Genome Sequence and Analysis of the Flavinogenic Yeast Candida membranifaciens IST 626
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Identification of Candida membranifaciens Isolates
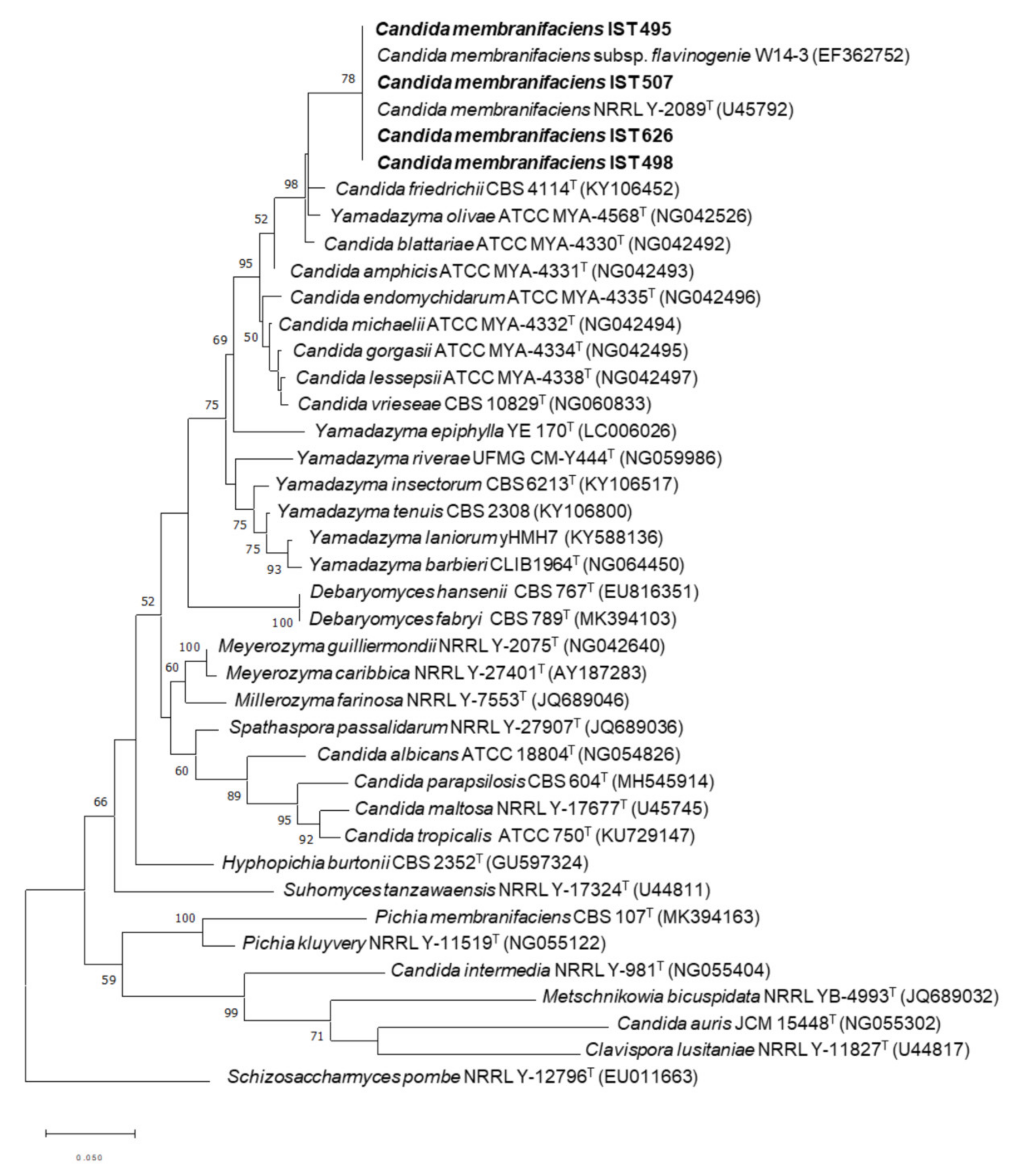
2.2. Phylogenetic Analysis
2.3. Yeast Strains Tested and Growth Media and Conditions Used
2.4. Quantification of Riboflavin
2.5. Genome Sequencing, Assembly, and Annotation
2.6. Analysis of Genome Content
2.7. Search for Putative Transcription Factor Binding Sites
3. Results and Discussion
3.1. Isolation and Identification of Candida membranifaciens Isolates
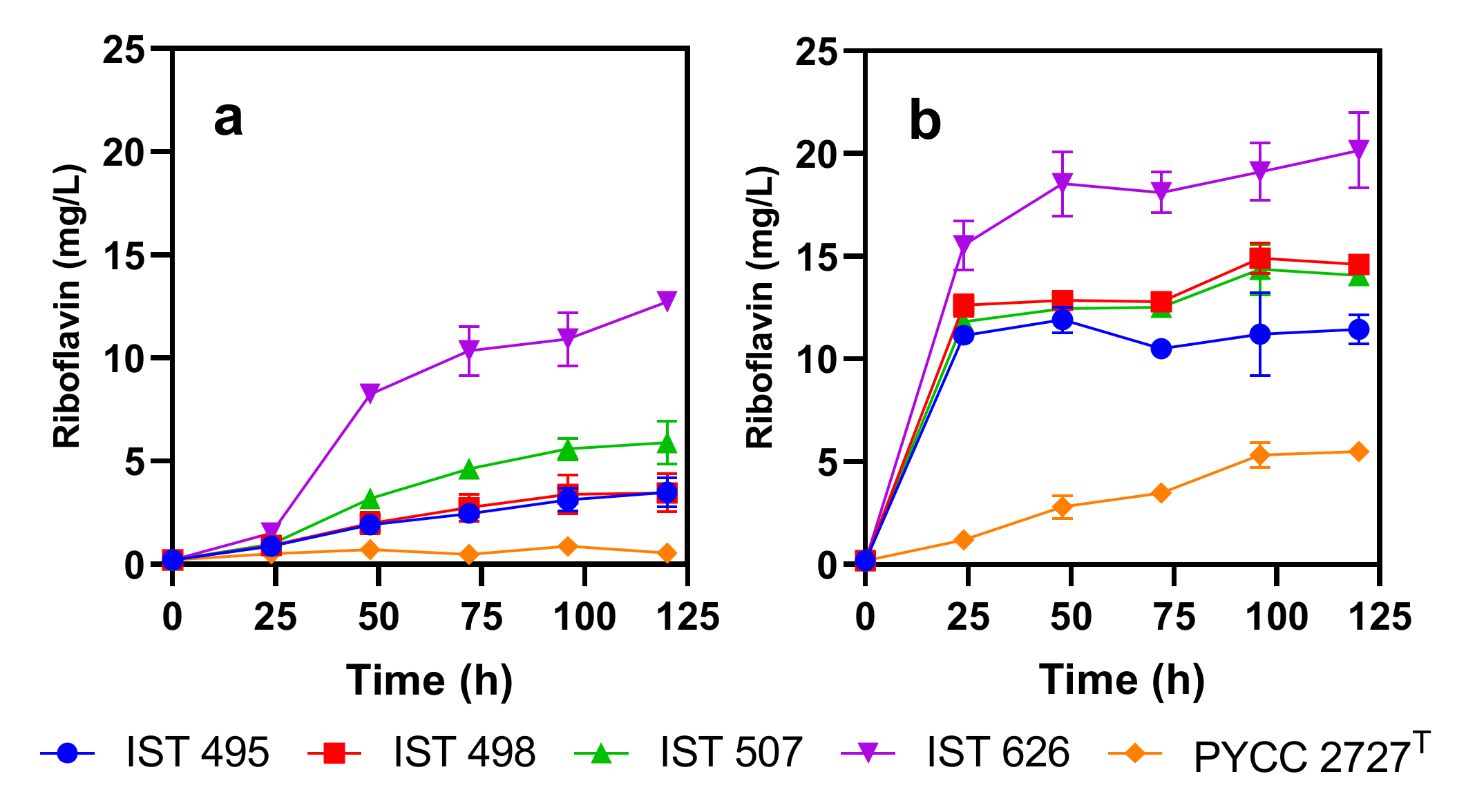
3.2. Candida membranifaciens Isolates Are Riboflavin Producers
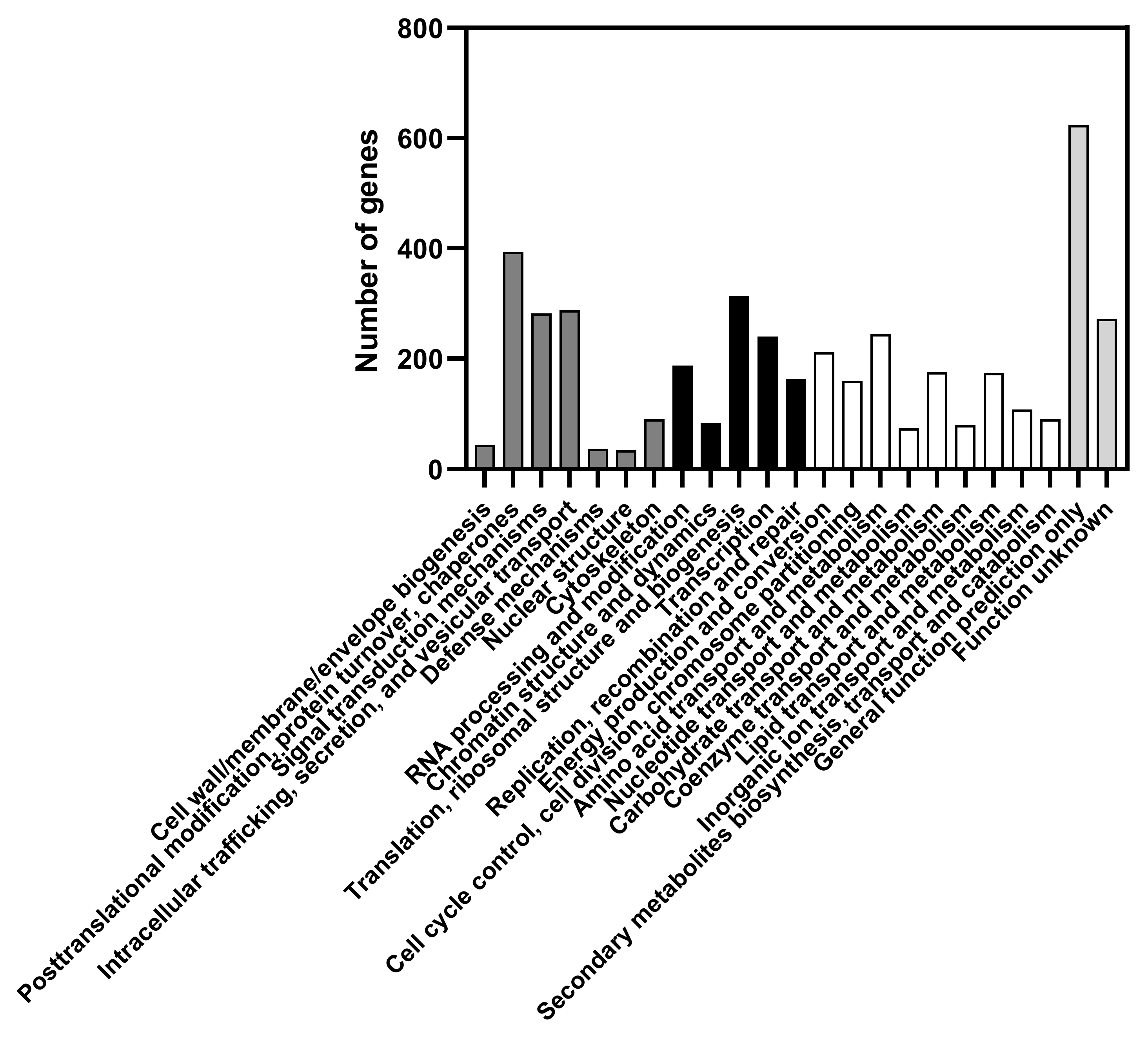
3.3. General features of Candida membranifaciens IST 626 Genome
3.4. Genomic Information and Corresponding Metabolic Traits
3.4.1. Proteins Associated with Riboflavin Production and Transport
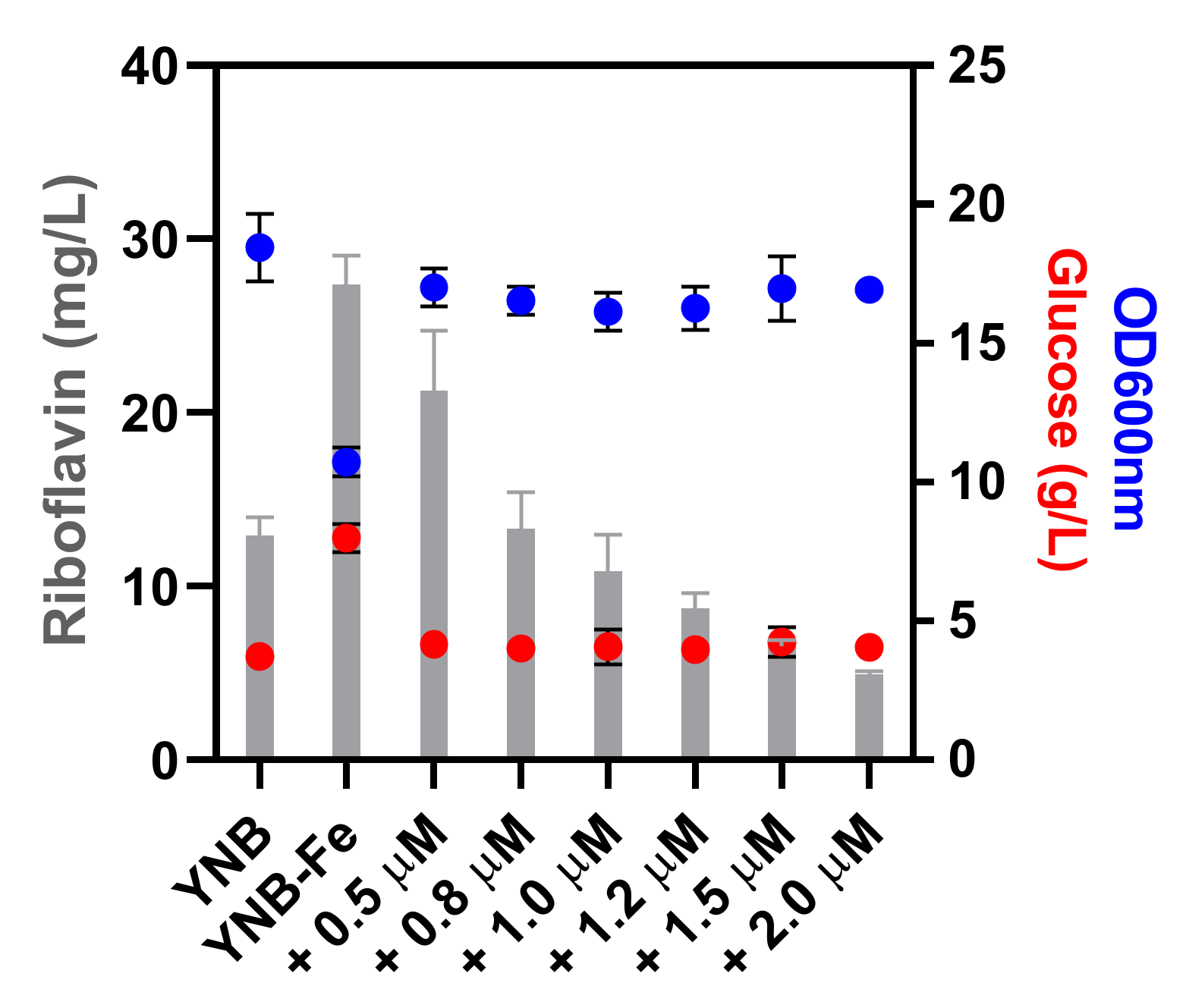
3.4.2. Proteins Associated with Iron Homeostasis and Regulation
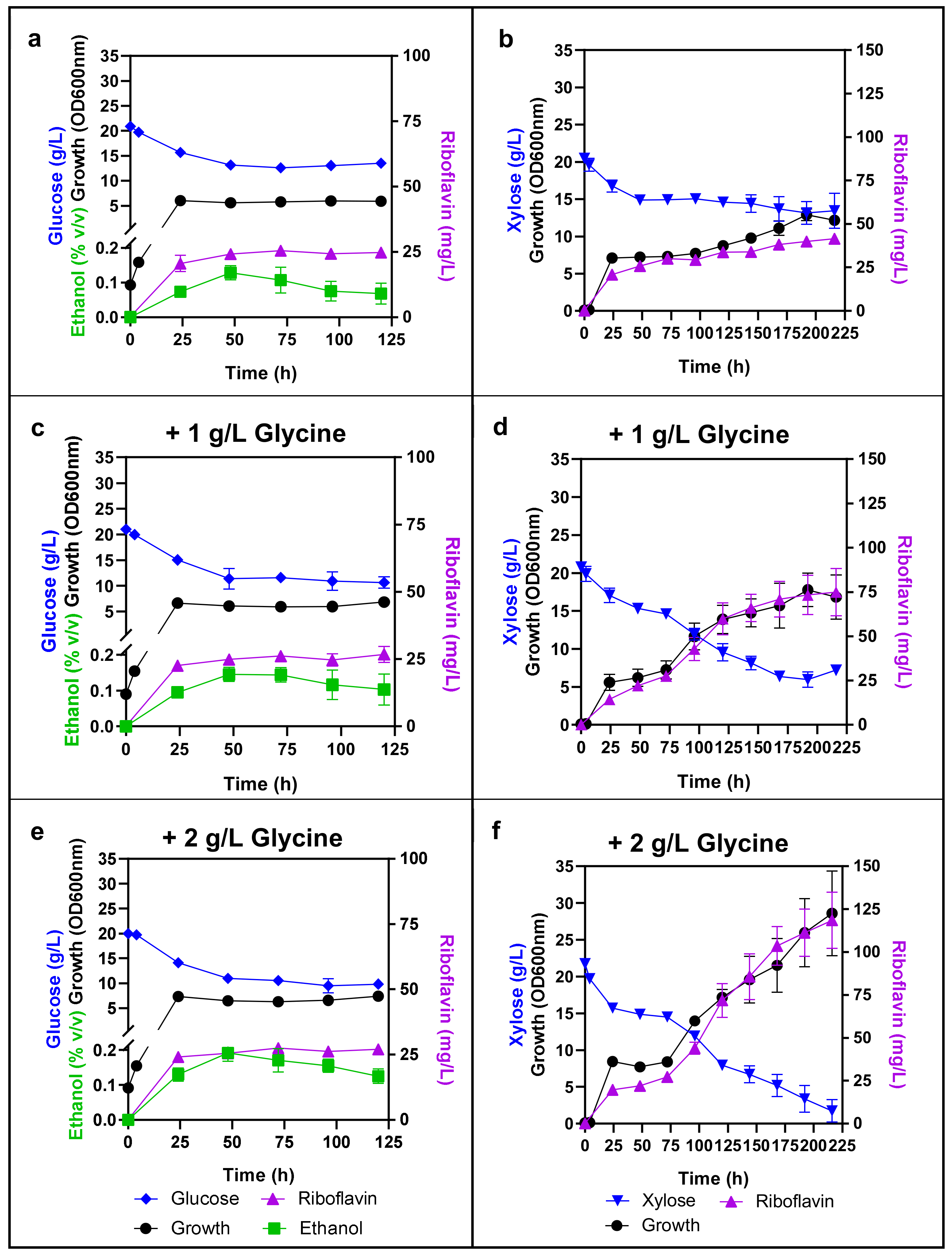
3.4.3. Proteins Associated with Sugar Transport and Metabolism
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lodder, J.; Kreger-van Rij, N.J.W. The Yeasts, a Taxonomic Study; North-Holland Publishing Co.: Amsterdam, The Netherlands, 1952. [Google Scholar]
- Wickerham, L.J.; Burton, K.A. A Clarification of the Relationship of Candida guilliermondii to Other Yeasts by a Study of Their Mating Types. J. Bacteriol. 1954, 68, 594–597. [Google Scholar] [CrossRef]
- Kurtzman, C.; Suzuki, M. Phylogenetic Analysis of Ascomycete Yeasts That Form Coenzyme Q-9 and the Proposal of the New Genera Babjeviella, Meyerozyma, Millerozyma, Priceomyces, and Scheffersomyces. Mycoscience 2010, 51, 2–14. [Google Scholar] [CrossRef]
- Lachance, M.A.; Boekhout, T.; Scorzetti, G.; Fell, J.W.; Kurtzman, C.P. Candida Berkhout (1923). In The Yeasts. A Taxonomic Study; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier: Amsterdam, The Netherlands, 2011; Volume 2, pp. 987–1278. ISBN 9780444521491. [Google Scholar]
- Suh, S.O.; Nguyen, N.H.; Blackwell, M. Nine New Candida Species near C. membranifaciens Isolated from Insects. Mycol. Res. 2005, 109, 1045–1056. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chi, Z.; Wang, X.; Ju, L.; Chi, Z.; Guo, N. Isolation and Characterization of Candida membranifaciens Subsp. flavinogenie W14-3, a Novel Riboflavin-Producing Marine Yeast. Microbiol. Res. 2008, 163, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Gudipati, V.; Koch, K.; Lienhart, W.D.; Macheroux, P. The Flavoproteome of the Yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta Proteins Proteom. 2014, 1844, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Schall, P.; Marutschke, L.; Grimm, B. The Flavoproteome of the Model Plant Arabidopsis thaliana. Int. J. Mol. Sci. 2020, 21, 5371. [Google Scholar] [CrossRef]
- Revuelta, J.L.; Ledesma-Amaro, R.; Jiménez, A. Industrial Production of Vitamin B2 by Microbial Fermentation. In Industrial Biotechnology of Vitamins, Biopigments, and Antioxidants; Vandamme, E.J., Revuelta, J.L., Eds.; Wiley-VCH: Weinheim, Germany, 2016; pp. 17–40. [Google Scholar]
- Schwechheimer, S.K.; Park, E.Y.; Revuelta, J.L.; Becker, J.; Wittmann, C. Biotechnology of Riboflavin. Appl. Microbiol. Biotechnol. 2016, 100, 2107–2119. [Google Scholar] [CrossRef]
- Averianova, L.A.; Balabanova, L.A.; Son, O.M.; Podvolotskaya, A.B.; Tekutyeva, L.A. Production of Vitamin B2 (Riboflavin) by Microorganisms: An Overview. Front. Bioeng. Biotechnol. 2020, 8, 570828. [Google Scholar] [CrossRef]
- Abbas, C.A.; Sibirny, A.A. Genetic Control of Biosynthesis and Transport of Riboflavin and Flavin Nucleotides and Construction of Robust Biotechnological Producers. Microbiol. Mol. Biol. Rev. 2011, 75, 321–360. [Google Scholar] [CrossRef]
- Nguyen, H.-V.; Gaillardin, C.; Neuveglise, C. Differentiation of Debaryomyces hansenii and Candida famata by RRNA Gene Intergenic Spacer Fingerprinting and Reassessment of Phylogenetic Relationships among D. Hansenii, C. famata, D. fabryi, C. flareri (=D. subglobosus) and D. prosopidis: Description of D. vietnamensis sp. nov. closely related to D. nepalensis. FEMS Yeast Res. 2009, 9, 641–662. [Google Scholar] [CrossRef][Green Version]
- Knight, S.A.B.; Lesuisse, E.; Stearman, R.; Klausner, R.D.; Dancis, A. Reductive Iron Uptake by Candida albicans: Role of Copper, Iron and the TUP1 Regulator. Microbiology 2002, 148, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Demain, A.L. Riboflavin Oversynthesis. Annu. Rev. Microbiol. 1972, 26, 369–388. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, D.A.; Logue, M.E.; Stajich, J.E.; Butler, G. A Fungal Phylogeny Based on 42 Complete Genomes Derived from Supertree and Combined Gene Analysis. BMC Evol. Biol. 2006, 6, 99. [Google Scholar] [CrossRef]
- Tanner, F.W.; Vojnovich, C.; Van Lanen, J.M. Riboflavin Production by Candida Species. Science 1945, 101, 180–181. [Google Scholar] [CrossRef]
- Petrovska, Y.; Lyzak, O.; Dmytruk, K.; Sibirny, A. Effect of Gene SFU1 on Riboflavin Synthesis in Flavinogenic Yeast Candida famata. Cytol. Genet. 2020, 54, 408–412. [Google Scholar] [CrossRef]
- Cecilia Mestre, M.; Rosa, C.A.; Safar, S.V.B.; Libkind, D.; Fontenla, S.B. Yeast Communities Associated with the Bulk-Soil, Rhizosphere and Ectomycorrhizosphere of a Nothofagus Pumilio Forest in Northwestern Patagonia, Argentina. FEMS Microbiol. Ecol. 2011, 78, 531–541. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Han, J.K.; Park, Y.H.; Park, J.H. Microorganisms and Process for the Production of Riboflavin by Fermentation. EP1426450A1, 9 June 2004. [Google Scholar]
- Heefner, D.L.; Weaver, C.A.; Yarus, M.J.; Burdzinski, L.A. Method for Producing Riboflavin with Candida famata. US5164303A, 17 November 1992. [Google Scholar]
- Chi, Z.; Wang, L.; Ju, L.; Chi, Z. Optimisation of Riboflavin Production by the Marine Yeast Candida membranifaciens Subsp. flavinogenie W14-3 Using Response Surface Methodology. Ann. Microbiol. 2008, 58, 677–681. [Google Scholar] [CrossRef]
- Dmytruk, K.V.; Yatsyshyn, V.Y.; Sybirna, N.O.; Fedorovych, D.V.; Sibirny, A.A. Metabolic Engineering and Classic Selection of the Yeast Candida famata (Candida flareri) for Construction of Strains with Enhanced Riboflavin Production. Metab. Eng. 2011, 13, 82–88. [Google Scholar] [CrossRef]
- Zaky, A.S.; Greetham, D.; Louis, E.J.; Tucker, G.A.; Du, C. A New Isolation and Evaluation Method for Marine-Derived Yeast Spp. with Potential Applications in Industrial Biotechnology. J. Microbiol. Biotechnol. 2016, 26, 1891–1907. [Google Scholar] [CrossRef]
- Hoffman, C.S. Preparation of yeast DNA. In Current Protocols in Molecular Biology; Ausubel, F., Brent, R., Kingston, R., Moore, D., Seidman, J., Smith, J., Struhl, K., Eds.; John Wiley & Sons, Inc.: New York, NY, USA, 1997; pp. 13.11.1–13.11.4. [Google Scholar]
- Kurtzman, C.P.; Robnett, C.J. Identification and Phylogeny of Ascomycetous Yeasts from Analysis of Nuclear Large Subunit (26S) Ribosomal DNA Partial Sequences. Antonie Van Leeuwenhoek 1998, 73, 331–371. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Silva, R.; Aguiar, T.Q.; Domingues, L. Blockage of the Pyrimidine Biosynthetic Pathway Affects Riboflavin Production in Ashbya gossypii. J. Biotechnol. 2015, 193, 37–40. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ledesma-Amaro, R.; Serrano-Amatriain, C.; Jiménez, A.; Revuelta, J.L. Metabolic Engineering of Riboflavin Production in Ashbya gossypii through Pathway Optimization. Microb. Cell Fact. 2015, 14, 163. [Google Scholar] [CrossRef] [PubMed]
- Grigoriev, I.V.; Nikitin, R.; Haridas, S.; Kuo, A.; Ohm, R.; Otillar, R.; Riley, R.; Salamov, A.; Zhao, X.; Korzeniewski, F.; et al. MycoCosm Portal: Gearing up for 1000 Fungal Genomes. Nucleic Acids Res. 2014, 42, D699–D704. [Google Scholar] [CrossRef]
- Koonin, E.V.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Krylov, D.M.; Makarova, K.S.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; Rao, B.S.; et al. A Comprehensive Evolutionary Classification of Proteins Encoded in Complete Eukaryotic Genomes. Genome Biol. 2004, 5, R7. [Google Scholar] [CrossRef]
- Ferreira, D.; Nobre, A.; Silva, M.L.; Faria-Oliveira, F.; Tulha, J.; Ferreira, C.; Lucas, C. XYLH Encodes a Xylose/H+ Symporter from the Highly Related Yeast Species Debaryomyces fabryi and Debaryomyces hansenii. FEMS Yeast Res. 2013, 13, 585–596. [Google Scholar] [CrossRef][Green Version]
- Saier, M.H.; Reddy, V.S.; Moreno-Hagelsieb, G.; Hendargo, K.J.; Zhang, Y.; Iddamsetty, V.; Lam, K.J.K.; Tian, N.; Russum, S.; Wang, J.; et al. The Transporter Classification Database (TCDB): 2021 Update. Nucleic Acids Res. 2021, 49, D461–D467. [Google Scholar] [CrossRef]
- Monteiro, P.T.; Oliveira, J.; Pais, P.; Antunes, M.; Palma, M.; Cavalheiro, M.; Galocha, M.; Godinho, C.P.; Martins, L.C.; Bourbon, N.; et al. YEASTRACT+: A Portal for Cross-Species Comparative Genomics of Transcription Regulation in Yeasts. Nucleic Acids Res. 2020, 48, D642–D649. [Google Scholar] [CrossRef]
- Godinho, C.P.; Palma, M.; Oliveira, J.; Mota, M.N.; Antunes, M.; Teixeira, M.C.; Monteiro, P.T.; Sá-Correia, I. The N.C.Yeastract and CommunityYeastract Databases to Study Gene and Genomic Transcription Regulation in Non-Conventional Yeasts. FEMS Yeast Res. 2021, 21, foab045. [Google Scholar] [CrossRef]
- Nordberg, H.; Cantor, M.; Dusheyko, S.; Hua, S.; Poliakov, I.S.; Shabalov, I.; Smirnova, T.; Grigoriev, I.V.; Dubchak, I. The Genome Portal of the Department of Energy Joint Genome Institute: 2014 Updates. Nucleic Acids Res. 2014, 42, D26–D31. [Google Scholar] [CrossRef] [PubMed]
- Satianpakiranakorn, P.; Khunnamwong, P.; Limtong, S. Yeast Communities of Secondary Peat Swamp Forests in Thailand and Their Antagonistic Activities against Fungal Pathogens Cause of Plant and Postharvest Fruit Diseases. PLoS ONE 2020, 15, e0230269. [Google Scholar] [CrossRef] [PubMed]
- Boonmak, C.; Khunnamwong, P.; Limtong, S. Yeast Communities of Primary and Secondary Peat Swamp Forests in Southern Thailand. Antonie Van Leeuwenhoek 2020, 113, 55–69. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, F.P.; Souza, A.C.; Magalhães-Guedes, K.T.; Dias, D.R.; Silva, C.F.; Schwan, R.F. Yeasts Diversity in Brazilian Cerrado Soils: Study of the Enzymatic Activities. Afr. J. Microbiol. Res. 2013, 7, 4176–4190. [Google Scholar] [CrossRef]
- Worst, D.J.; Gerrits, M.M.; Vandenbroucke-Grauls, C.M.J.E.; Kusters, J.G. Helicobacter pylori RibBA-Mediated Riboflavin Production Is Involved in Iron Acquisition. J. Bacteriol. 1998, 180, 1473. [Google Scholar] [CrossRef]
- Crossley, R.A.; Gaskin, D.J.H.; Holmes, K.; Mulholland, F.; Wells, J.M.; Kelly, D.J.; Van Vliet, A.H.M.; Walton, N.J. Riboflavin Biosynthesis Is Associated with Assimilatory Ferric Reduction and Iron Acquisition by Campylobacter jejuni. Appl. Environ. Microbiol. 2007, 73, 7819–7825. [Google Scholar] [CrossRef]
- Philpott, C.C.; Protchenko, O. Response to Iron Deprivation in Saccharomyces cerevisiae. Eukaryot. Cell 2008, 7, 20. [Google Scholar] [CrossRef]
- Holmes-Hampton, G.P.; Jhurry, N.D.; McCormick, S.P.; Lindahl, P.A. Iron Content of Saccharomyces cerevisiae Cells Grown under Iron-Deficient and Iron-Overload Conditions. Biochemistry 2013, 52, 105. [Google Scholar] [CrossRef]
- Ramos-Alonso, L.; Romero, A.M.; Martínez-Pastor, M.T.; Puig, S. Iron Regulatory Mechanisms in Saccharomyces cerevisiae. Front. Microbiol. 2020, 11, 2222. [Google Scholar] [CrossRef]
- Schwechheimer, S.K.; Becker, J.; Peyriga, L.; Portais, J.C.; Wittmann, C. Metabolic Flux Analysis in Ashbya gossypii Using 13C-Labeled Yeast Extract: Industrial Riboflavin Production under Complex Nutrient Conditions. Microb. Cell Fact. 2018, 17, 162. [Google Scholar] [CrossRef]
- Villas-Bôas, S.G.; Åkesson, M.; Nielsen, J. Biosynthesis of Glyoxylate from Glycine in Saccharomyces cerevisiae. FEMS Yeast Res. 2005, 5, 703–709. [Google Scholar] [CrossRef] [PubMed]
- Grant, C.M.; MacIver, F.H.; Dawes, I.W. Glutathione Synthetase Is Dispensable for Growth under Both Normal and Oxidative Stress Conditions in the Yeast Saccharomyces cerevisiae Due to an Accumulation of the Dipeptide Gamma-Glutamylcysteine. Mol. Biol. Cell 1997, 8, 1699–1707. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, D.A.; Dawes, I.W. Genetics of the Synthesis of Serine from Glycine and the Utilization of Glycine as Sole Nitrogen Source by Saccharomyces cerevisiae. Genetics 1995, 140, 1213–1222. [Google Scholar] [CrossRef] [PubMed]
- Martins, L.C.; Palma, M.; Angelov, A.; Nevoigt, E.; Liebl, W.; Sá-Correia, I. Complete Utilization of the Major Carbon Sources Present in Sugar Beet Pulp Hydrolysates by the Oleaginous Red Yeasts Rhodotorula toruloides and R. mucilaginosa. J. Fungi 2021, 7, 215. [Google Scholar] [CrossRef]
- Wohlbach, D.J.; Kuo, A.; Sato, T.K.; Potts, K.M.; Salamov, A.A.; LaButti, K.M.; Sun, H.; Clum, A.; Pangilinan, J.L.; Lindquist, E.A.; et al. Comparative Genomics of Xylose-Fermenting Fungi for Enhanced Biofuel Production. Proc. Natl. Acad. Sci. USA 2011, 108, 13212–13217. [Google Scholar] [CrossRef]
- Shen, X.X.; Opulente, D.A.; Kominek, J.; Zhou, X.; Steenwyk, J.L.; Buh, K.V.; Haase, M.A.B.; Wisecaver, J.H.; Wang, M.; Doering, D.T.; et al. Tempo and Mode of Genome Evolution in the Budding Yeast Subphylum. Cell 2018, 175, 1533–1545.e20. [Google Scholar] [CrossRef]
- Haase, M.A.B.; Kominek, J.; Langdon, Q.K.; Kurtzman, C.P.; Hittinger, C.T. Genome Sequence and Physiological Analysis of Yamadazyma laniorum f.a. sp. nov. and a Reevaluation of the Apocryphal Xylose Fermentation of Its Sister Species, Candida tenuis. FEMS Yeast Res. 2017, 17, fox019. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Kiryutin, B.; Koonin, E.V.; Krylov, D.M.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; et al. The COG Database: An Updated Version Includes Eukaryotes. BMC Bioinform. 2003, 4, 41. [Google Scholar] [CrossRef]
- Sa, N.; Rawat, R.; Thornburg, C.; Walker, K.D.; Roje, S. Identification and Characterization of the Missing Phosphatase on the Riboflavin Biosynthesis Pathway in Arabidopsis thaliana. Plant J. 2016, 88, 705–716. [Google Scholar] [CrossRef]
- Skrzypek, M.S.; Binkley, J.; Binkley, G.; Miyasato, S.R.; Simison, M.; Sherlock, G. The Candida Genome Database (CGD): Incorporation of Assembly 22, Systematic Identifiers and Visualization of High Throughput Sequencing Data. Nucleic Acids Res. 2017, 45, D592. [Google Scholar] [CrossRef]
- Liu, S.; Hu, W.; Wang, Z.; Chen, T. Production of Riboflavin and Related Cofactors by Biotechnological Processes. Microb. Cell Fact. 2020, 19, 31. [Google Scholar] [CrossRef] [PubMed]
- Dmytruk, K.; Lyzak, O.; Yatsyshyn, V.; Kluz, M.; Sibirny, V.; Puchalski, C.; Sibirny, A. Construction and Fed-Batch Cultivation of Candida famata with Enhanced Riboflavin Production. J. Biotechnol. 2014, 172, 11–17. [Google Scholar] [CrossRef]
- Dmytruk, K.V.; Voronovsky, A.Y.; Sibirny, A.A. Insertion Mutagenesis of the Yeast Candida famata (Debaryomyces hansenii) by Random Integration of Linear DNA Fragments. Curr. Genet. 2006, 50, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Noble, S.M. Post-Transcriptional Regulation of the Sef1 Transcription Factor Controls the Virulence of Candida albicans in Its Mammalian Host. PLoS Pathog. 2012, 8, e1002956. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Pande, K.; French, S.D.; Tuch, B.B.; Noble, S.M. An Iron Homeostasis Regulatory Circuit with Reciprocal Roles in Candida albicans Commensalism and Pathogenesis. Cell Host Microbe 2011, 10, 118–135. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; Prasad, H.K.; Sinha, I.; Agarwal, N.; Natarajan, K. Cap2-HAP Complex Is a Critical Transcriptional Regulator That Has Dual but Contrasting Roles in Regulation of Iron Homeostasis in Candida albicans. J. Biol. Chem. 2011, 286, 25154–25170. [Google Scholar] [CrossRef]
- Sibirny, A.A.; Boretsky, Y.R. Pichia guilliermondii. In Yeast Biotechnology: Diversity and Applications; Springer: Berlin/Heidelberg, Germany, 2009; pp. 113–134. ISBN 9781402082917. [Google Scholar]
- Tsyrulnyk, A.O.; Andreieva, Y.A.; Ruchala, J.; Fayura, L.R.; Dmytruk, K.V.; Fedorovych, D.V.; Sibirny, A.A. Expression of Yeast Homolog of the Mammal BCRP Gene Coding for Riboflavin Efflux Protein Activates Vitamin B2 Production in the Flavinogenic Yeast Candida famata. Yeast 2020, 37, 467–473. [Google Scholar] [CrossRef]
- Enright, A.J.; Van Dongen, S.; Ouzounis, C.A. An Efficient Algorithm for Large-Scale Detection of Protein Families. Nucleic Acids Res. 2002, 30, 1575–1584. [Google Scholar] [CrossRef]
- Reihl, P.; Stolz, J. The Monocarboxylate Transporter Homolog Mch5p Catalyzes Riboflavin (Vitamin B2) Uptake in Saccharomyces cerevisiae. J. Biol. Chem. 2005, 280, 39809–39817. [Google Scholar] [CrossRef]
- Fourie, R.; Kuloyo, O.O.; Mochochoko, B.M.; Albertyn, J.; Pohl, C.H. Iron at the Centre of Candida albicans Interactions. Front. Cell. Infect. Microbiol. 2018, 8, 185. [Google Scholar] [CrossRef]
- Duval, C.; Macabiou, C.; Garcia, C.; Lesuisse, E.; Camadro, J.; Auchère, F. The Adaptive Response to Iron Involves Changes in Energetic Strategies in the Pathogen Candida albicans. Microbiologyopen 2020, 9, e970. [Google Scholar] [CrossRef] [PubMed]
- Yun, C.W.; Bauler, M.; Moore, R.E.; Klebba, P.E.; Philpott, C.C. The Role of the FRE Family of Plasma Membrane Reductases in the Uptake of Siderophore-Iron in Saccharomyces cerevisiae. J. Biol. Chem. 2001, 276, 10218–10223. [Google Scholar] [CrossRef]
- Wang, T.P.; Quintanar, L.; Severance, S.; Solomon, E.I.; Kosman, D.J. Targeted Suppression of the Ferroxidase and Iron Trafficking Activities of the Multicopper Oxidase Fet3p from Saccharomyces cerevisiae. J. Biol. Inorg. Chem. 2003, 8, 611–620. [Google Scholar] [CrossRef] [PubMed]
- Almeida, R.S.; Wilson, D.; Hube, B. Candida albicans Iron Acquisition within the Host. FEMS Yeast Res. 2009, 9, 1000–1012. [Google Scholar] [CrossRef]
- Weissman, Z.; Shemer, R.; Kornitzer, D. Deletion of the Copper Transporter CaCCC2 Reveals Two Distinct Pathways for Iron Acquisition in Candida albicans. Mol. Microbiol. 2002, 44, 1551–1560. [Google Scholar] [CrossRef]
- Huffman, D.L.; O’Halloran, T.V. Energetics of Copper Trafficking between the Atx1 Metallochaperone and the Intracellular Copper Transporter, Ccc2. J. Biol. Chem. 2000, 275, 18611–18614. [Google Scholar] [CrossRef] [PubMed]
- Dix, D.R.; Bridgham, J.T.; Broderius, M.A.; Byersdorfer, C.A.; Eide, D.J. The FET4 Gene Encodes the Low Affinity Fe(I1) Transport Protein of Saccharomyces cerevisiae. J. Biol. Chem. 1994, 269, 26092–26099. [Google Scholar] [CrossRef]
- Heymann, P.; Ernst, J.F.; Winkelmann, G. Identification and Substrate Specificity of a Ferrichrome-Type Siderophore Transporter (Arn1p) in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 2006, 186, 221–227. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kim, D.; Yukl, E.T.; Moënne-Loccoz, P.; Ortiz De Montellano, P.R. Fungal Heme Oxygenases: Functional Expression and Characterization of Hmx1 from Saccharomyces cerevisiae and CaHmx1 from Candida albicans. Biochemistry 2006, 45, 14772–14780. [Google Scholar] [CrossRef]
- Weissman, Z.; Kornitzer, D. A Family of Candida Cell Surface Haem-Binding Proteins Involved in Haemin and Haemoglobin-Iron Utilization. Mol. Microbiol. 2004, 53, 1209–1220. [Google Scholar] [CrossRef]
- Ror, S.; Panwar, S.L. Sef1-Regulated Iron Regulon Responds to Mitochondria-Dependent Iron-Sulfur Cluster Biosynthesis in Candida albicans. Front. Microbiol. 2019, 10, 1528. [Google Scholar] [CrossRef] [PubMed]
- Blazhenko, O.V. Glutathione Deficiency Leads to Riboflavin Oversynthesis in the Yeast Pichia guilliermondii. Curr. Microbiol. 2014, 69, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Palma, M.; Goffeau, A.; Spencer-Martins, I.; Baret, P.V. A Phylogenetic Analysis of the Sugar Porters in Hemiascomycetous Yeasts. J. Mol. Microbiol. Biotechnol. 2007, 12, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Baruffini, E.; Goffrini, P.; Donnini, C.; Lodi, T. Galactose Transport in Kluyveromyces lactis: Major Role of the Glucose Permease Hgt1. FEMS Yeast Res. 2006, 6, 1235–1242. [Google Scholar] [CrossRef]
| Feature | |
|---|---|
| Total reads | 17,196,026 |
| No. of scaffolds (≥2000 bp) | 56 |
| Coverage | 224x |
| N50 (bp) | 667,103 bp |
| L50 | 7 |
| Maximum contig length (bp) | 1,053,043 bp |
| Minimum contig length (bp) | 2057 bp |
| Assembly size (bp) | 11,508,125 bp |
| GC content (%) | 32.15% |
| Protein ID | Homolog in C. albicans | Amino Acid Identity (%) | Molecular Function (*) | Putative TFs Based on Predicted Binding Sites |
|---|---|---|---|---|
| 30281 | RIB1 | 74% | GTP cyclohydrolase II | Efg1, Tye7, Brg1, Hmo1, Nrg1, Rap1, Hap43/Hap5, Upc2, Wor3, Mrr1, Ace2, Hap5, Rtg1, Rtg3, Zcf29 |
| 15396 | RIB2 | 67% | Pseudouridine synthase | Efg1, Tye7, Ndt80, Skn7 (**) |
| 15025 | RIB3 | 77% | 3,4-dihydroxy-2-butanone-4-phosphate synthase (DHBP synthase) | Efg1, Brg1, Nrg1, Rim101, Hap43/Hap5, Wor3, Mrr1, Hap5, Rgt1, Rtg3, Zcf29, Skn7 |
| 34729 | RIB4 | 77% | Lumazine synthase (6,7-dimethyl-8-ribityllumazine synthase synthase) | Efg1, Tye7, Ndt80, Nrg1, Upc2, Wor3, Mrr1, Rgt1, Ace2, Zcf29 |
| 13130 | RIB5 | 69% | Riboflavin synthase | Efg1, Brg1, Nrg1, Rap1, Hap43/Hap5, Mrr1, Rgt1, Ace2, Hap5, Zcf29, Skn7, Cta8 |
| 18055 | RIB7 | 53% | 5-amino-6-(5-phosphoribosylamino)uracil reductase | Brg1, Hap43/Hap5, Mrr1 (**) |
| 25813 | FMN1 | 50% | Riboflavin kinase | Upc2, Wor3, Mrr1, Hap5, Zcf29 (**) |
| 24533 | FAD1 | 51% | Flavin adenine dinucleotide (FAD) synthetase | Nrg1, Tbf1, Wor3 (**) |
| 34484 (***) | RHR2 | 84% | Predicted haloacid-halidohydrolase | Efg1, Tye7, Ndt80, Brg1, Nrg1, Rap1, Rim101, Hap43/Hap5, Wor1, Mrr1, Mig1, Rgt1, Zcf29, Skn7 |
| 23627 (***) | DOG1 | 26% | Predicted haloacid-halidohydrolase | Brg1, Mrr1, Hap5 (**) |
| Protein ID | Homolog in C. albicans | Amino Acid Identity (%) | Molecular Function (*) | |
|---|---|---|---|---|
| Regulation of iron metabolism | 24521 | Sfu1 | 62% | Zinc finger, GATA-type transcription factor |
| 23730 | Hap43 | 52% | Basic-leucine zipper domain | |
| 13665 | Sef1 | 67% | Zn(2)-C6 fungal-type DNA-binding domain | |
| 29747 | Aft2 | 41% | Hypothetical transcription factor | |
| 18845 | Hap5 | 79% | Histone-like transcription factor | |
| High-affinity reductive system | 16867 | Fre10 | 45% | Ferric reductase (**) |
| 34201 | Cfl2 | 43% | Ferric reductase (**) | |
| 10230 | Fre9 | 51% | Ferric reductase (**) | |
| 23390 | Cfl2 | 24% | Ferric reductase (**) | |
| 14512 | Frp2 | 39% | Ferric reductase (**) | |
| 31983 | orf19.867 | 26% | Ferric reductase (**) | |
| 20041 | orf19.4843 | 50% | Ferric reductase | |
| 34362 | Fet31 | 69% | Multicopper oxidase | |
| 12903 | Fet33 | 58% | Multicopper oxidase | |
| 29609 | Ftr1 | 74% | High-affinity iron transporter | |
| 35518 | Ccc2 | 49% | Cu2+ transporter P-type ATPase | |
| 34023 | Crp1 | 55% | Cu2+ transporter P-type ATPase | |
| 14754 | Atx1 | 60% | Copper metallochaperone | |
| Siderophore uptake system | 14012 | Sit1 | 58% | Siderophore iron transporter |
| 28384 | Sit1 | 45% | Siderophore iron transporter | |
| 29780 | Sit1 | 26% | Siderophore iron transporter | |
| Heme or hemoglobin utilization system | 28257 | Hmx1 | 64% | Heme oxygenase |
| 25343 | Pga10 (Rbt51) | 52% | Glycosylphosphatidylinositol (GPI)-modified cell wall protein | |
| 30576 | Pga7 | 57% | Glycosylphosphatidylinositol (GPI)-modified cell wall protein | |
| Iron-sulfur assembly | 13591 | Isu1 | 64% | Iron-sulfur cluster assembly |
| 17237 | Nfu2 (orf19.6283) | 75% | Iron-sulfur cluster binding activity | |
| 28973 | Nfu1 (orf19.2067) | 70% | Iron-sulfur cluster assembly | |
| 26437 | Isa1 | 52% | Iron-sulfur cluster assembly | |
| 32180 | Isa2 | 56% | Iron-sulfur cluster assembly | |
| 24028 | Dre2 | 39% | Iron-sulfur cluster assembly | |
| 25680 | Yfh1 | 46% | Mitochondrial matrix protein frataxin, involved in Fe/S protein biosynthesis | |
| 14713 | Atm1 | 72% | mitochondrial ABC transporter; transport of iron-sulfur cluster precursors | |
| 18779 | Nar1 | 54% | Putative cytosolic iron-sulfur (FeS) protein assembly machinery protein | |
| 28349 | Nbp35 | 65% | ATPase activity, iron-sulfur cluster binding activity and role in iron-sulfur cluster assembly | |
| 15021 | Nbp35 | 84% | ATPase activity, iron-sulfur cluster binding activity and role in iron-sulfur cluster assembly | |
| 12204 | Nbp35 | 77% | ATPase activity, iron-sulfur cluster binding activity and role in iron-sulfur cluster assembly |
| Protein ID | Homolog in C. albicans | Amino Acid Identity (%) | Molecular Function (*) | Putative TFs Based on Predicted Binding Sites |
|---|---|---|---|---|
| 17681 | orf19.3120 | 56% | Riboflavin excretase. ATP-binding cassette ABC transporter. | Efg1, Tye7, Rap1, Rim101, Cph2, Hap43/Hap5, Mrr1, Ace2, Hap5, Zcf29, Skn7, Cta8, Cph2 |
| 5199 | orf19.6263 | 55% | Carbohydrate transport and metabolism|Monocarboxylate transporter | Efg1/Tye7, Ndt80, Wbr3, Mrr1, Rgt1, Hap5, Zcf29, Cta8, Cph2 |
| 31581 | orf19.6263 | 34% | Carbohydrate transport and metabolism|Monocarboxylate transporter | Tec1, Efg1/Tye7, Efg1, Brg1, Nrg1, Rap1, Hsm1/Cph2, Hap43/Hap5, Wor3, Mrr1, Mig1, Hap5, Rgt1/Rgt3, Zcf29, Cta8, Cph2 |
| 31756 | orf19.5720 | 52% | Carbohydrate transport and metabolism|Monocarboxylate transporter | Nrg1, Wor2, Hap5, Cta8, Cph2 |
| 31951 | orf19.6263 | 23% | Carbohydrate transport and metabolism|Monocarboxylate transporter | Efg1, Efg1/Tye7, Rap1, Mrr1, Skn7, Cta8, Cph2 |
| 34553 | orf19.5720 | 48% | Carbohydrate transport and metabolism|Monocarboxylate transporter | Tec1, Efg1, Ndt80, Brg1, Rap1, Hap43/Hap5, Wor2, Mrr1, Zcf29, Skn7, Cta8, Cph2 |
| 34676 | orf19.2751 | 44% | Carbohydrate transport and metabolism|Monocarboxylate transporter | Efg1/Tye7, Ndt80, Brg1, Rap1, Tfb1, Rim101, Hap43/Hap5, Upc2, Wor1, Mrr1, Zcf29, Skn7, Cta8, Cph2 |
| 31444 | orf19.6263 | 53% | Carbohydrate transport and metabolism|Monocarboxylate transporter | Tec1, Efg1/Tye7, Nrg1, Cbf1, Upc2, Wor7, Mrr1, Mig1, Hap5, Skn7, Cta8, Cph2 |
| 33236 | orf19.4337 | 45% | Carbohydrate transport and metabolism|Monocarboxylate transporter | Tec1, Brg1, Rap1, Wor2, Mrr1, Hap5, Zcf29, Cta8, Skn7, Cph2 |
| 33235 | orf19.4337 | 46% | Carbohydrate transport and metabolism|Monocarboxylate transporter | Brg1, rap1, Rim101, Wor3, Mrr1, Rgt1, Ace2, Hap5, Zcf29, Cta8, Cph2 |
| 19539 | orf19.4337 | 27% | Carbohydrate transport and metabolism|Monocarboxylate transporter | Efg1, Efg1/Tye7, Brg1, Nrg1, Rap1, Hsm1/Cph2, Hap43/Hap5, Upc2, Wor3, Wor2, Mrr1, Mig1, Rgt1, Ace2, Hap5, Zcf20, Skn7, Cta8, Cph2 |
| 15793 | orf19.6209 | 56% | Carbohydrate transport and metabolism|Monocarboxylate transporter | Mac1, Tec1, Efg1, Efg1/Tye7, Ndt80, Brg1, Rap1, Hap43/Hap5, Wor3, Mrr1, Mig1, Rgt1, Hap5, Zcf29, Skn7, Cta8, Cph2 |
| 26147 | orf19.6209 | 56% | Carbohydrate transport and metabolism|Monocarboxylate transporter | Tec1, Efg1/Tye7, Efg1, Ndt80, Brg1, Nrg1, Rap1, Tbf1, Hsm1/cph2, Wor, Mrr1, Rgt1, Hap5, Zcf29, Skn7, Cta8, Cph2 |
| Annotation Description (Transporter Classification) | Number of Transporters |
|---|---|
| Maltotriose/maltose:H+ symporter (2.A.1.1.10) | 1 |
| General α-glucoside:H+ symporter (2.A.1.1.11) | 7 |
| Glucose/xylose: H+ symporter (2.A.1.1.51) | 2 |
| Xylose facilitator (2.A.1.1.40) | 3 |
| Glucose/xylose facilitator (2.A.1.1.67) | 3 |
| High affinity glucose transporter (2.A.1.1.39) | 4 |
| Glucose/Mannose/Galactose/Fructose:H+ symporter (2.A.1.1.43) | 2 |
| Lactose/Galactose:H+ symporter (2.A.1.1.9) | 6 |
| Glycerol:H+ symporter (2.A.1.1.38) | 3 |
| Glycerol uptake permease (Glycerol:H+ symporter) (2.A.1.1.73) | 2 |
| Quinate:H+ symporter (2.A.1.1.7) | 6 |
| Myoinositol:H+ symporter (2.A.1.1.8) | 1 |
| Hexose sensor (2.A.1.1.64) | 1 |
| Glucose Transporter/Sensor (2.A.1.1.68) | 2 |
| Sugar/Polyol transporter (2.A.1.1.69) | 2 |
| Total | 45 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palma, M.; Mondo, S.; Pereira, M.; Vieira, É.; Grigoriev, I.V.; Sá-Correia, I. Genome Sequence and Analysis of the Flavinogenic Yeast Candida membranifaciens IST 626. J. Fungi 2022, 8, 254. https://doi.org/10.3390/jof8030254
Palma M, Mondo S, Pereira M, Vieira É, Grigoriev IV, Sá-Correia I. Genome Sequence and Analysis of the Flavinogenic Yeast Candida membranifaciens IST 626. Journal of Fungi. 2022; 8(3):254. https://doi.org/10.3390/jof8030254
Chicago/Turabian StylePalma, Margarida, Stephen Mondo, Mariana Pereira, Érica Vieira, Igor V. Grigoriev, and Isabel Sá-Correia. 2022. "Genome Sequence and Analysis of the Flavinogenic Yeast Candida membranifaciens IST 626" Journal of Fungi 8, no. 3: 254. https://doi.org/10.3390/jof8030254
APA StylePalma, M., Mondo, S., Pereira, M., Vieira, É., Grigoriev, I. V., & Sá-Correia, I. (2022). Genome Sequence and Analysis of the Flavinogenic Yeast Candida membranifaciens IST 626. Journal of Fungi, 8(3), 254. https://doi.org/10.3390/jof8030254








