO2 in drainage fluid allows for rapid detection of infectious complications during the follow-up period after abdominal surgery. Infection 1994, 22, 386–389. [Google Scholar] [CrossRef]
- Simmen, H.P.; Blaser, J. Analysis of pH and pO2 in abscesses, peritoneal fluid, and drainage fluid in the presence or absence of bacterial infection during and after abdominal surgery. Am. J. Surg. 1993, 166, 24–27. [Google Scholar] [CrossRef]
- Grahl, N.; Kowalski, C.H.; Cramer, R.A. Detection of low oxygen microenvironments in a murine model of invasive pulmonary aspergillosis using Pimonidazole. Methods Mol. Biol. 2021, 2260, 197–205. [Google Scholar] [CrossRef]
- Grahl, N.; Cramer, R.A., Jr. Regulation of hypoxia adaptation: An overlooked virulence attribute of pathogenic fungi? Med. Mycol. 2010, 48, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Tarrand, J.J.; Han, X.Y.; Kontoyiannis, D.P.; May, G.S. Aspergillus hyphae in infected tissue: Evidence of physiologic adaptation and effect on culture recovery. J. Clin. Microbiol. 2005, 43, 382–386. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Brock, M.; Jouvion, G.; Droin-Bergère, S.; Dussurget, O.; Nicola, M.A.; Ibrahim-Granet, O. Bioluminescent Aspergillus fumigatus, a new tool for drug efficiency testing and in vivo monitoring of invasive aspergillosis. Appl. Environ. Microbiol. 2008, 74, 7023–7035. [Google Scholar] [CrossRef] [PubMed]
- Willger, S.D.; Puttikamonkul, S.; Kim, K.H.; Burritt, J.B.; Grahl, N.; Metzler, L.J.; Barbuch, R.; Bard, M.; Lawrence, C.B.; Cramer, R.A., Jr. A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus. PLoS Pathog. 2008, 4, e1000200. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim-Granet, O.; Jouvion, G.; Hohl, T.M.; Droin-Bergère, S.; Philippart, F.; Kim, O.Y.; Adib-Conquy, M.; Schwendener, R.; Cavaillon, J.M.; Brock, M. In vivo bioluminescence imaging and histopathopathologic analysis reveal distinct roles for resident and recruited immune effector cells in defense against invasive aspergillosis. BMC Microbiol. 2010, 10, 105. [Google Scholar] [CrossRef]
- Hsu, J.L.; Khan, M.A.; Sobel, R.A.; Jiang, X.; Clemons, K.V.; Nguyen, T.T.; Stevens, D.A.; Martinez, M.; Nicolls, M.R. Aspergillus fumigatus invasion increases with progressive airway ischemia. PLoS ONE 2013, 8, e77136. [Google Scholar] [CrossRef]
- Kowalski, C.H.; Beattie, S.R.; Fuller, K.K.; McGurk, E.A.; Tang, Y.W.; Hohl, T.M.; Obar, J.J.; Cramer, R.A., Jr. Heterogeneity among isolates reveals that fitness in low oxygen correlates with Aspergillus fumigatus virulence. mBio 2016, 7, e01515-16. [Google Scholar] [CrossRef]
- Dhingra, S.; Buckey, J.C.; Cramer, R.A. Hyperbaric oxygen reduces Aspergillus fumigatus proliferation in vitro and influences in vivo disease outcomes. Antimicrob. Agents Chemother. 2018, 62, e01953-17. [Google Scholar] [CrossRef]
- Willger, S.D.; Cornish, E.J.; Chung, D.; Fleming, B.A.; Lehmann, M.M.; Puttikamonkul, S.; Cramer, R.A. Dsc orthologs are required for hypoxia adaptation, triazole drug responses, and fungal virulence in Aspergillus fumigatus. Eukaryot. Cell 2012, 11, 1557–1567. [Google Scholar] [CrossRef]
- Chung, D.; Barker, B.M.; Carey, C.C.; Merriman, B.; Werner, E.R.; Lechner, B.E.; Dhingra, S.; Cheng, C.; Xu, W.; Blosser, S.J.; et al. ChIP-seq and in vivo transcriptome analyses of the Aspergillus fumigatus SREBP SrbA reveals a new regulator of the fungal hypoxia response and virulence. PLoS Pathog. 2014, 10, e1004487. [Google Scholar] [CrossRef]
- Blosser, S.J.; Merriman, B.; Grahl, N.; Chung, D.; Cramer, R.A. Two C4-sterol methyl oxidases (Erg25) catalyse ergosterol intermediate demethylation and impact environmental stress adaptation in Aspergillus fumigatus. Microbiology 2014, 160 Pt 11, 2492–2506. [Google Scholar] [CrossRef]
- Vaknin, Y.; Hillmann, F.; Iannitti, R.; Ben Baruch, N.; Sandovsky-Losica, H.; Shadkchan, Y.; Romani, L.; Brakhage, A.; Kniemeyer, O.; Osherov, N. Identification and characterization of a novel Aspergillus fumigatus rhomboid family putative protease, RbdA, involved in hypoxia sensing and virulence. Infect. Immun. 2016, 84, 1866–1878. [Google Scholar] [CrossRef] [PubMed]
- Kroll, K.; Shekhova, E.; Mattern, D.J.; Thywissen, A.; Jacobsen, I.D.; Strassburger, M.; Heinekamp, T.; Shelest, E.; Brakhage, A.A.; Kniemeyer, O. The hypoxia-induced dehydrogenase HorA is required for coenzyme Q10 biosynthesis, azole sensitivity and virulence of Aspergillus fumigatus. Mol. Microbiol. 2016, 101, 92–108. [Google Scholar] [CrossRef]
- Barker, B.M.; Kroll, K.; Vödisch, M.; Mazurie, A.; Kniemeyer, O.; Cramer, R.A. Transcriptomic and proteomic analyses of the Aspergillus fumigatus hypoxia response using an oxygen-controlled fermenter. BMC Genom. 2012, 13, 62. [Google Scholar] [CrossRef]
- Losada, L.; Barker, B.M.; Pakala, S.; Pakala, S.; Joardar, V.; Zafar, N.; Mounaud, S.; Fedorova, N.; Nierman, W.C.; Cramer, R.A. Large-scale transcriptional response to hypoxia in Aspergillus fumigatus observed using RNAseq identifies a novel hypoxia regulated ncRNA. Mycopathologia 2014, 178, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Kale, S.D.; Ayubi, T.; Chung, D.; Tubau-Juni, N.; Leber, A.; Dang, H.X.; Karyala, S.; Hontecillas, R.; Lawrence, C.B.; Cramer, R.A.; et al. Modulation of immune signaling and metabolism highlights host and fungal transcriptional responses in mouse models of invasive pulmonary aspergillosis. Sci. Rep. 2017, 7, 17096. [Google Scholar] [CrossRef] [PubMed]
- Vödisch, M.; Scherlach, K.; Winkler, R.; Hertweck, C.; Braun, H.P.; Roth, M.; Haas, H.; Werner, E.R.; Brakhage, A.A.; Kniemeyer, O. Analysis of the Aspergillus fumigatus proteome reveals metabolic changes and the activation of the pseurotin A biosynthesis gene cluster in response to hypoxia. J. Proteome Res. 2011, 10, 2508–2524. [Google Scholar] [CrossRef]
- Kroll, K.; Pähtz, V.; Hillmann, F.; Vaknin, Y.; Schmidt-Heck, W.; Roth, M.; Jacobsen, I.D.; Osherov, N.; Brakhage, A.A.; Kniemeyer, O. Identification of hypoxia-inducible target genes of Aspergillus fumigatus by transcriptome analysis reveals cellular respiration as an important contributor to hypoxic survival. Eukaryot. Cell 2014, 13, 1241–1253. [Google Scholar] [CrossRef]
- Shekhova, E.; Ivanova, L.; Krüger, T.; Stroe, M.C.; Macheleidt, J.; Kniemeyer, O.; Brakhage, A.A. Redox proteomic analysis reveals oxidative modifications of proteins by increased levels of intracellular reactive oxygen species during hypoxia adaptation of Aspergillus fumigatus. Proteomics 2019, 19, e1800339. [Google Scholar] [CrossRef]
- Rees, C.A.; Stefanuto, P.H.; Beattie, S.R.; Bultman, K.M.; Cramer, R.A.; Hill, J.E. Sniffing out the hypoxia volatile metabolic signature of Aspergillus fumigatus. J. Breath Res. 2017, 11, 036003. [Google Scholar] [CrossRef]
- Binder, U.; Maurer, E.; Lackner, M.; Lass-Flörl, C. Effect of reduced oxygen on the antifungal susceptibility of clinically relevant aspergilli. Antimicrob. Agents Chemother. 2015, 59, 1806–1810. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, C.H.; Morelli, K.A.; Schultz, D.; Nadell, C.D.; Cramer, R.A. Fungal biofilm architecture produces hypoxic microenvironments that drive antifungal resistance. Proc. Natl. Acad. Sci. USA 2020, 117, 22473–22483. [Google Scholar] [CrossRef] [PubMed]
- Maurer, E.; Aigner, M.; Lass-Flörl, C.; Binder, U. Hypoxia decreases diagnostic biomarkers for Aspergillosis in vitro. J. Fungi 2019, 5, 61. [Google Scholar] [CrossRef] [PubMed]
- Wezensky, S.J.; Cramer, R.A., Jr. Implications of hypoxic microenvironments during invasive aspergillosis. Med. Mycol. 2011, 49 (Suppl. S1), S120–S124. [Google Scholar] [CrossRef]
- Grahl, N.; Shepardson, K.M.; Chung, D.; Cramer, R.A. Hypoxia and fungal pathogenesis: To air or not to air? Eukaryot. Cell 2012, 11, 560–570. [Google Scholar] [CrossRef]
- Hillmann, F.; Shekhova, E.; Kniemeyer, O. Insights into the cellular responses to hypoxia in filamentous fungi. Curr. Genet. 2015, 61, 441–455. [Google Scholar] [CrossRef]
- Chung, H.; Lee, Y.H. Hypoxia: A double-edged sword during fungal pathogenesis? Front. Microbiol. 2020, 11, 1920. [Google Scholar] [CrossRef]
- Bao, B.; Groves, K.; Zhang, J.; Handy, E.; Kennedy, P.; Cuneo, G.; Supuran, C.T.; Yared, W.; Rajopadhye, M.; Peterson, J.D. In vivo imaging and quantification of carbonic anhydrase IX expression as an endogenous biomarker of tumor hypoxia. PLoS ONE 2012, 7, e50860. [Google Scholar] [CrossRef]
- Carvalho, A.; Cunha, C.; Iannitti, R.G.; De Luca, A.; Giovannini, G.; Bistoni, F.; Romani, L. Inflammation in aspergillosis: The good, the bad, and the therapeutic. Ann. N. Y. Acad. Sci. 2012, 1273, 52–59. [Google Scholar] [CrossRef]
- Fröhlich, S.; Boylan, J.; McLoughlin, P. Hypoxia-induced inflammation in the lung: A potential therapeutic target in acute lung injury? Am. J. Respir. Cell Mol. Biol. 2013, 48, 271–279. [Google Scholar] [CrossRef]
- Rider, P.; Kaplanov, I.; Romzova, M.; Bernardis, L.; Braiman, A.; Voronov, E.; Apte, R.N. The transcription of the alarmin cytokine interleukin-1 alpha is controlled by hypoxia inducible factors 1 and 2 alpha in hypoxic cells. Front. Immunol. 2012, 3, 290. [Google Scholar] [CrossRef] [PubMed]
- Caffrey, A.K.; Lehmann, M.M.; Zickovich, J.M.; Espinosa, V.; Shepardson, K.M.; Watschke, C.P.; Hilmer, K.M.; Thammahong, A.; Barker, B.M.; Rivera, A.; et al. IL-1α signaling is critical for leukocyte recruitment after pulmonary Aspergillus fumigatus challenge. PLoS Pathog. 2015, 11, e1004625. [Google Scholar] [CrossRef]
- Gabay, C.; Lamacchia, C.; Palmer, G. IL-1 pathways in inflammation and human diseases. Nat. Rev. Rheumatol. 2010, 6, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Shepardson, K.M.; Ngo, L.Y.; Aimanianda, V.; Latgé, J.P.; Barker, B.M.; Blosser, S.J.; Iwakura, Y.; Hohl, T.M.; Cramer, R.A. Hypoxia enhances innate immune activation to Aspergillus fumigatus through cell wall modulation. Microbes Infect. 2013, 15, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Fliesser, M.; Wallstein, M.; Kurzai, O.; Einsele, H.; Löffler, J. Hypoxia attenuates anti-Aspergillus fumigatus immune responses initiated by human dendritic cells. Mycoses 2016, 59, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Hutchens, M.; Luker, G.D. Applications of bioluminescence imaging to the study of infectious diseases. Cell. Microbiol. 2007, 9, 2315–2322. [Google Scholar] [CrossRef]
- Contag, C.H.; Contag, P.R.; Mullins, J.I.; Spilman, S.D.; Stevenson, D.K.; Benaron, D.A. Photonic detection of bacterial pathogens in living hosts. Mol. Microbiol. 1995, 18, 593–603. [Google Scholar] [CrossRef]
- Campbell, A.K. Chemiluminescence. Principles and Applications in Biology and Medicine; Ellis Horwood Lld and VCH Verlagsgesellschaft mbH: Chichester, UK, 1988. [Google Scholar]
- Robertson, J.B.; Stowers, C.C.; Boczko, E.; Johnson, C.H. Real-time luminescence monitoring of cell-cycle and respiratory oscillations in yeast. Proc. Natl. Acad. Sci. USA 2008, 105, 17988–17993. [Google Scholar] [CrossRef]
- Hastings, J.W.; McElroy, W.D.; Coulombre, J. The effect of oxygen upon the immobilization reaction in firefly luminescence. J. Cell. Comp. Physiol. 1953, 42, 137–150. [Google Scholar] [CrossRef]
- Poelmans, J.; Himmelreich, U.; Vanherp, L.; Zhai, L.; Hillen, A.; Holvoet, B.; Belderbos, S.; Brock, M.; Maertens, J.; Vande Velde, G.; et al. A multimodal imaging approach enables in vivo assessment of antifungal treatment in a mouse model of invasive pulmonary aspergillosis. Antimicrob. Agents Chemother. 2018, 62, e00240-18. [Google Scholar] [CrossRef]
- Davies, G.; Rolle, A.M.; Maurer, A.; Spycher, P.R.; Schillinger, C.; Solouk-Saran, D.; Hasenberg, M.; Weski, J.; Fonslet, J.; Dubois, A.; et al. Towards translational immunoPET/MR imaging of invasive pulmonary aspergillosis: The humanised monoclonal antibody JF5 detects Aspergillus lung infections in vivo. Theranostics 2017, 7, 3398–3414. [Google Scholar] [CrossRef] [PubMed]
- Thornton, C.R. Molecular imaging of invasive pulmonary aspergillosis using immunoPET/MRI: The future looks bright. Front. Microbiol. 2018, 9, 691. [Google Scholar] [CrossRef] [PubMed]
- Henneberg, S.; Hasenberg, A.; Maurer, A.; Neumann, F.; Bornemann, L.; Gonzalez-Menendez, I.; Kraus, A.; Hasenberg, M.; Thornton, C.R.; Pichler, B.J.; et al. Antibody-guided in vivo imaging of Aspergillus fumigatus lung infections during antifungal azole treatment. Nat. Commun. 2021, 12, 1707. [Google Scholar] [CrossRef] [PubMed]
- Kent, B.D.; Mitchell, P.D.; McNicholas, W.T. Hypoxemia in patients with COPD: Cause, effects, and disease progression. Int. J. Chronic Obstr. Pulm. Dis. 2011, 6, 199–208. [Google Scholar] [CrossRef]
- Montgomery, S.T.; Mall, M.A.; Kicic, A.; Stick, S.M.; Arest, C.F. Hypoxia and sterile inflammation in cystic fibrosis airways: Mechanisms and potential therapies. Eur. Respir. J. 2017, 49, 1600903. [Google Scholar] [CrossRef]
- Fritzsching, B.; Zhou-Suckow, Z.; Trojanek, J.B.; Schubert, S.C.; Schatterny, J.; Hirtz, S.; Agrawal, R.; Muley, T.; Kahn, N.; Sticht, C.; et al. Hypoxic epithelial necrosis triggers neutrophilic inflammation via IL-1 receptor signaling in cystic fibrosis lung disease. Am. J. Respir. Crit. Care Med. 2015, 191, 902–913. [Google Scholar] [CrossRef]
- Bignell, E.; Negrete-Urtasun, S.; Calcagno, A.M.; Haynes, K.; Arst, H.N.; Rogers, T., Jr. The Aspergillus pH-responsive transcription factor PacC regulates virulence. Mol. Microbiol. 2005, 55, 1072–1084. [Google Scholar] [CrossRef]
- Barda, O.; Maor, U.; Sadhasivam, S.; Bi, Y.; Zakin, V.; Prusky, D.; Sionov, E. The pH-responsive transcription factor PacC governs pathogenicity and ochratoxin A biosynthesis in Aspergillus carbonarius. Front. Microbiol. 2020, 11, 210. [Google Scholar] [CrossRef]
- Amich, J.; Vicentefranqueira, R.; Mellado, E.; Ruiz-Carmuega, A.; Leal, F.; Calera, J.A. The ZrfC alkaline zinc transporter is required for Aspergillus fumigatus virulence and its growth in the presence of the Zn/Mn-chelating protein calprotectin. Cell. Microbiol. 2014, 16, 548–564. [Google Scholar] [CrossRef]
- Amich, J.; Calera, J.A. Zinc acquisition: A key aspect in Aspergillus fumigatus virulence. Mycopathologia 2014, 178, 379–385. [Google Scholar] [CrossRef]
- Berezin, M.Y.; Guo, K.; Akers, W.; Northdurft, R.E.; Culver, J.P.; Teng, B.; Vasalatiy, O.; Barbacow, K.; Gandjbakhche, A.; Griffiths, G.L.; et al. Near-infrared fluorescence lifetime pH-sensitive probes. Biophys. J. 2011, 100, 2063–2072. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ghosh, S.K.; Kim, P.; Zhang, X.A.; Yun, S.H.; Moore, A.; Lippard, S.J.; Medarova, Z. A novel imaging approach for early detection of prostate cancer based on endogenous zinc sensing. Cancer Res. 2010, 70, 6119–6127. [Google Scholar] [CrossRef] [PubMed]
- Schaffner, A.; Douglas, H.; Braude, A. Selective protection against conidia by mononuclear and against mycelia by polymorphonuclear phagocytes in resistance to Aspergillus. Observations on these two lines of defense in vivo and in vitro with human and mouse phagocytes. J. Clin. Investig. 1982, 69, 617–631. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim-Granet, O.; Philippe, B.; Boleti, H.; Boisvieux-Ulrich, E.; Grenet, D.; Stern, M.; Latgé, J.P. Phagocytosis and intracellular fate of Aspergillus fumigatus conidia in alveolar macrophages. Infect. Immun. 2003, 71, 891–903. [Google Scholar] [CrossRef]
- Philippe, B.; Ibrahim-Granet, O.; Prévost, M.C.; Gougerot-Pocidalo, M.A.; Sanchez Perez, M.; Van der Meeren, A.; Latgé, J.P. Killing of Aspergillus fumigatus by alveolar macrophages is mediated by reactive oxidant intermediates. Infect. Immun. 2003, 71, 3034–3042. [Google Scholar] [CrossRef] [PubMed]
- Volling, K.; Thywissen, A.; Brakhage, A.A.; Saluz, H.P. Phagocytosis of melanized Aspergillus conidia by macrophages exerts cytoprotective effects by sustained PI3K/Akt signalling. Cell. Microbiol. 2011, 13, 1130–1148. [Google Scholar] [CrossRef]
- Mircescu, M.M.; Lipuma, L.; van Rooijen, N.; Pamer, E.G.; Hohl, T.M. Essential role for neutrophils but not alveolar macrophages at early time points following Aspergillus fumigatus infection. J. Infect. Dis. 2009, 200, 647–656. [Google Scholar] [CrossRef]
- Tanaka, R.J.; Boon, N.J.; Vrcelj, K.; Nguyen, A.; Vinci, C.; Armstrong-James, D.; Bignell, E. In silico modeling of spore inhalation reveals fungal persistence following low dose exposure. Sci. Rep. 2015, 5, 13958. [Google Scholar] [CrossRef]
- Pollmächer, J.; Figge, M.T. Agent-based model of human alveoli predicts chemotactic signaling by epithelial cells during early Aspergillus fumigatus infection. PLoS ONE 2014, 9, e111630. [Google Scholar] [CrossRef]
- Bonnett, C.R.; Cornish, E.J.; Harmsen, A.G.; Burritt, J.B. Early neutrophil recruitment and aggregation in the murine lung inhibit germination of Aspergillus fumigatus conidia. Infect. Immun. 2006, 74, 6528–6539. [Google Scholar] [CrossRef]
- Feldman, M.B.; Dutko, R.A.; Wood, M.A.; Ward, R.A.; Leung, H.M.; Snow, R.F.; De La Flor, D.J.; Yonker, L.M.; Reedy, J.L.; Tearney, G.J.; et al. Aspergillus fumigatus cell wall promotes apical airway epithelial recruitment of human neutrophils. Infect. Immun. 2020, 88, e00813-19. [Google Scholar] [CrossRef] [PubMed]
- Crapo, J.D.; Barry, B.E.; Gehr, P.; Bachofen, M.; Weibel, E.R. Cell number and cell characteristics of the normal human lung. Am. Rev. Respir. Dis. 1982, 126, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Bertuzzi, M.; Hayes, G.E.; Bignell, E.M. Microbial uptake by the respiratory epithelium: Outcomes for host and pathogen. FEMS Microbiol. Rev. 2019, 43, 145–161. [Google Scholar] [CrossRef] [PubMed]
- Ewald, J.; Rivieccio, F.; Radosa, L.; Schuster, S.; Brakhage, A.A.; Kaleta, C. Dynamic optimization reveals alveolar epithelial cells as key mediators of host defense in invasive aspergillosis. PLoS Comput. Biol. 2021, 17, e1009645. [Google Scholar] [CrossRef]
- Blickensdorf, M.; Timme, S.; Figge, M.T. Hybrid agent-based modeling of Aspergillus fumigatus Infection to quantitatively investigate the role of Pores of Kohn in human alveoli. Front. Microbiol. 2020, 11, 1951. [Google Scholar] [CrossRef]
- Pollmächer, J.; Figge, M.T. Deciphering chemokine properties by a hybrid agent-based model of Aspergillus fumigatus infection in human alveoli. Front. Microbiol. 2015, 6, 503. [Google Scholar] [CrossRef]
- Wasylnka, J.A.; Moore, M.M. Uptake of Aspergillus fumigatus conidia by phagocytic and nonphagocytic cells in vitro: Quantitation using strains expressing green fluorescent protein. Infect. Immun. 2002, 70, 3156–3163. [Google Scholar] [CrossRef]
- Han, X.; Yu, R.; Zhen, D.; Tao, S.; Schmidt, M.; Han, L. β-1,3-Glucan-induced host phospholipase D activation is involved in Aspergillus fumigatus internalization into type II human pneumocyte A549 cells. PLoS ONE 2011, 6, e21468. [Google Scholar] [CrossRef]
- Wasylnka, J.A.; Moore, M.M. Aspergillus fumigatus conidia survive and germinate in acidic organelles of A549 epithelial cells. J. Cell Sci. 2003, 116 Pt 8, 1579–1587. [Google Scholar] [CrossRef]
- Oosthuizen, J.L.; Gomez, P.; Ruan, J.; Hackett, T.L.; Moore, M.M.; Knight, D.A.; Tebbutt, S.J. Dual organism transcriptomics of airway epithelial cells interacting with conidia of Aspergillus fumigatus. PLoS ONE 2011, 6, e20527. [Google Scholar] [CrossRef]
- Gomez, P.; Hackett, T.L.; Moore, M.M.; Knight, D.A.; Tebbutt, S.J. Functional genomics of human bronchial epithelial cells directly interacting with conidia of Aspergillus fumigatus. BMC Genom. 2010, 11, 358. [Google Scholar] [CrossRef] [PubMed]
- Paris, S.; Boisvieux-Ulrich, E.; Crestani, B.; Houcine, O.; Taramelli, D.; Lombardi, L.; Latgé, J.P. Internalization of Aspergillus fumigatus conidia by epithelial and endothelial cells. Infect. Immun. 1997, 65, 1510–1514. [Google Scholar] [CrossRef] [PubMed]
- Seidel, C.; Moreno-Velásquez, S.D.; Ben-Ghazzi, N.; Gago, S.; Read, N.D.; Bowyer, P. Phagolysosomal survival enables non-lytic hyphal escape and ramification through lung epithelium during Aspergillus fumigatus infection. Front. Microbiol. 2020, 11, 1955. [Google Scholar] [CrossRef] [PubMed]
- Amitani, R.; Kawanami, R. Interaction of Aspergillus with human respiratory mucosa: A study with organ culture model. Med. Mycol. 2009, 47 (Suppl. S1), S127–S131. [Google Scholar] [CrossRef] [PubMed]
- Ben-Ghazzi, N.; Moreno-Velásquez, S.; Seidel, C.; Thomson, D.; Denning, D.W.; Read, N.D.; Bowyer, P.; Gago, S. Characterisation of Aspergillus fumigatus endocytic trafficking within airway epithelial cells using high-resolution automated quantitative confocal microscopy. J. Fungi 2021, 7, 454. [Google Scholar] [CrossRef]
- Keizer, E.M.; Wösten, H.; de Cock, H. EphA2-dependent internalization of A. fumigatus conidia in A549 lung cells is modulated by DHN-melanin. Front. Microbiol. 2020, 11, 534118. [Google Scholar] [CrossRef]
- Fernandes, J.; Hamidi, F.; Leborgne, R.; Beau, R.; Castier, Y.; Mordant, P.; Boukkerou, A.; Latgé, J.P.; Pretolani, M. Penetration of the human pulmonary epithelium by Aspergillus fumigatus hyphae. J. Infect. Dis. 2018, 218, 1306–1313. [Google Scholar] [CrossRef]
- Bidula, S.; Kenawy, H.; Ali, Y.M.; Sexton, D.; Schwaeble, W.J.; Schelenz, S. Role of ficolin-A and lectin complement pathway in the innate defense against pathogenic Aspergillus species. Infect. Immun. 2013, 81, 1730–1740. [Google Scholar] [CrossRef]
- Jepsen, C.S.; Dubey, L.K.; Colmorten, K.B.; Moeller, J.B.; Hammond, M.A.; Nielsen, O.; Schlosser, A.; Templeton, S.P.; Sorensen, G.L.; Holmskov, U. FIBCD1 binds Aspergillus fumigatus and regulates lung epithelial response to cell wall components. Front. Immunol. 2018, 9, 1967. [Google Scholar] [CrossRef]
- Khosravi, A.R.; Alheidary, S.; Nikaein, D.; Asghari, N. Aspergillus fumigatus conidia stimulate lung epithelial cells (TC-1 JHU-1) to produce IL-12, IFNγ, IL-13 and IL-17 cytokines: Modulatory effect of propolis extract. J. Mycol. Med. 2018, 28, 594–598. [Google Scholar] [CrossRef]
- Beisswenger, C.; Hess, C.; Bals, R. Aspergillus fumigatus conidia induce interferon-β signalling in respiratory epithelial cells. Eur. Respir. J. 2012, 39, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Balloy, V.; Sallenave, J.M.; Wu, Y.; Touqui, L.; Latgé, J.P.; Si-Tahar, M.; Chignard, M. Aspergillus fumigatus-induced interleukin-8 synthesis by respiratory epithelial cells is controlled by the phosphatidylinositol 3-kinase, p38 MAPK, and ERK1/2 pathways and not by the toll-like receptor-MyD88 pathway. J. Biol. Chem. 2008, 283, 30513–30521. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liu, R.; Noordhoek, J.A.; Kauffman, H.F. Interaction of airway epithelial cells (A549) with spores and mycelium of Aspergillus fumigatus. J. Infect. 2005, 51, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Jhingran, A.; Kasahara, S.; Shepardson, K.M.; Junecko, B.A.; Heung, L.J.; Kumasaka, D.K.; Knoblaugh, S.E.; Lin, X.; Kazmierczak, B.I.; Reinhart, T.A.; et al. Compartment-specific and sequential role of MyD88 and CARD9 in chemokine induction and innate defense during respiratory fungal infection. PLoS Pathog. 2015, 11, e1004589. [Google Scholar] [CrossRef]
- Okaa, U.J.; Bertuzzi, M.; Fortune-Grant, R.; Thomson, D.D.; Moyes, D.L.; Naglik, J.R.; Bignell, E. Aspergillus fumigatus drives tissue damage via iterative assaults upon mucosal integrity and immune homeostasis. bioRxiv 2021. [CrossRef]
- Osherov, N. Interaction of the pathogenic mold Aspergillus fumigatus with lung epithelial cells. Front. Microbiol. 2012, 3, 346. [Google Scholar] [CrossRef]
- Bigot, J.; Guillot, L.; Guitard, J.; Ruffin, M.; Corvol, H.; Balloy, V.; Hennequin, C. Bronchial epithelial cells on the front line to fight lung infection-causing Aspergillus fumigatus. Front. Immunol. 2020, 11, 1041. [Google Scholar] [CrossRef]
- Bertuzzi, M.; Howell, G.J. Single-cell analysis of fungal uptake in cultured airway epithelial cells using differential fluorescent staining and imaging flow cytometry. Methods Mol. Biol. 2021, 2260, 83–109. [Google Scholar] [CrossRef]
- Chaudhary, N.; Datta, K.; Askin, F.B.; Staab, J.F.; Marr, K.A. Cystic fibrosis transmembrane conductance regulator regulates epithelial cell response to Aspergillus and resultant pulmonary inflammation. Am. J. Respir. Crit. Care Med. 2012, 185, 301–310. [Google Scholar] [CrossRef]
- Nemecek, J.C.; Wüthrich, M.; Klein, B.S. Global control of dimorphism and virulence in fungi. Science 2006, 312, 583–588. [Google Scholar] [CrossRef]
- Köhler, J.R.; Casadevall, A.; Perfect, J. The spectrum of fungi that infects humans. Cold Spring Harb. Perspect. Med. 2014, 5, a019273. [Google Scholar] [CrossRef] [PubMed]
- Sephton-Clark, P.; Voelz, K. Spore germination of pathogenic filamentous fungi. Adv. Appl. Microbiol. 2018, 102, 117–157. [Google Scholar] [CrossRef] [PubMed]
- Hohl, T.M.; Van Epps, H.L.; Rivera, A.; Morgan, L.A.; Chen, P.L.; Feldmesser, M.; Pamer, E.G. Aspergillus fumigatus triggers inflammatory responses by stage-specific beta-glucan display. PLoS Pathog. 2005, 1, e30. [Google Scholar] [CrossRef] [PubMed]
- Steele, C.; Rapaka, R.R.; Metz, A.; Pop, S.M.; Williams, D.L.; Gordon, S.; Kolls, J.K.; Brown, G.D. The beta-glucan receptor dectin-1 recognizes specific morphologies of Aspergillus fumigatus. PLoS Pathog. 2005, 1, e42. [Google Scholar] [CrossRef]
- Gersuk, G.M.; Underhill, D.M.; Zhu, L.; Marr, K.A. Dectin-1 and TLRs permit macrophages to distinguish between different Aspergillus fumigatus cellular states. J. Immunol. 2006, 176, 3717–3724. [Google Scholar] [CrossRef]
- Bozza, S.; Gaziano, R.; Spreca, A.; Bacci, A.; Montagnoli, C.; di Francesco, P.; Romani, L. Dendritic cells transport conidia and hyphae of Aspergillus fumigatus from the airways to the draining lymph nodes and initiate disparate Th responses to the fungus. J. Immunol. 2002, 168, 1362–1371. [Google Scholar] [CrossRef]
- Netea, M.G.; Warris, A.; Van der Meer, J.W.; Fenton, M.J.; Verver-Janssen, T.J.; Jacobs, L.E.; Andresen, T.; Verweij, P.E.; Kullberg, B.J. Aspergillus fumigatus evades immune recognition during germination through loss of toll-like receptor-4-mediated signal transduction. J. Infect. Dis. 2003, 188, 320–326. [Google Scholar] [CrossRef]
- Gazendam, R.P.; van Hamme, J.L.; Tool, A.T.; Hoogenboezem, M.; van den Berg, J.M.; Prins, J.M.; Vitkov, L.; van de Veerdonk, F.L.; van den Berg, T.K.; Roos, D.; et al. Human neutrophils use different mechanisms to kill Aspergillus fumigatus conidia and hyphae: Evidence from phagocyte defects. J. Immunol. 2016, 196, 1272–1283. [Google Scholar] [CrossRef]
- Alekseeva, L.; Huet, D.; Féménia, F.; Mouyna, I.; Abdelouahab, M.; Cagna, A.; Guerrier, D.; Tichanné-Seltzer, V.; Baeza-Squiban, A.; Chermette, R.; et al. Inducible expression of beta defensins by human respiratory epithelial cells exposed to Aspergillus fumigatus organisms. BMC Microbiol. 2009, 9, 33. [Google Scholar] [CrossRef]
- Loeffler, J.; Haddad, Z.; Bonin, M.; Romeike, N.; Mezger, M.; Schumacher, U.; Kapp, M.; Gebhardt, F.; Grigoleit, G.U.; Stevanović, S.; et al. Interaction analyses of human monocytes co-cultured with different forms of Aspergillus fumigatus. J. Med. Microbiol. 2009, 58 Pt 1, 49–58. [Google Scholar] [CrossRef]
- Shah, A.; Kannambath, S.; Herbst, S.; Rogers, A.; Soresi, S.; Carby, M.; Reed, A.; Mostowy, S.; Fisher, M.C.; Shaunak, S.; et al. Calcineurin orchestrates lateral transfer of Aspergillus fumigatus during macrophage cell death. Am. J. Respir. Crit. Care Med. 2016, 194, 1127–1139. [Google Scholar] [CrossRef] [PubMed]
- Levitz, S.M.; Diamond, R.D. Mechanisms of resistance of Aspergillus fumigatus conidia to killing by neutrophils in vitro. J. Infect. Dis. 1985, 152, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Ellett, F.; Jorgensen, J.; Frydman, G.H.; Jones, C.N.; Irimia, D. Neutrophil interactions stimulate evasive hyphal branching by Aspergillus fumigatus. PLoS Pathog. 2017, 13, e1006154. [Google Scholar] [CrossRef] [PubMed]
- Schaffner, A.; Douglas, H.; Braude, A.I.; Davis, C.E. Killing of Aspergillus spores depends on the anatomical source of the macrophage. Infect. Immun. 1983, 42, 1109–1115. [Google Scholar] [CrossRef]
- Cornish, E.J.; Hurtgen, B.J.; McInnerney, K.; Burritt, N.L.; Taylor, R.M.; Jarvis, J.N.; Wang, S.Y.; Burritt, J.B. Reduced nicotinamide adenine dinucleotide phosphate oxidase-independent resistance to Aspergillus fumigatus in alveolar macrophages. J. Immunol. 2008, 180, 6854–6867. [Google Scholar] [CrossRef]
- Grimm, M.J.; Vethanayagam, R.R.; Almyroudis, N.G.; Dennis, C.G.; Khan, A.N.; D’Auria, A.C.; Singel, K.L.; Davidson, B.A.; Knight, P.R.; Blackwell, T.S.; et al. Monocyte- and macrophage-targeted NADPH oxidase mediates antifungal host defense and regulation of acute inflammation in mice. J. Immunol. 2013, 190, 4175–4184. [Google Scholar] [CrossRef]
- Botterel, F.; Gross, K.; Ibrahim-Granet, O.; Khoufache, K.; Escabasse, V.; Coste, A.; Cordonnier, C.; Escudier, E.; Bretagne, S. Phagocytosis of Aspergillus fumigatus conidia by primary nasal epithelial cells in vitro. BMC Microbiol. 2008, 8, 97. [Google Scholar] [CrossRef]
- Escobar, N.; Ordonez, S.R.; Wösten, H.A.; Haas, P.J.; de Cock, H.; Haagsman, H.P. Hide, keep quiet, and keep low: Properties that make Aspergillus fumigatus a successful lung pathogen. Front. Microbiol. 2016, 7, 438. [Google Scholar] [CrossRef]
- Richard, N.; Marti, L.; Varrot, A.; Guillot, L.; Guitard, J.; Hennequin, C.; Imberty, A.; Corvol, H.; Chignard, M.; Balloy, V. Human bronchial epithelial cells inhibit Aspergillus fumigatus germination of extracellular conidia via FleA recognition. Sci. Rep. 2018, 8, 15699. [Google Scholar] [CrossRef]
- Araujo, R.; Rodrigues, A.G. Variability of germinative potential among pathogenic species of Aspergillus. J. Clin. Microbiol. 2004, 42, 4335–4337. [Google Scholar] [CrossRef]
- Gresnigt, M.S.; Becker, K.L.; Leenders, F.; Alonso, M.F.; Wang, X.; Meis, J.F.; Bain, J.M.; Erwig, L.P.; van de Veerdonk, F.L. Differential kinetics of Aspergillus nidulans and Aspergillus fumigatus phagocytosis. J. Innate Immun. 2018, 10, 145–160. [Google Scholar] [CrossRef] [PubMed]
- Paulussen, C.; Hallsworth, J.E.; Álvarez-Pérez, S.; Nierman, W.C.; Hamill, P.G.; Blain, D.; Rediers, H.; Lievens, B. Ecology of aspergillosis: Insights into the pathogenic potency of Aspergillus fumigatus and some other Aspergillus species. Microb. Biotechnol. 2017, 10, 296–322. [Google Scholar] [CrossRef] [PubMed]
- Keizer, E.M.; Valdes, I.D.; Forn-Cuni, G.; Klijn, E.; Meijer, A.H.; Hillmann, F.; Wösten, H.; de Cock, H. Variation of virulence of five Aspergillus fumigatus isolates in four different infection models. PLoS ONE 2021, 16, e0252948. [Google Scholar] [CrossRef] [PubMed]
- Caffrey-Carr, A.K.; Kowalski, C.H.; Beattie, S.R.; Blaseg, N.A.; Upshaw, C.R.; Thammahong, A.; Lust, H.E.; Tang, Y.W.; Hohl, T.M.; Cramer, R.A.; et al. Interleukin 1α is critical for resistance against highly virulent Aspergillus fumigatus isolates. Infect. Immun. 2017, 85, e00661-17. [Google Scholar] [CrossRef]
- Knox, B.P.; Blachowicz, A.; Palmer, J.M.; Romsdahl, J.; Huttenlocher, A.; Wang, C.C.; Keller, N.P.; Venkateswaran, K. Characterization of Aspergillus fumigatus isolates from air and surfaces of the international space station. mSphere 2016, 1, e00227-16. [Google Scholar] [CrossRef]
- Young, A.M.; Palmer, A.E. Methods to illuminate the role of Salmonella effector proteins during infection: A review. Front. Cell. Infect. Microbiol. 2017, 7, 363. [Google Scholar] [CrossRef]
- Knox, B.P.; Deng, Q.; Rood, M.; Eickhoff, J.C.; Keller, N.P.; Huttenlocher, A. Distinct innate immune phagocyte responses to Aspergillus fumigatus conidia and hyphae in zebrafish larvae. Eukaryot. Cell 2014, 13, 1266–1277. [Google Scholar] [CrossRef]
- Knox, B.P.; Huttenlocher, A.; Keller, N.P. Real-time visualization of immune cell clearance of Aspergillus fumigatus spores and hyphae. Fungal Genet. Biol. 2017, 105, 52–54. [Google Scholar] [CrossRef]
- Rosowski, E.E.; Knox, B.P.; Archambault, L.S.; Huttenlocher, A.; Keller, N.P.; Wheeler, R.T.; Davis, J.M. The zebrafish as a model host for invasive fungal infections. J. Fungi 2018, 4, 136. [Google Scholar] [CrossRef]
- Koch, B.; Hajdamowicz, N.H.; Lagendijk, E.; Ram, A.; Meijer, A.H. Aspergillus fumigatus establishes infection in zebrafish by germination of phagocytized conidia, while Aspergillus niger relies on extracellular germination. Sci. Rep. 2019, 9, 12791. [Google Scholar] [CrossRef]
- Rosowski, E.E. Illuminating macrophage contributions to host-pathogen interactions in vivo: The power of zebrafish. Infect. Immun. 2020, 88, e00906-19. [Google Scholar] [CrossRef] [PubMed]
- Thrikawala, S.; Rosowski, E.E. Infection of zebrafish larvae with Aspergillus spores for analysis of host-pathogen interactions. J. Vis. Exp. JoVE 2020, 159, e61165. [Google Scholar] [CrossRef] [PubMed]
- Meeker, N.D.; Trede, N.S. Immunology and zebrafish: Spawning new models of human disease. Dev. Comp. Immunol. 2008, 32, 745–757. [Google Scholar] [CrossRef] [PubMed]
- Hamied, A.; Alnedawy, Q.; Correia, A.; Hacker, C.; Ramsdale, M.; Hashimoto, H.; Kudoh, T. Identification and characterization of highly fluorescent pigment cells in embryos of the Arabian killifish (Aphanius dispar). iScience 2020, 23, 101674. [Google Scholar] [CrossRef]
| Light Sheet Fluorescence Microscopy (LSFM) | Fluorescence Tomography/Bioluminescence Tomography | Imaging Flow Cytometry (IFC) | Zebrafish Model of Aspergillosis | |
|---|---|---|---|---|
| Advantages |
|
|
|
|
| Disadvantages |
|
|
|
|
| Processing |
|
|
|
|
| Key Study | Amich et al. (2020) [15] | Gresnigt et al. (2016) [16] | Bertuzzi et al. (2022) [17] | Rosowski et al. (2018) [18] |
| Alternatives | Histology: labour intensive and practically unfeasible for whole lungs, potentially resulting in sampling bias;Confocal laser-scanning microscopy: requires same clearing process as LSFM and has similar disadvantages but may provide higher spatial resolution; however, the imaging depth is limited to approx. 1mm fluorescence signal-pending;ImmunoPET/MRI: similar to fluorescence tomography but primarily being explored as a diagnostic tool. | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ortiz, S.C.; Pennington, K.; Thomson, D.D.; Bertuzzi, M. Novel Insights into Aspergillus fumigatus Pathogenesis and Host Response from State-of-the-Art Imaging of Host–Pathogen Interactions during Infection. J. Fungi 2022, 8, 264. https://doi.org/10.3390/jof8030264
Ortiz SC, Pennington K, Thomson DD, Bertuzzi M. Novel Insights into Aspergillus fumigatus Pathogenesis and Host Response from State-of-the-Art Imaging of Host–Pathogen Interactions during Infection. Journal of Fungi. 2022; 8(3):264. https://doi.org/10.3390/jof8030264
Chicago/Turabian StyleOrtiz, Sébastien C., Katie Pennington, Darren D. Thomson, and Margherita Bertuzzi. 2022. "Novel Insights into Aspergillus fumigatus Pathogenesis and Host Response from State-of-the-Art Imaging of Host–Pathogen Interactions during Infection" Journal of Fungi 8, no. 3: 264. https://doi.org/10.3390/jof8030264
APA StyleOrtiz, S. C., Pennington, K., Thomson, D. D., & Bertuzzi, M. (2022). Novel Insights into Aspergillus fumigatus Pathogenesis and Host Response from State-of-the-Art Imaging of Host–Pathogen Interactions during Infection. Journal of Fungi, 8(3), 264. https://doi.org/10.3390/jof8030264






