Role of Two G-Protein α Subunits in Vegetative Growth, Cell Wall Integrity, and Virulence of the Entomopathogenic Fungus Metarhizium robertsii
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strains and Culture
2.2. Sequence Analysis
2.3. Subcellular Localization of MrGPA2 and MrGPA4
2.4. Gene Deletion and Complementation
2.5. Phenotype Assays
2.6. Quantitative RT-PCR (RT-qPCR)
2.7. Statistical Analysis
3. Results
3.1. Characteristics of MrGPA2 and MrGPA4 from M. robertsii
3.2. MrGPA2 and MrGPA4 Are Located in Cytoplasm
3.3. Gene Knockout and Complementation
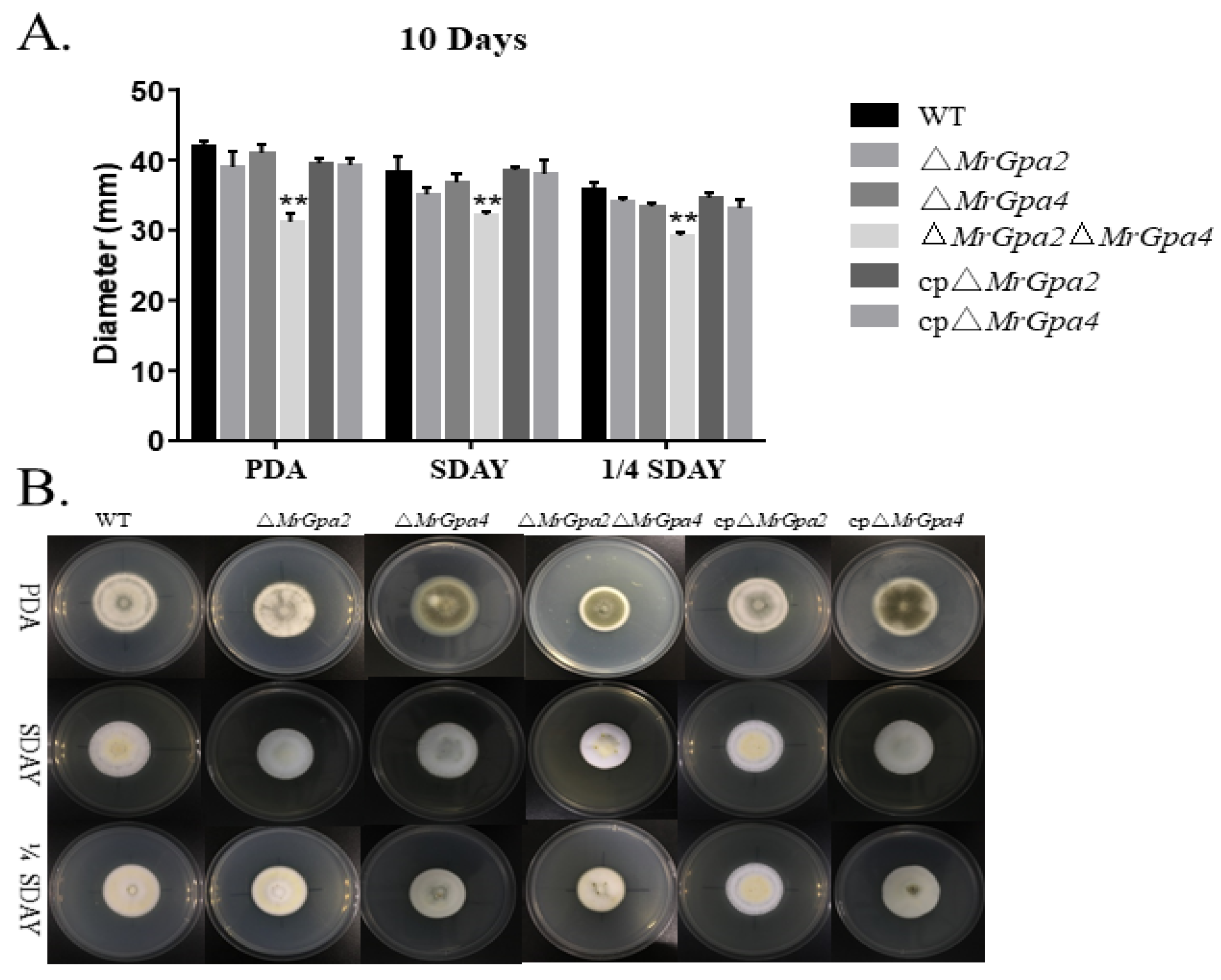
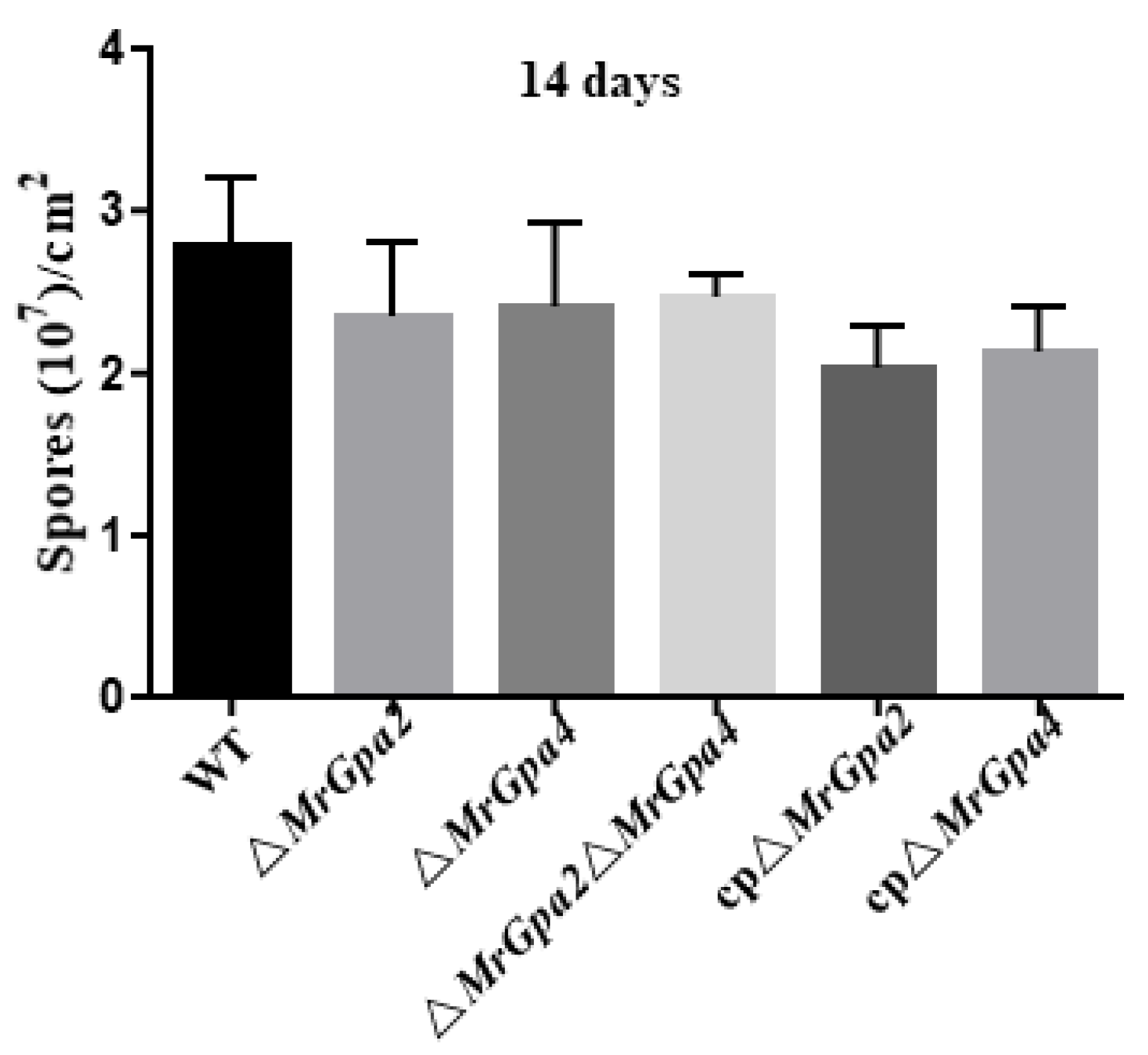
3.4. ∆MrGpa2∆MrGpa4 Strain Leads to a Decrease in Vegetative Growth but Had No Effect on Conidiation
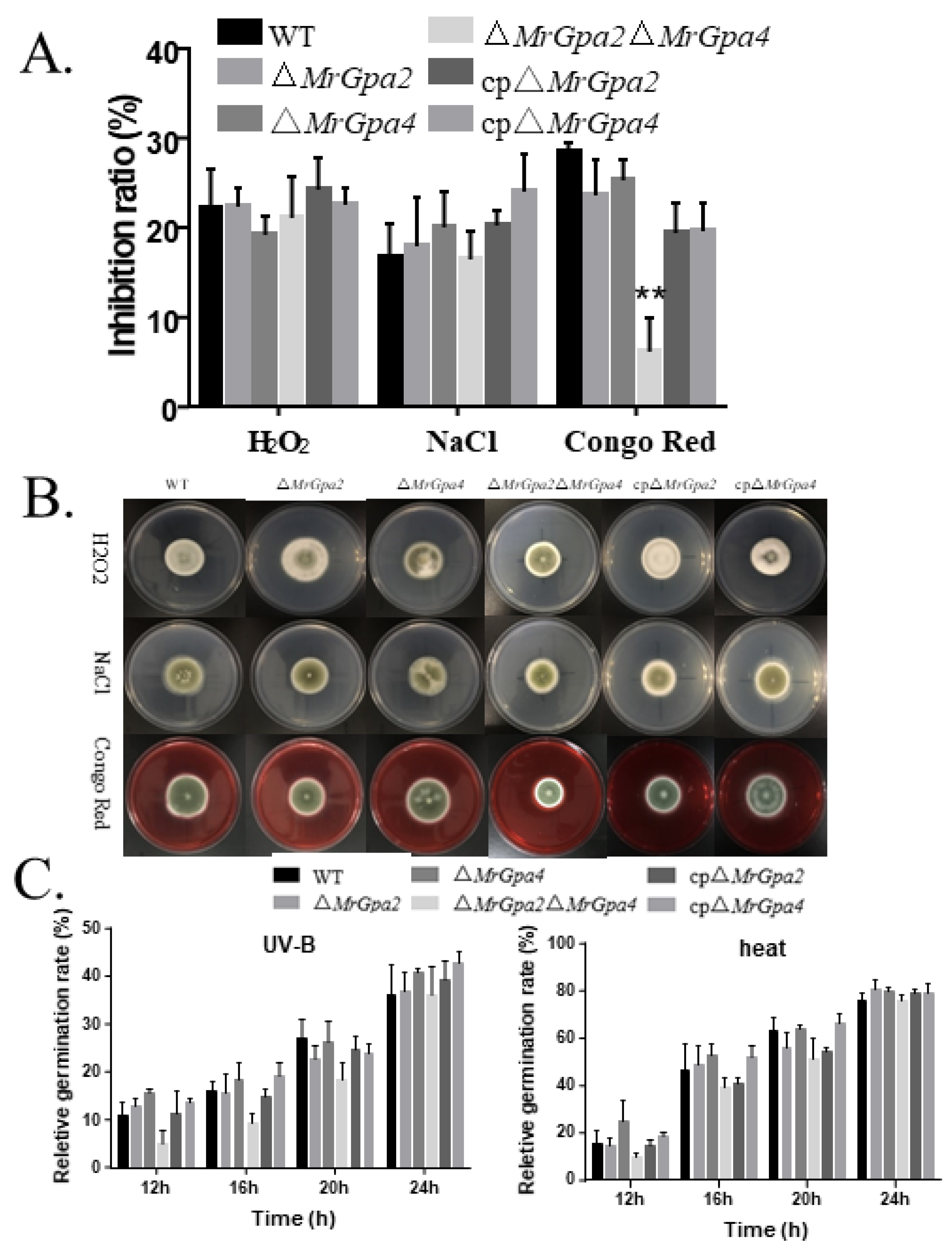
3.5. Deletion of Both MrGpa2 and MrGpa4 Enhanced Cell Wall Integrity
3.6. ∆MrGpa2∆MrGpa4 Strain Displayed an Increased Survival Rate of the Larvae in Bioassays
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhu, X.G.; Tong, S.M.; Ying, S.H.; Feng, M.G. Antioxidant activities of four superoxide dismutases in Metarhizium robertsii and their contributions to pest control potential. Appl. Microbiol. Biotechnol. 2018, 102, 9221–9230. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, S. Insect Pathogenic Fungi: Genomics, Molecular Interactions, and Genetic Improvements. Annu. Rev. Entomol. 2017, 62, 73–90. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.X.; Mouhoumed, A.Z.; Tong, S.M.; Ying, S.H.; Feng, M.G. BrlA and abaA govern virulence-required dimorphic switch, conidiation, and pathogenicity in a fungal insect pathogen. mSystems 2019, 4, e00140-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakravorty, D.; Assmann, S.M. G protein subunit phosphorylation as a regulatory mechanism in heterotrimeric G protein signaling in mammals, yeast, and plants. Biochem. J. 2018, 475, 3331–3357. [Google Scholar] [CrossRef] [PubMed]
- Park, H.S.; Kim, M.J.; Yu, J.H.; Shin, K.S. Heterotrimeric G-protein signalers and RGSs in Aspergillus fumigatus. Pathogens 2020, 9, 902. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, K.; Qin, Q.; Lin, G.; Hu, T.; Xu, Z.; Wang, S. G protein alpha subunit GpaB is required for asexual development, aflatoxin biosynthesis and pathogenicity by regulating cAMP signaling in Aspergillus flavus. Toxins 2018, 10, 117. [Google Scholar] [CrossRef] [Green Version]
- Guo, L.; Yang, L.; Liang, C.; Wang, J.; Liu, L.; Huang, J. The G-protein subunits FGA2 and FGB1 play distinct roles in development and pathogenicity in the banana fungal pathogen Fusarium oxysporum f. sp. cubense. Physiol. Mol. Plant Pathol. 2016, 93, 29–38. [Google Scholar] [CrossRef]
- Li, Y.; Que, Y.; Liu, Y.; Yue, X.; Meng, X.; Zhang, Z.; Wang, Z. The putative G gamma subunit gene MGG1 is required for conidiation, appressorium formation, mating and pathogenicity in Magnaporthe oryzae. Curr. Genet. 2015, 61, 641–651. [Google Scholar] [CrossRef]
- Tong, Y.; Wu, H.; Liu, Z.; Wang, Z.; Huang, B. G-protein subunit Gα(i) in mitochondria, MrGPA1, affects conidiation, stress resistance, and virulence of entomopathogenic fungus Metarhizium robertsii. Front. Microbiol. 2020, 11, 1251. [Google Scholar] [CrossRef]
- Harashima, T.; Heitman, J. G alpha subunit Gpa2 recruits kelch repeat subunits that inhibit receptor-G protein coupling during cAMP-induced dimorphic transitions in Saccharomyces cerevisiae. Mol. Biol. Cell 2005, 16, 4557–4571. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Wright, S.J.; Krystofova, S.; Park, G.; Borkovich, K.A. Heterotrimeric G protein signaling in filamentous fungi. Annu. Rev. Microbiol. 2007, 61, 423–452. [Google Scholar] [CrossRef] [PubMed]
- Won, S.; Michkov, A.V.; Krystofova, S.; Garud, A.V.; Borkovich, K.A. Genetic and physical interactions between G alpha subunits and components of the G beta gamma dimer of heterotrimeric G proteins in Neurospora crassa. Eukaryot. Cell 2012, 11, 1239–1248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gronover, C.S.; Kasulke, D.; Tudzynski, P.; Tudzynski, B. The role of G protein alpha subunits in the infection process of the gray mold fungus Botrytis cinerea. Mol. Plant Microbe Interact. 2001, 14, 1293–1302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, L.; Yang, Y.; Yang, L.; Wang, F.; Wang, G.; Huang, J. Functional analysis of the G-protein α subunits FGA1 and FGA3 in the banana pathogen Fusarium oxysporum f. sp. cubense. Physiol. Mol. Plant Pathol. 2016, 94, 75–82. [Google Scholar] [CrossRef]
- Gao, Q.; Jin, K.; Ying, S.H.; Zhang, Y.; Xiao, G.; Shang, Y.; Duan, Z.; Hu, X.; Xie, X.Q.; Zhou, G.; et al. Genome sequencing and comparative transcriptomics of the model entomopathogenic fungi Metarhizium anisopliae and M. acridum. PLoS Genet. 2011, 7, e1001264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, W.; Pei, Y.; Bidochka, M.J. Transformation of Metarhizium anisopliae mediated by Agrobacterium tumefaciens. Can. J. Microbiol. 2006, 52, 623–626. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, B.; Cen, K.; Lu, Y.; Zhang, S.; Wang, C. Diverse effect of phosphatidylcholine biosynthetic genes on phospholipid homeostasis, cell autophagy and fungal developments in Metarhizium robertsii. Environ. Microbiol. 2018, 20, 293–304. [Google Scholar] [CrossRef]
- Fang, W.; Bidochka, M.J. Expression of genes involved in germination, conidiogenesis and pathogenesis in Metarhizium anisopliae using quantitative real-time RT-PCR. Mycol. Res. 2006, 110 Pt 10, 1165–1171. [Google Scholar] [CrossRef]
- Huang, W.; Shang, Y.; Chen, P.; Gao, Q.; Wang, C. MrpacC regulates sporulation, insect cuticle penetration and immune evasion in Metarhizium robertsii. Environ. Microbiol. 2015, 17, 994–1008. [Google Scholar] [CrossRef]
- Jin, K.; Peng, G.; Liu, Y.; Xia, Y. The acid trehalase, ATM1, contributes to the in vivo growth and virulence of the entomopathogenic fungus, Metarhizium acridum. Fungal. Genet. Biol. 2015, 77, 61–67. [Google Scholar] [CrossRef]
- Liu, Q.; Ying, S.H.; Li, J.G.; Tian, C.G.; Feng, M.G. Insight into the transcriptional regulation of Msn2 required for conidiation, multi-stress responses and virulence of two entomopathogenic fungi. Fungal. Genet. Biol. 2013, 54, 42–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ming, Y.; Wei, Q.; Jin, K.; Xia, Y. MaSnf1, a sucrose non-fermenting protein kinase gene, is involved in carbon source utilization, stress tolerance, and virulence in Metarhizium acridum. Appl. Microbiol. Biotechnol. 2014, 98, 10153–10164. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Kang, Q.; Lu, Y.; Bai, L.; Wang, C. Unveiling the biosynthetic puzzle of destruxins in Metarhizium species. Proc. Natl. Acad. Sci. USA 2012, 109, 1287–1292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; St Leger, R.J. A collagenous protective coat enables Metarhizium anisopliae to evade insect immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 6647–6652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, J.S.; Selvaraj, V.; Gunupuru, L.R.; Kharwar, R.N.; Sarma, B.K. Plant G-protein signaling cascade and host defense. 3 Biotech 2020, 10, 219. [Google Scholar] [CrossRef]
- Marrari, Y.; Crouthamel, M.; Irannejad, R.; Wedegaertner, P.B. Assembly and trafficking of heterotrimeric G proteins. Biochemistry 2007, 46, 7665–7677. [Google Scholar] [CrossRef] [Green Version]
- Nair, R.; Kiran, A.; Saini, D. G protein Signaling, Journeys Beyond the Plasma Membrane. J. Indian Inst. Sci. 2017, 97, 95–108. [Google Scholar] [CrossRef]
- Kou, Y.; Naqvi, N.I. Surface sensing and signaling networks in plant pathogenic fungi. Semin. Cell Dev. Biol. 2016, 57, 84–92. [Google Scholar] [CrossRef]
- Yu, D.; Xie, R.; Wang, Y.; Xie, T.; Xu, L.; Huang, B. The G-protein coupled receptor GPRK contributes to fungal development and full virulence in Metarhizium robertsii. J. Invertebr. Pathol. 2021, 183, 107627. [Google Scholar] [CrossRef]
- Shang, J.; Shang, Y.; Tang, G.; Wang, C. Identification of a key G-protein coupled receptor in mediating appressorium formation and fungal virulence against insects. Sci. China Life Sci. 2021, 64, 466–477. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tong, Y.; Wu, H.; He, L.; Qu, J.; Liu, Z.; Wang, Y.; Chen, M.; Huang, B. Role of Two G-Protein α Subunits in Vegetative Growth, Cell Wall Integrity, and Virulence of the Entomopathogenic Fungus Metarhizium robertsii. J. Fungi 2022, 8, 132. https://doi.org/10.3390/jof8020132
Tong Y, Wu H, He L, Qu J, Liu Z, Wang Y, Chen M, Huang B. Role of Two G-Protein α Subunits in Vegetative Growth, Cell Wall Integrity, and Virulence of the Entomopathogenic Fungus Metarhizium robertsii. Journal of Fungi. 2022; 8(2):132. https://doi.org/10.3390/jof8020132
Chicago/Turabian StyleTong, Youmin, Hao Wu, Lili He, Jiaojiao Qu, Zhenbang Liu, Yulong Wang, Mingjun Chen, and Bo Huang. 2022. "Role of Two G-Protein α Subunits in Vegetative Growth, Cell Wall Integrity, and Virulence of the Entomopathogenic Fungus Metarhizium robertsii" Journal of Fungi 8, no. 2: 132. https://doi.org/10.3390/jof8020132
APA StyleTong, Y., Wu, H., He, L., Qu, J., Liu, Z., Wang, Y., Chen, M., & Huang, B. (2022). Role of Two G-Protein α Subunits in Vegetative Growth, Cell Wall Integrity, and Virulence of the Entomopathogenic Fungus Metarhizium robertsii. Journal of Fungi, 8(2), 132. https://doi.org/10.3390/jof8020132




