Casein Kinase 2 Mediates Degradation of Transcription Factor Pcf1 during Appressorium Formation in the Rice Blast Fungus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. RNA Isolation and Gene Expression Level Identification
2.3. Subcellular Localization and Fluorescence Intensity Determination
2.4. Affinity Purification and Mass Spectrometry Analysis
2.5. Yeast Two-Hybrid Assay
2.6. Co-Immunoprecipitation
2.7. Ubiquitin Detection
2.8. Statistical Analysis
3. Results
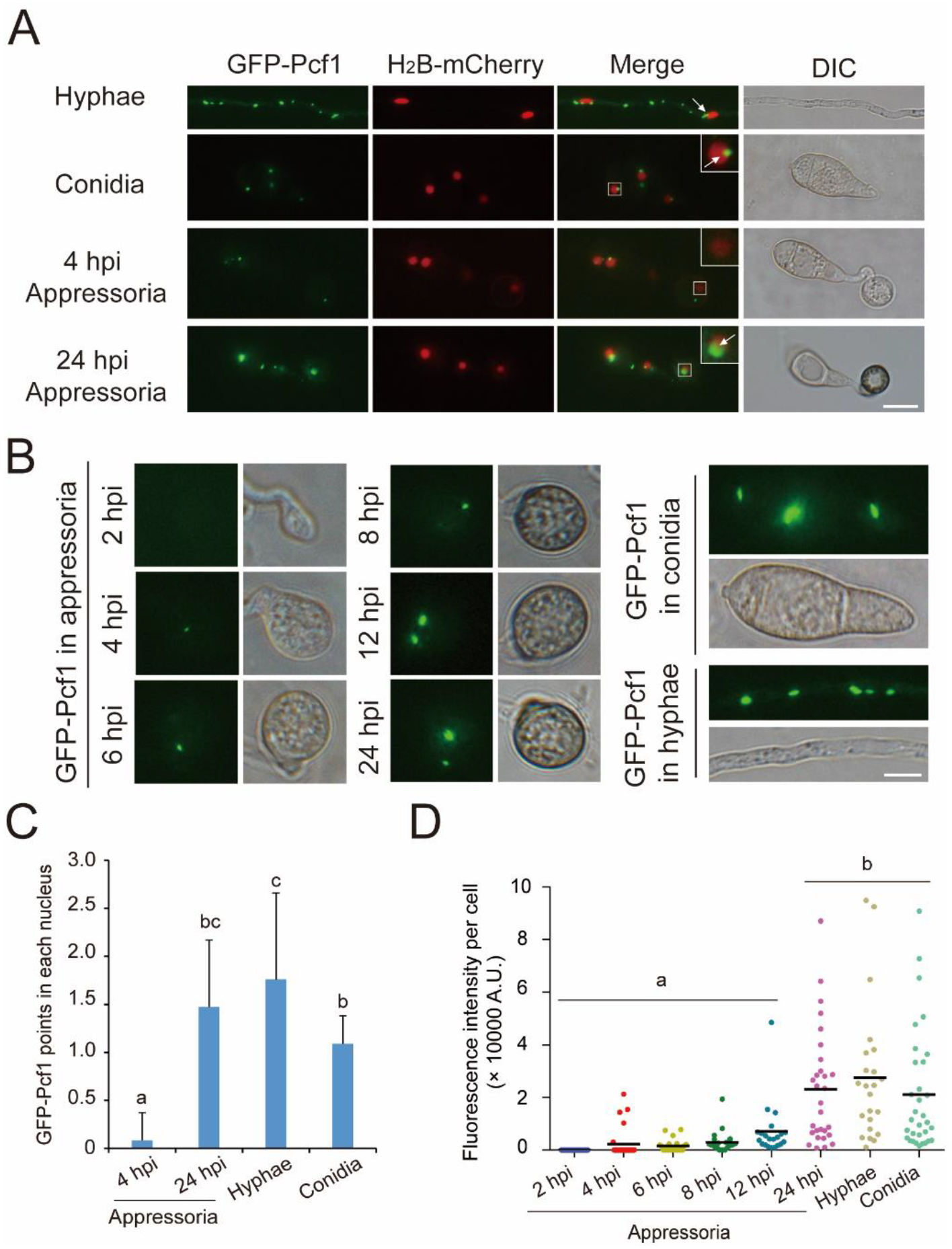
3.1. Expression of PCF1 Is Repressed in Conidia and Incipient Appressoria
3.2. Pcf1 Is Degraded in the Nuclei of Incipient Appressoria
3.3. Identification of Proteins Interacting with Pcf1
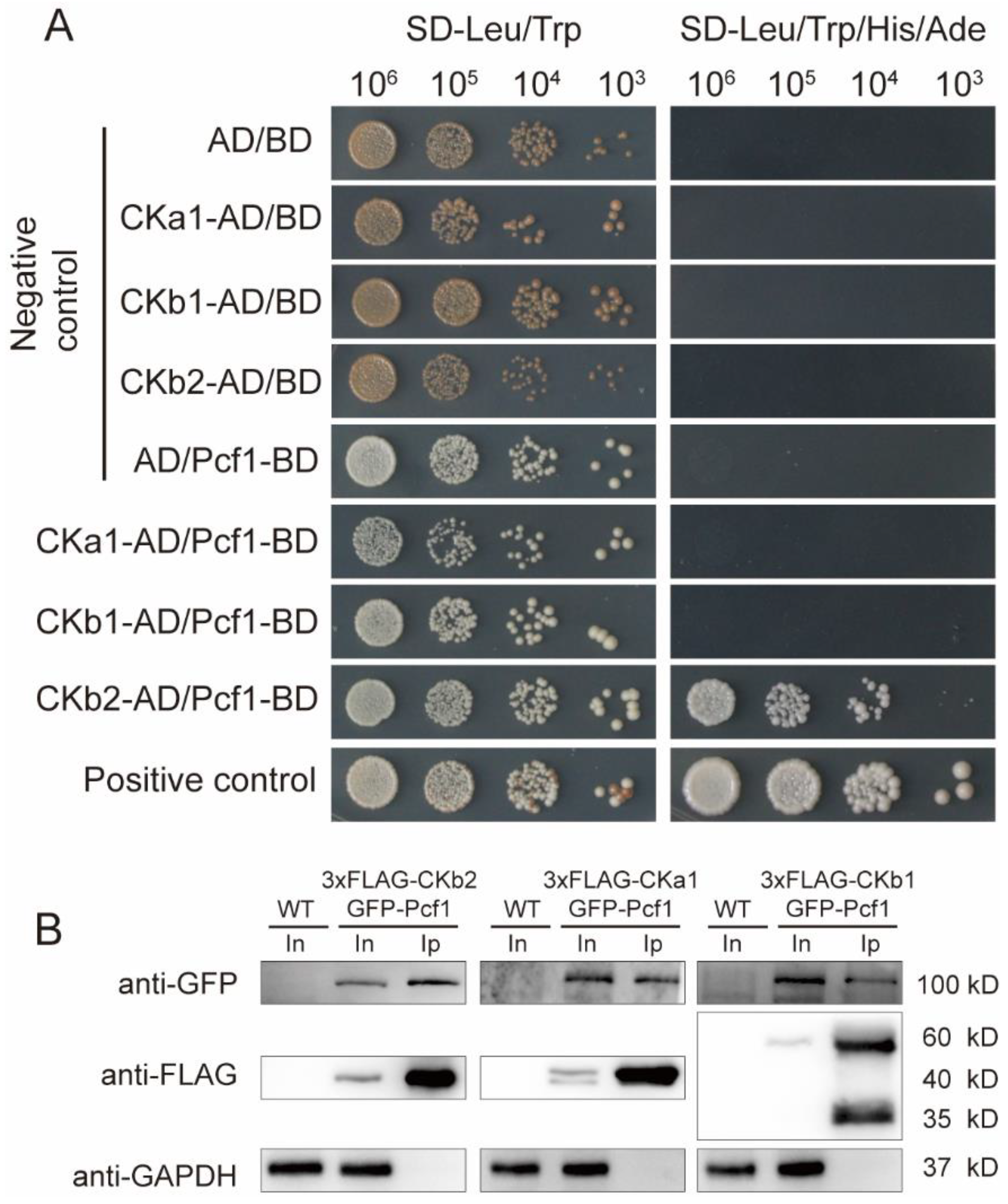
3.4. Pcf1 Interacts with CK2 through CKb2 Subunit
3.5. Pcf1 Is Ubiquitinated in the Hyphae
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ryder, L.S.; Dagdas, Y.F.; Kershaw, M.J.; Venkataraman, C.; Madzvamuse, A.; Yan, X.; Cruz-Mireles, N.; Soanes, D.M.; Oses-Ruiz, M.; Styles, V.; et al. A sensor kinase controls turgor-driven plant infection by the rice blast fungus. Nature 2019, 574, 423–427. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Chen, N.; Zhu, X.; Su, Z.; Wang, J.; Lu, J.; Liu, X.; Lin, F. The casein kinase MoYck1 regulates development, autophagy, and virulence in the rice blast fungus. Virulence 2019, 10, 719–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cooper, J.; Donofrio, N. Regulators unite to enable plant entry. Nat. Microbiol. 2021, 6, 1349–1350. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ning, G.; Huang, L.; Zhao, Y.; Dong, B.; Lu, J.; Lin, F. Calpains are involved in asexual and sexual development, cell wall integrity and pathogenicity of the rice blast fungus. Sci. Rep. 2016, 6, 31204. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Xu, Y.; Chen, J.; Luo, Y.; Lv, Y.; Su, J.; Kershaw, M.J.; Li, W.; Wang, J.; Yin, J.; et al. MoSnt2-dependent deacetylation of histone H3 mediates MoTor-dependent autophagy and plant infection by the rice blast fungus Magnaporthe oryzae. Autophagy 2018, 14, 1543–1561. [Google Scholar] [CrossRef] [Green Version]
- Oh, Y.; Donofrio, N.; Pan, H.; Coughlan, S.; Brown, D.E.; Meng, S.; Mitchell, T.; Dean, R.A. Transcriptome analysis reveals new insight into appressorium formation and function in the rice blast fungus Magnaporthe oryzae. Genome Biol. 2008, 9, R85. [Google Scholar] [CrossRef] [Green Version]
- Oh, Y.; Franck, W.L.; Han, S.O.; Shows, A.; Gokce, E.; Muddiman, D.C.; Dean, R.A. Polyubiquitin is required for growth, development and pathogenicity in the rice blast fungus Magnaporthe oryzae. PLoS ONE 2012, 7, e42868. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Chen, N.; Zhu, X.; Liang, S.; Li, L.; Wang, J.; Lu, J.; Lin, F.; Liu, X. F-box proteins MoFwd1, MoCdc4 and MoFbx15 regulate development and pathogenicity in the rice blast fungus Magnaporthe oryzae. Environ. Microbiol. 2019, 21, 3027–3045. [Google Scholar] [CrossRef]
- Shi, H.; Chen, G.; Chen, Y.; Dong, B.; Lu, J.; Liu, X.; Lin, F. MoRad6-mediated ubiquitination pathways are essential for development and pathogenicity in Magnaporthe oryzae. Environ. Microbiol. 2016, 18, 4170–4187. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, N.; Zheng, Y.; Yue, J.; Bhadauria, V.; Peng, Y.; Chen, Q. Ubiquitination in the rice blast fungus Magnaporthe oryzae: From development and pathogenicity to stress responses. Phytopathol. Res. 2022, 4, 1. [Google Scholar] [CrossRef]
- Zheng, W.; Zhou, J.; He, Y.; Xie, Q.; Chen, A.; Zheng, H.; Shi, L.; Zhao, X.; Zhang, C.; Huang, Q.; et al. Retromer is essential for autophagy-dependent plant infection by the rice blast fungus. PLoS Genet. 2015, 11, e1005704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, Y.; Ramos-Pamplona, M.; Naqvi, N.I. Autophagy-assisted glycogen catabolism regulates asexual differentiation in Magnaporthe oryzae. Autophagy 2009, 5, 33–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, X.; Li, L.; Wu, M.; Liang, S.; Shi, H.; Liu, X.; Lin, F. Current opinions on autophagy in pathogenicity of fungi. Virulence 2019, 10, 481–489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; Liu, X.; Feng, X.; Min, H.; Lin, F. An autophagy gene, MgATG5, is required for cell differentiation and pathogenesis in Magnaporthe oryzae. Curr. Genet. 2009, 55, 461–473. [Google Scholar] [CrossRef]
- Liu, X.; Zhao, Y.; Zhu, X.; Zeng, X.; Huang, L.; Dong, B.; Su, Z.; Wang, Y.; Lu, J.; Lin, F. Autophagy-related protein MoAtg14 is involved in differentiation, development and pathogenicity in the rice blast fungus Magnaporthe oryzae. Sci. Rep. 2017, 7, 40018. [Google Scholar] [CrossRef] [Green Version]
- Borgo, C.; D’Amore, C.; Sarno, S.; Salvi, M.; Ruzzene, M. Protein kinase CK2: A potential therapeutic target for diverse human diseases. Signal Transduct. Target. Ther. 2021, 6, 183. [Google Scholar] [CrossRef]
- Masłyk, M.; Kochanowicz, E.; Zieliński, R.; Kubiński, K.; Hellman, U.; Szyszka, R. Yeast surviving factor Svf1 as a new interacting partner, regulator and in vitro substrate of protein kinase CK2. Mol. Cell. Biochem. 2008, 312, 61–69. [Google Scholar] [CrossRef]
- Filhol, O.; Giacosa, S.; Wallez, Y.; Cochet, C. Protein kinase CK2 in breast cancer: The CK2β regulatory subunit takes center stage in epithelial plasticity. Cell. Mol. Life Sci. 2015, 72, 3305–3322. [Google Scholar] [CrossRef]
- Guerra, B.; Issinger, O.G. Protein kinase CK2 and its role in cellular proliferation, development and pathology. Electrophoresis 1999, 20, 391–408. [Google Scholar] [CrossRef]
- Unger, G.M.; Davis, A.T.; Slaton, J.W.; Ahmed, K. Protein kinase CK2 as regulator of cell survival: Implications for cancer therapy. Curr. Cancer Drug Targets 2004, 4, 77–84. [Google Scholar] [CrossRef]
- Glover, C.V.; Bidwai, A.P.; Reed, J.C. Structure and function of Saccharomyces cerevisiae casein kinase II. Mol. Cell. Biol. 1994, 40, 481–488. [Google Scholar]
- Fernández-Sáiz, V.; Targosz, B.-S.; Lemeer, S.; Eichner, R.; Langer, C.; Bullinger, L.; Reiter, C.; Slotta-Huspenina, J.; Schroeder, S.; Knorn, A.-M.; et al. SCFFbxo9 and CK2 direct the cellular response to growth factor withdrawal via Tel2/Tti1 degradation and promote survival in multiple myeloma. Nat. Cell Biol. 2013, 15, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Franck, N.; Le Seyec, J.; Guguen-Guillouzo, C.; Erdtmann, L. Hepatitis C virus NS2 protein is phosphorylated by the protein kinase CK2 and targeted for degradation to the proteasome. J. Virol. 2005, 79, 2700–2708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, T., Jr.; Delhase, M.; Hoffmann, A.; Karin, M. CK2 Is a C-Terminal IkappaB Kinase Responsible for NF-kappaB Activation during the UV Response. Mol. Cell 2003, 12, 829–839. [Google Scholar] [CrossRef]
- Scaglioni, P.P.; Yung, T.M.; Choi, S.C.; Baldini, C.; Konstantinidou, G.; Pandolfi, P.P. CK2 mediates phosphorylation and ubiquitin-mediated degradation of the PML tumor suppressor. Mol. Cell. Biochem. 2008, 316, 149–154. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, D.; Chen, Y.; Ye, W.; Lin, Q.; Lu, G.; Ebbole, D.J.; Olsson, S.; Wang, Z. Magnaporthe oryzae CK2 accumulates in nuclei, nucleoli, at septal pores and forms a large ring structure in appressoria, and is involved in rice blast pathogenesis. Front. Cell. Infect. Microbiol. 2019, 9, 113. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, D.; Liu, D.; Li, Y.; Li, H.; Xie, Y.; Wang, Z.; Hansen, B.O.; Olsson, S. Conserved eukaryotic kinase CK2 chaperone intrinsically disordered protein interactions. Appl. Environ. Microbiol. 2020, 86, e02191-19. [Google Scholar] [CrossRef]
- Yu, R.; Shen, X.; Liu, M.; Liu, X.; Yin, Z.; Li, X.; Feng, W.; Hu, J.; Zhang, H.; Zheng, X.; et al. The rice blast fungus MoRgs1 functioning in cAMP signaling and pathogenicity is regulated by casein kinase MoCk2 phosphorylation and modulated by membrane protein MoEmc2. PLoS Pathog. 2021, 17, e1009657. [Google Scholar] [CrossRef]
- Galhano, R.; Illana, A.; Ryder, L.S.; Rodríguez-Romero, J.; Demuez, M.; Badaruddin, M.; Martinez-Rocha, A.L.; Soanes, D.M.; Studholme, D.J.; Talbot, N.J.; et al. Tpc1 is an important Zn(II)2Cys6 transcriptional regulator required for polarized growth and virulence in the rice blast fungus. PLoS Pathog. 2017, 13, e1006516. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Cao, H.; Zhang, L.; Huang, P.; Lin, F. Systematic analysis of Zn2Cys6 transcription factors required for development and pathogenicity by high-throughput gene knockout in the rice blast fungus. PLoS Pathog. 2014, 10, e1004432. [Google Scholar] [CrossRef]
- Li, X.; Han, X.; Liu, Z.; He, C. The function and properties of the transcriptional regulator COS1 in Magnaporthe oryzae. Fungal. Biol. 2013, 117, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Park, S.Y.; Kim, K.S.; Rho, H.S.; Chi, M.H.; Choi, J.; Park, J.; Kong, S.; Park, J.; Goh, J.; et al. Homeobox transcription factors are required for conidiation and appressorium development in the rice blast fungus Magnaporthe oryzae. PLoS Genet. 2009, 5, e1000757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, W.; Ru, Y.; Hong, L.; Zhu, Q.; Zuo, R.; Guo, X.; Wang, J.; Zhang, H.; Zheng, X.; Wang, P.; et al. System-wide characterization of bZIP transcription factor proteins involved in infection-related morphogenesis of Magnaporthe oryzae. Environ. Microbiol. 2015, 17, 1377–1396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Odenbach, D.; Breth, B.; Thines, E.; Weber, R.W.; Anke, H.; Foster, A.J. The transcription factor Con7p is a central regulator of infection-related morphogenesis in the rice blast fungus Magnaporthe grisea. Mol. Microbiol. 2007, 64, 293–307. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Chen, Y.; Du, Y.; Dong, Y.; Guo, W.; Zhai, S.; Zhang, H.; Dong, S.; Zhang, Z.; Wang, Y.; et al. The bZIP transcription factor MoAP1 mediates the oxidative stress response and is critical for pathogenicity of the rice blast fungus Magnaporthe oryzae. PLoS Pathog. 2011, 7, e1001302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breth, B.; Odenbach, D.; Yemelin, A.; Schlinck, N.; Schröder, M.; Bode, M.; Antelo, L.; Andresen, K.; Thines, E.; Foster, A.J. The role of the Tra1p transcription factor of Magnaporthe oryzae in spore adhesion and pathogenic development. Fungal. Genet. Biol. 2013, 57, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Lu, J.; Khan, I.A.; Zhang, L.; He, R.; Lin, F. A novel Magnaporthe oryzae gene MCG1, encoding an extracellular globular protein, affects conidial germination and appressorial formation. Int. J. Agric. Technol. 2011, 7, 1647–1660. [Google Scholar]
- Cao, H.; Huang, P.; Zhang, L.; Shi, Y.; Sun, D.; Yan, Y.; Liu, X.; Dong, B.; Chen, G.; Snyder, J.H.; et al. Characterization of 47 Cys2 -His2 zinc finger proteins required for the development and pathogenicity of the rice blast fungus Magnaporthe oryzae. New Phytol. 2016, 211, 1035–1051. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Liu, W.; Wang, C.; Xu, Q.; Wang, Y.; Ding, S.; Xu, J. A MADS-box transcription factor MoMcm1 is required for male fertility, microconidium production and virulence in Magnaporthe oryzae. Mol. Microbiol. 2011, 80, 33–53. [Google Scholar] [CrossRef]
- Chao, C.T.; Ellingboe, A.H. Selection for mating competence in Magnaporthe grisea pathogenic to rice. Can. J. Bot. 1991, 69, 2130–2134. [Google Scholar] [CrossRef]
- Sun, D.; Cao, H.; Shi, Y.; Huang, P.; Dong, B.; Liu, X.; Lin, F.; Lu, J. The regulatory factor X protein MoRfx1 is required for development and pathogenicity in the rice blast fungus Magnaporthe oryzae. Mol. Plant Pathol. 2017, 18, 1075–1088. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Li, L.; Cai, Y.; Wu, X.; Shi, H.; Liang, S.; Qu, Y.; Naqvi, N.I.; Del Poeta, M.; Dong, B.; et al. A VASt-domain protein regulates autophagy, membrane tension, and sterol homeostasis in rice blast fungus. Autophagy 2021, 17, 2939–2961. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Zhang, C. Data Processing System (DPS) software with experimental design, statistical analysis and data mining developed for use in entomological research. Insect Sci. 2013, 20, 254–260. [Google Scholar] [CrossRef]
- Luo, Y.; Zhang, H.; Qi, L.; Zhang, S.; Zhou, X.; Zhang, Y.; Xu, J. FgKin1 kinase localizes to the septal pore and plays a role in hyphal growth, ascospore germination, pathogenesis, and localization of Tub1 beta-tubulins in Fusarium graminearum. New Phytol. 2014, 204, 943–954. [Google Scholar] [CrossRef]
- Jacob, S.; Foster, A.J.; Yemelin, A.; Thines, E. Histidine kinases mediate differentiation, stress response, and pathogenicity in Magnaporthe oryzae. MicrobiologyOpen 2014, 3, 668–687. [Google Scholar] [CrossRef]
- Pavlopoulou, A.; Kossida, S. Phylogenetic analysis of the eukaryotic RNA (cytosine-5)-methyltransferases. Genomics 2009, 93, 350–357. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Zhai, S.; Sun, Y.; Li, M.; Dong, Y.; Wang, X.; Zhang, H.; Zheng, X.; Wang, P.; Zhang, Z. MoTup1 is required for growth, conidiogenesis and pathogenicity of Magnaporthe oryzae. Mol. Plant Pathol. 2015, 16, 799–810. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, H.; Shen, X. The ubiquitin-proteasome system and its potential application in hepatocellular carcinoma therapy. Cancer Lett. 2016, 379, 245–252. [Google Scholar] [CrossRef]
- Qi, S.; Dong, J.; Xu, Z.; Cheng, X.; Zhang, W.; Qin, J. PROTAC: An effective targeted protein degradation strategy for cancer therapy. Front. Pharmacol. 2021, 12, 692574. [Google Scholar] [CrossRef]
- Anjago, W.M.; Zhou, T.; Zhang, H.; Shi, M.; Yang, T.; Zheng, H.; Wang, Z. Regulatory network of genes associated with stimuli sensing, signal transduction and physiological transformation of appressorium in Magnaporthe oryzae. Mycology 2018, 9, 211–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selvaraj, P.; Shen, Q.; Yang, F.; Naqvi, N.I. Cpk2, a catalytic subunit of cyclic AMP-PKA, Regulates growth and pathogenesis in rice blast. Front. Microbiol. 2017, 8, 2289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turrà, D.; Segorbe, D.; Di Pietro, A. Protein kinases in plant-pathogenic fungi: Conserved regulators of infection. Annu. Rev. Phytopathol. 2014, 52, 267–288. [Google Scholar] [CrossRef] [PubMed]
- He, N.; An, B.; Xie, Q.; Yan, X.; Feng, H.; Luo, H.; He, C. Synergistic deletion of RGS1 and COS1 may reduce the pathogenicity of Magnaporthe oryzae. Arch. Microbiol. 2019, 201, 807–816. [Google Scholar] [CrossRef]
- Park, S.Y.; Choi, J.; Lim, S.E.; Lee, G.W.; Park, J.; Kim, Y.; Kong, S.; Kim, S.R.; Rho, H.S.; Jeon, J.; et al. Global expression profiling of transcription factor genes provides new insights into pathogenicity and stress responses in the rice blast fungus. PLoS Pathog. 2013, 9, e1003350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, C.H.; Chen, S.; Shirsekar, G.; Zhou, B.; Khang, C.H.; Songkumarn, P.; Afzal, A.J.; Ning, Y.; Wang, R.; Bellizzi, M.; et al. The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant Cell 2012, 24, 4748–4762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Song, L.; Zhang, H.; Lin, Y.; Shen, X.; Guo, J.; Su, M.; Shi, G.; Wang, Z.; Lu, G. Rice ubiquitin-conjugating enzyme OsUBC26 is essential for immunity to the blast fungus Magnaporthe oryzae. Mol. Plant Pathol. 2021, 22, 1613–1623. [Google Scholar] [CrossRef]


| Protein | Locus | Description | % PSMs * | Reference |
|---|---|---|---|---|
| CKa1/CK2a | MGG_03696 | Casein kinase II subunit alpha | 23 | [26] |
| CKb1/CK2b1 | MGG_00446 | Casein kinase II subunit beta-1 | 19 | [26] |
| CKb2/CK2b2 | MGG_05651 | Casein kinase II subunit beta-2 | 1 | [26] |
| Yck1 | MGG_02829 | Casein kinase I | 6 | [2] |
| Rad3 | MGG_12633 | The ortholog of protein kinase Rad3 in S. pombe | 3 | |
| Kin1 | MGG_01279 | Camkl Kin1 protein kinase | 1 | [45] |
| Hik3 | MGG_12530 | Histidine kinase | 1 | [46] |
| Hik1 | MGG_11174 | Histidine kinase | 1 | [46] |
| Sik1 | MGG_07915 | Pre-rRNA processing nucleolar protein Sik1 | 7 | |
| Nop58 | MGG_07008 | Nucleolar protein Nop-58 | 5 | |
| Ssr4 | MGG_00174 | SWI/SNF and RSC complexes subunit Ssr4 | 2 | |
| Swi3 | MGG_01720 | The SWI/SNF chromatin remodeling complex | 4 | |
| Nop2 | MGG_01292 | Nucleolar protein Nop2 | 2 | [47] |
| Arp8 | MGG_05229 | INO80 chromatin remodeling complex subunit (Arp8) | 1 | |
| Ies1 | MGG_08312 | INO80 chromatin remodeling complex | 1 | |
| RuvB-like helicase 1 | MGG_03958 | INO80 chromatin remodeling complex | 2 | |
| Fzc53 | MGG_09829 | Zn(2)-Cys(6) zinc finger domain protein | 1 | [30] |
| Tup1 | (MGG_08829) | Transcriptional repressor Rco-1 | 1 | [48] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, P.; Li, Y.; Wang, J.; Wang, Q.; Huang, Z.; Liu, X.; Lin, F.; Lu, J. Casein Kinase 2 Mediates Degradation of Transcription Factor Pcf1 during Appressorium Formation in the Rice Blast Fungus. J. Fungi 2022, 8, 144. https://doi.org/10.3390/jof8020144
Huang P, Li Y, Wang J, Wang Q, Huang Z, Liu X, Lin F, Lu J. Casein Kinase 2 Mediates Degradation of Transcription Factor Pcf1 during Appressorium Formation in the Rice Blast Fungus. Journal of Fungi. 2022; 8(2):144. https://doi.org/10.3390/jof8020144
Chicago/Turabian StyleHuang, Pengyun, Yan Li, Jing Wang, Qing Wang, Zhicheng Huang, Xiaohong Liu, Fucheng Lin, and Jianping Lu. 2022. "Casein Kinase 2 Mediates Degradation of Transcription Factor Pcf1 during Appressorium Formation in the Rice Blast Fungus" Journal of Fungi 8, no. 2: 144. https://doi.org/10.3390/jof8020144
APA StyleHuang, P., Li, Y., Wang, J., Wang, Q., Huang, Z., Liu, X., Lin, F., & Lu, J. (2022). Casein Kinase 2 Mediates Degradation of Transcription Factor Pcf1 during Appressorium Formation in the Rice Blast Fungus. Journal of Fungi, 8(2), 144. https://doi.org/10.3390/jof8020144







