Multi-Gene Phylogenetic Analyses Revealed Five New Species and Two New Records of Distoseptisporales from China
Abstract
1. Introduction
2. Materials and Methods
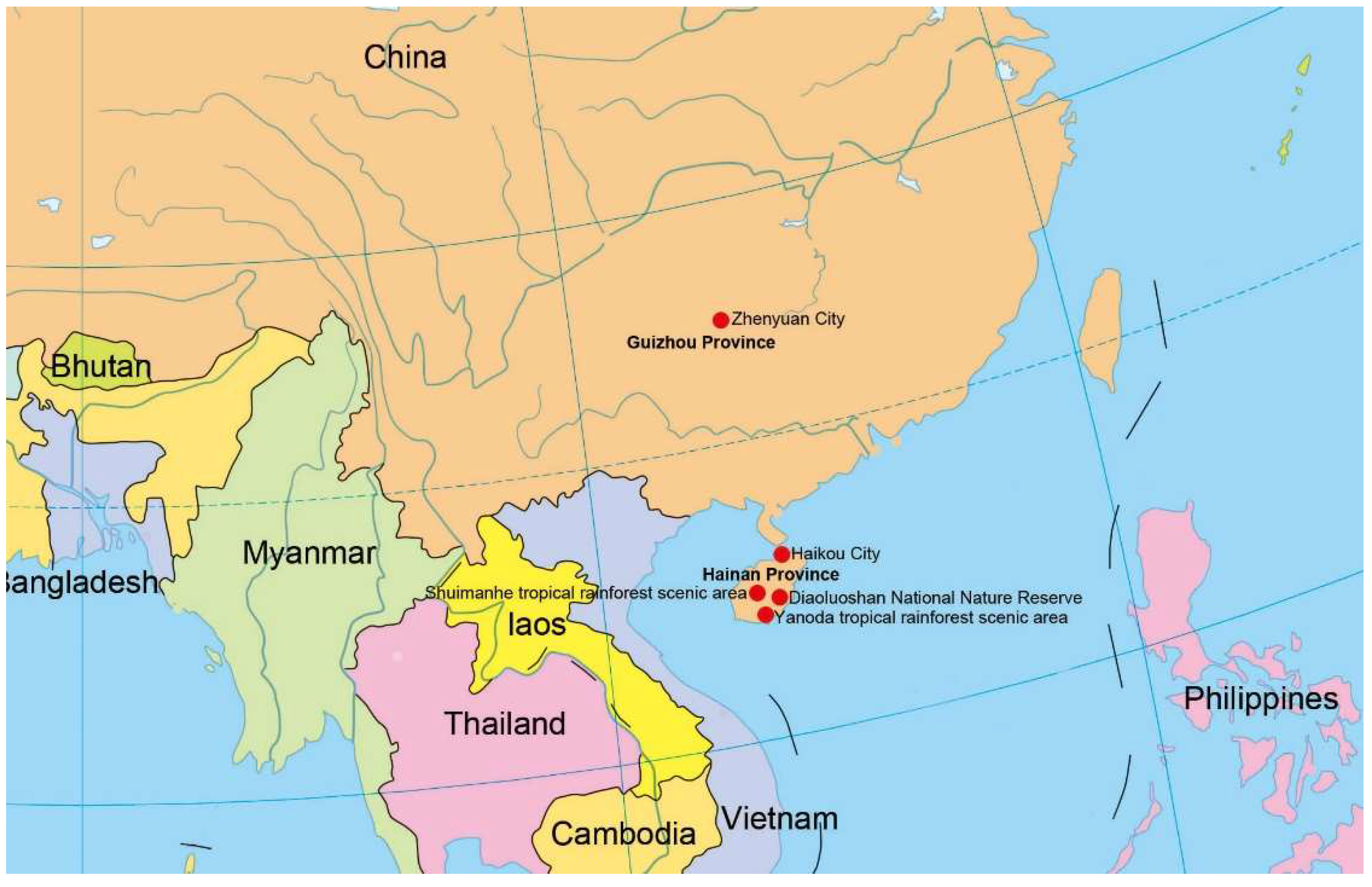
2.1. Sample Collection, Specimen Examination, and Isolation
2.2. DNA Extraction, PCR Amplification, and Sequencing
2.3. Phylogenetic Analyses
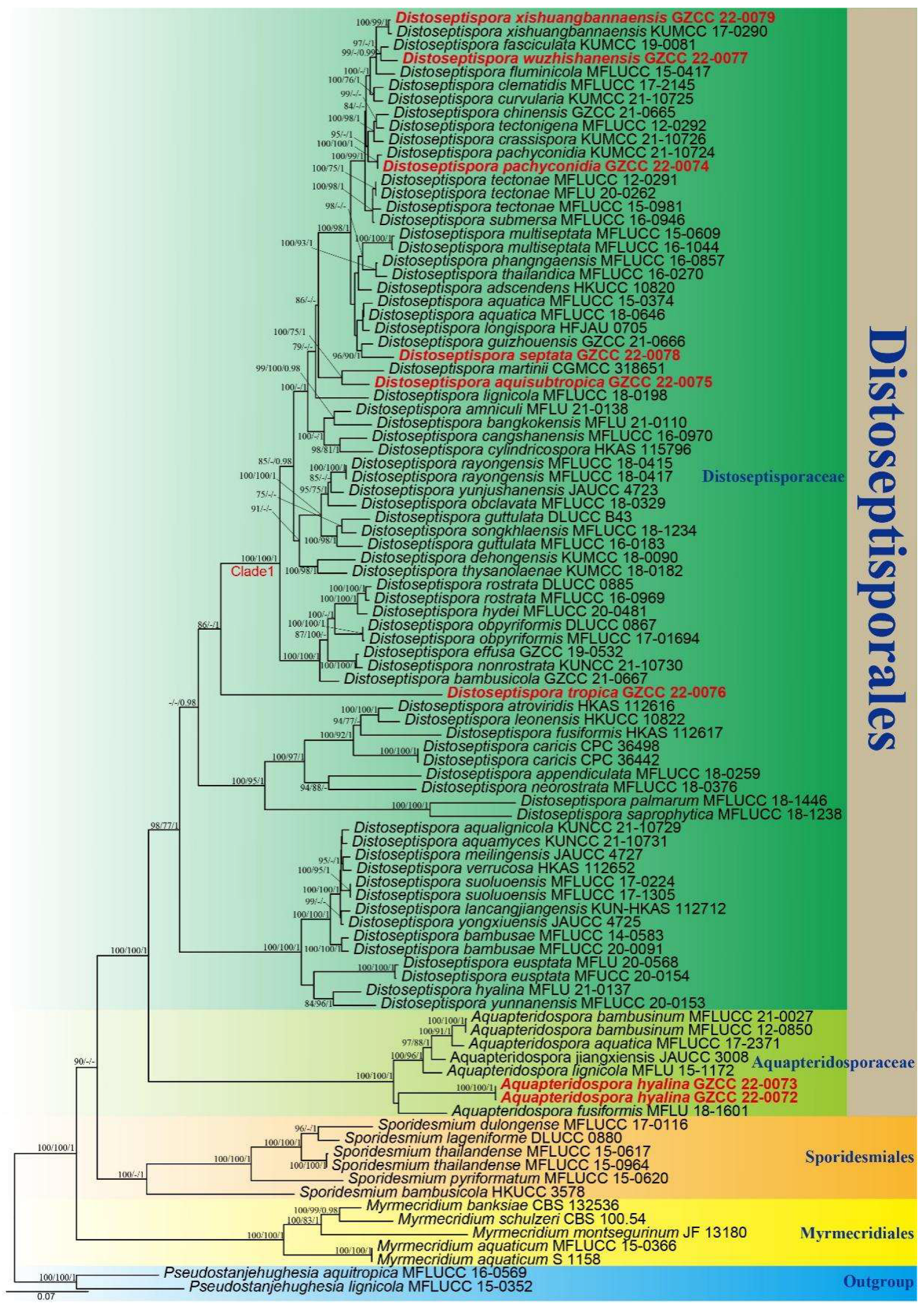
3. Phylogenetic Results
4. Taxonomy
5. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hyde, K.D.; Bao, D.F.; Hongsanan, S.; Chethana, K.; Yang, J.; Suwannarach, N. Evolution of freshwater Diaporthomycetidae (Sordariomycetes) provides evidence for five new orders and six new families. Fungal Divers. 2021, 107, 71–105. [Google Scholar] [CrossRef]
- Luo, Z.L.; Hyde, K.D.; Liu, J.K.; Maharachchikumbura, S.S.N.; Jeewon, R.; Bao, D.F.; Bhat, D.J.; Lin, C.G.; Li, W.L.; Yang, J.; et al. Freshwater Sordariomycetes. Fungal Divers. 2019, 99, 451–660. [Google Scholar] [CrossRef]
- Dong, W.; Hyde, K.D.; Jeewon, R.; Doilom, M.; Yu, X.D.; Wang, G.N.; Liu, N.G.; Hu, D.M.; Nalumpang, S.; Zhang, H. Towards a natural classification of annulatascaceae-like taxa II: Introducing five new genera and eighteen new species from freshwater. Mycosphere 2021, 12, 1–88. [Google Scholar] [CrossRef]
- Yang, J.; Maharachchikumbura, S.S.N.; Hyde, K.D.; Bhat, D.J.; McKenzie, E.H.C.; Bahkali, A.H.; Gareth Jones, E.B.G.; Liu, Z.Y. Aquapteridospora lignicola gen. et sp. nov., a new hyphomycetous taxon (Sordariomycetes) from wood submerged in a freshwater stream. Cryptogam. Mycol. 2015, 36, 469–478. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Al-Ani, L.K.T.; Tedersoo, L.; Haelewaters, D.; Rajeshkumar, K.C.; Zhao, R.L.; Aptroot, A.; Leontyev, D.V.; Saxena, R.K.; et al. Outline of Fungi and fungus-like taxa. Mycosphere 2020, 11, 1060–1456. [Google Scholar] [CrossRef]
- Dai, D.Q.; Phookamsak, R.; Wijayawardene, N.N.; Li, W.J.; Bhat, D.J.; Xu, J.C.; Taylor, J.E.; Hyde, K.D.; Chukeatirote, E. Bambusicolous fungi. Fungal Divers. 2016, 82, 1–105. [Google Scholar] [CrossRef]
- Yang, J.; Liu, L.L.; Jones, E.B.G.; Li, W.L.; Hyde, K.D.; Liu, Z.Y. Morphological variety in Distoseptispora and introduction of six novel species. J. Fungi. 2021, 7, 945. [Google Scholar] [CrossRef]
- Bao, D.F.; Hyde, K.D.; McKenzie, E.H.C.; Jeewon, R.; Su, H.Y.; Nalumpang, S.; Luo, Z.L. Biodiversity of lignicolous freshwater hyphomycetes from China and Thailand and description of sixteen species. J. Fungi. 2021, 7, 669. [Google Scholar] [CrossRef] [PubMed]
- Su, H.Y.; Hyde, K.D.; Maharachchikumbura, S.; Ariyawansa, H.A.; Luo, Z.L.; Promputtha, I.; Tian, Q.; Lin, C.G.; Shang, Q.J.; Zhao, Y.C.; et al. The families Distoseptisporaceae fam. nov., Kirschsteiniotheliaceae, Sporormiaceae and Torulaceae, with new species from freshwater in Yunnan Province, China. Fungal Divers. 2016, 80, 375–409. [Google Scholar] [CrossRef]
- Yang, J.; Maharachchikumbura, S.; Liu, J.K.; Hyde, K.D.; Jones, E.B.G.; Al-Sadi, A.M.; Liu, Z.Y. Pseudostanjehughesia aquitropica gen. et sp. nov. and Sporidesmium sensu lato species from freshwater habitats. Mycol. Prog. 2018, 17, 591–616. [Google Scholar] [CrossRef]
- Monkai, J.; Boonmee, S.; Ren, G.; Wei, D.; Phookamsak, R.; Mortimer, P.E. Distoseptispora hydei sp. nov. (Distoseptisporaceae), a novel lignicolous fungus on decaying bamboo in Thailand. Phytotaxa 2020, 459, 93–107. [Google Scholar] [CrossRef]
- Shen, H.W.; Bao, D.F.; Hyde, K.D.; Su, H.Y.; Bhat, D.J.; Luo, Z.L. Two novel species and two new records of Distoseptispora from freshwater habitats in China and Thailand. MycoKeys 2021, 84, 79–101. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.W.; Ma, Y.R.; Li, Z.; Zhang, X.G. Acrodictys-like wood decay fungi from southern China, with two new families Acrodictyaceae and Junewangiaceae. Sci. Rep. 2017, 7, 7888. [Google Scholar] [CrossRef] [PubMed]
- Hyde, K.D.; Hongsanan, S.; Jeewon, R.; Bhat, D.J.; McKenzie, E.H.C.; Jones, E.B.G.; Phookamsak, R.; Ariyawansa, H.; Boonmee, S.; Zhao, Q.; et al. Fungal diversity notes 367–490: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016, 80, 1–270. [Google Scholar] [CrossRef]
- Hyde, K.D.; Tennakoon, D.S.; Jeewon, R.; Bhat, D.J.; Maharachchikumbura, S.; Rossi, W.; Leonardi, M.; Lee, H.B.; Mun, H.Y.; Houbraken, J.; et al. Fungal diversity notes 1036–1150: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2019, 96, 1–242. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, R.; Qing, Y.; Yang, H.; Li, C.X.; Wang, G.N.; Zhang, D.; Ning, P. Polyphasic identification of Distoseptispora with six new species from freshwater. J. Fungi 2022, 8, 1063. [Google Scholar] [CrossRef]
- Maharachchikumbura, S.S.N.; Hyde, K.D.; Jones, E.B.G.; McKenzie, E.H.C.; Bhat, D.J.; Dayarathne, M.C.; Huang, S.K.; Norphanphoun, C.; Senanayake, I.C.; Perera, R.H.; et al. Families of Sordariomycetes. Mycosphere 2016, 79, 1–317. [Google Scholar] [CrossRef]
- Luo, Z.L.; Hyde, K.D.; Liu, J.K.; Bhat, D.J.; Su, H.; Bao, D.F.; Li, W.L. Lignicolous freshwater fungi from China II: Novel Distoseptispora (Distoseptisporaceae) species from northwestern Yunnan Province and a suggested unified method for studying lignicolous freshwater fungi. Mycosphere 2018, 9, 444–461. [Google Scholar] [CrossRef]
- Phookamsak, R.; Hyde, K.D.; Jeewon, R.; Bhat, D.J.; Jones, E.B.G.; Maharachchikumbura, S.; Raspé, O.; Karunarathna, S.C.; Wanasinghe, D.; Hongsanan, S.; et al. Fungal diversity notes 929–1035: Taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Divers. 2019, 95, 1–273. [Google Scholar] [CrossRef]
- Tibpromma, S.; Hyde, K.D.; Mckenzie, E.; Bhat, D.J.; Phillips, A.J.L.; Wanasinghe, D.N.; Samarakoon, M.C.; Jayawardena, R.S. Fungal diversity notes 840–928: Micro-fungi associated with Pandanaceae. Fungal Divers. 2018, 100, 1–160. [Google Scholar] [CrossRef]
- Li, W.L.; Liu, Z.P.; Zhang, T.; Dissanayake, A.J.; Luo, Z.L.; Su, H.Y.; Liu, J.K. Additions to Distoseptispora (Distoseptisporaceae) associated with submerged decaying wood in China. Phytotaxa 2021, 520, 75–86. [Google Scholar] [CrossRef]
- Shoemaker, R.A.; White, G.P. Lasiosphaeria caesariata with Sporidesmium hormiscioides and L. triseptata with S. adscendens. Sydowia 1985, 38, 278–283. [Google Scholar]
- McKenzie, E.H.C. Dematiaceous hyphomycetes on Pandanaceae. 5. Sporidesmium sensu lato. Mycotaxon 1995, 56, 9–29. [Google Scholar]
- Hyde, K.D. Mycosphere notes 325-344–Novel species and records of fungal taxa from around the world. Mycosphere 2021, 12, 1101–1156. [Google Scholar] [CrossRef]
- Sun, Y.; Goonasekara, I.D.; Thambugala, K.M.; Jayawardena, R.S.; Wang, Y.; Hyde, K.D. Distoseptispora bambusae sp. nov. (Distoseptisporaceae) on bamboo from China and Thailand. Biodivers. Data J. 2020, 8, e53678. [Google Scholar] [CrossRef]
- Phukhamsakda, C.; McKenzie, E.H.C.; Phillips, A.; Jones, E.B.G.; Bhat, D.J.; Stadler, M.; Bhunjun, C.S.; Wanasinghe, D.N.; Thongbai, B.; Camporesi, E.; et al. Microfungi associated with Clematis (Ranunculaceae) with an integrated approach to delimiting species boundaries. Fungal Divers. 2020, 102, 1–203. [Google Scholar] [CrossRef]
- Phukhamsakda, C.; Nilsson, R.H.; Bhunjun, C.S.; Farias, A.R.G.D.; Sun, Y.; Wijesinghe, S.N.; Raza, M.; Bao, D.F.; Lu, L.; Tibpromma, S.; et al. The numbers of fungi: Contributions from traditional taxonomic studies and challenges of metabarcoding. Fungal Divers. 2022, 114, 327–386. [Google Scholar] [CrossRef]
- Zhai, Z.J.; Yan, J.Q.; Li, W.W.; Gao, Y.; Hu, H.J.; Zhou, J.P.; Song, H.Y.; Hu, D.M. Three novel species of Distoseptispora (Distoseptisporaceae) isolated from bamboo in Jiangxi Province, China. MycoKeys 2022, 88, 35–54. [Google Scholar] [CrossRef]
- Crous, P.; Wingfield, M.; Lombard, L.; Roets, F.; Swart, W.; Alvarado, P.; Carnegie, A.; Moreno, G.; Luangsa-Ard, J.; Thangavel, R.; et al. Fungal Planet description sheets: 951–1041. Pers. Mol. Phylogeny Evol. Fungi. 2019, 43, 223–425. [Google Scholar] [CrossRef]
- Senanayake, I.C.; Rathnayaka, A.R.; Marasinghe, D.S.; Calabon, M.S.; Gentekaki, E.; Lee, H.B.; Hurdeal, V.G.; Pem, D.; Dis-sanayake, L.S.; Wijesinghe, S.N.; et al. Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere 2020, 11, 2678–2754. [Google Scholar] [CrossRef]
- Jayasiri, S.C.; Hyde, K.D.; Ariyawansa, H.A.; Bhat, J.; Buyck, B.; Cai, L.; Dai, Y.C.; Abd-Elsalam, K.A.; Ertz, D.; Hidayat, I.; et al. The Faces of Fungi database: Fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 2015, 74, 3–18. [Google Scholar] [CrossRef]
- Index Fungorum. Available online: http://www.indexfungorum.org/names/names.asp. (accessed on 5 May 2021).
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed]
- White, T.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols—A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Rehner, S.A.; Samuels, G.J. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol. Res. 1994, 98, 625–634. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Katoh, K.; Standley, D.M. Evolution. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Daniel, G.P.; Daniel, G.B.; Miguel, R.J.; Florentino, F.R.; David, P. ALTER: Program-oriented conversion of DNA and protein alignments. Nucleic Acids Res. 2010, 38, W14–W18. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods); Version 4; Sinauer Associates: Sunderland, UK, 2003. [Google Scholar]
- Hillis, D.M.; Bull, J.J. An Empirical Test of Bootstrapping as a Method for Assessing Confidence in Phylogenetic Analysis. Syst. Biol. 1993, 42, 182–192. [Google Scholar] [CrossRef]
- Nylander, J.A.A.; Zoology, S.; Posada, D.; Mrmodeltest, R.; Os, F. MrModeltest2 v. 2.3 (Program for Selecting DNA Substitution Models Using PAUP*); Evolutionary Biology Centre: Uppsala, Sweden, 2008. [Google Scholar]
- Huelsenbeck, J.P.; Ronquist, F.J.B. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Dong, W.; Hyde, K.D.; Maharachchikumbura, S.S.; Hongsanan, S.; Bhat, D.J.; Al-Sadi, A.M.; Zhang, D. Towards a natural classification of Annulatascaceae-like taxa: Introducing Atractosporales ord. nov. and six new families. Fungal Divers. 2017, 85, 75–110. [Google Scholar] [CrossRef]
- Crous, P.W.; Summerell, B.A.; Shivas, R.G.; Burgess, T.I.; Decock, C.A.; Dreyer, L.L.; Granke, L.L.; Guest, D.I.; Hardy, G.; Hausbeck, M.; et al. Fungal Planet description sheets: 107–127. Persoonia 2012, 28, 138–182. [Google Scholar] [CrossRef] [PubMed]
- Arzanlou, M.; Groenewald, J.Z.; Gams, W.; Braun, U.; Shin, H.D.; Crous, P.W. Phylogenetic and morphotaxonomic revision of Ramichloridium and allied genera. Stud. Mycol. 2007, 58, 57–93. [Google Scholar] [CrossRef] [PubMed]
- Réblová, M.; Fournier, J.; Štěpánek, V. Two new lineages of aquatic ascomycetes: Atractospora gen. nov. and Rubellisphaeria gen. et sp. nov., and a sexual morph of Myrmecridium montsegurinum sp. nov. Mycol. Prog. 2016, 15, 21. [Google Scholar] [CrossRef]
- Chethana, K.W.T.; Manawasinghe, I.S.; Hurdeal, V.G.; Bhunjun, C.S.; Appadoo, M.A.; Gentekaki, E.; Raspé, O.; Promputtha, I.; Hyde, K.D. What are fungal species and how to delineate them? Fungal Divers. 2021, 109, 1–25. [Google Scholar] [CrossRef]
- Maharachchikumbura, S.S.N.; Chen, Y.; Ariyawansa, H.A.; Hyde, K.D.; Haelewaters, D.; Perera, R.H.; Samara-koon, M.C.; Wanasinghe, D.N.; Bustamante, D.E.; Liu, J.K.; et al. Integrative approaches for species delimitation in Ascomycota. Fungal Divers. 2021, 109, 155–179. [Google Scholar] [CrossRef]
- Pem, D.; Jeewon, R.; Chethana, K.W.T.; Hongsanan, S.; Doilom, M.; Suwannarach, N.; Hyde, K.D. Species concepts of Dothideomycetes: Classification, phylogenetic inconsistencies and taxonomic standardization. Fungal Divers. 2021, 109, 283–319. [Google Scholar] [CrossRef]
- Shenoy, B.D.; Jeewon, R.; Wu, W.P.; Bhat, D.J.; Hyde, K.D. Ribosomal and RPB2 DNA sequence analyses suggest that Sporidesmium and morphologically similar genera are polyphyletic. Mycol. Res. 2006, 110, 916–928. [Google Scholar] [CrossRef]
- Bhunjun, C.S.; Niskanen, T.; Suwannarach, N.; Wannathes, N.; Chen, Y.-J.; McKenzie, E.H.C.; Maharachchikumbura, S.S.N.; Buyck, B.; Zhao, C.-L.; Fan, Y.-G.; et al. The numbers of fungi: Are the most speciose genera truly diverse? Fungal Divers. 2022, 114, 387–462. [Google Scholar] [CrossRef]
- Hyde, K.D.; Jeewon, R.; Chen, Y.J.; Bhunjun, C.S.; Calabon, M.S.; Jiang, H.B.; Lin, C.G.; Norphanphoun, C.; Sysouphanthong, P.; Pem, D.; et al. The numbers of fungi: Is the descriptive curve flattening? Fungal Divers. 2020, 103, 219–271. [Google Scholar] [CrossRef]
- Dong, W.; Wang, B.; Hyde, K.D.; McKenzie, E.H.C.; Raja, H.A.; Tanaka, K.; Abdel-Wahab, M.A.; Abdel-Aziz, F.A.; Doilom, M.; Phookamsak, R.; et al. Freshwater Dothideomycetes. Fungal Divers. 2020, 105, 319–575. [Google Scholar] [CrossRef]
- Peng, S.Q.; Liu, Y.L.; Huang, J.E.; Li, X.H.; Yan, X.Y.; Song, H.Y.; Gao, Y.; Zhai, Z.J.; Liu, Y.Q.; Hu, D.M. Aquapteridospora jiangxiensis, a new aquatic hyphomycetous fungus from a freshwater habitat in China. Arch. Microbiol. 2022, 204, 378. [Google Scholar] [CrossRef] [PubMed]







| Locus | Primer | Initial Denaturation | Denaturation | Annealing | Elongation | Final Extension | Hold |
|---|---|---|---|---|---|---|---|
| LSU | ITS5/ITS4 | 94 °C/3 min | 94 °C/45 s | 56 °C/50 s | 72 °C/1 min | 72 °C/10 min | 4 °C |
| 40 cycles | |||||||
| ITS | LR0R/LR5 | 94 °C/3 min | 94 °C/45 s | 56 °C/50 s | 72 °C/1 min | 72 °C/10 min | |
| 40 cycles | |||||||
| tef1α | 983F/2218R | 94 °C/3 min | 94 °C/30 s | 56 °C/50 s | 72 °C/1 min | 72 °C/10 min | |
| 30 cycles | |||||||
| rpb2 | frpb2-5f/frpb2-7cr | 95 °C/5 min | 95 °C/15 s | 56 °C/50 s | 72 °C/1 min | 72 °C/10 min | |
| 35 cycles | |||||||
| Taxon | Strain | GenBank Accessions | Reference | |||
|---|---|---|---|---|---|---|
| LSU | ITS | tef1α | rpb2 | |||
| Aquapteridospora fusiformis | MFLU 18-1601T | MK849798 | MK828652 | MN194056 | – | Luo et al. (2019) |
| A. aquatica | MFLUCC 17-2371T | MW287767 | MW286493 | – | – | Dong et al. (2021) |
| A. bambusinum | MFLUCC 12-0850T | KU863149 | KU940161 | KU940213 | Dai et al. (2016) | |
| A. bambusinum | MFLUCC 21-0027 | MZ412526 | MZ412514 | MZ442688 | – | Bao et al. (2021) |
| A. hyalina | GZCC 22-0072T | ON527945 | ON527937 | ON533681 | ON533689 | This study |
| A. hyalina | GZCC 22-0073 | ON527948 | ON527940 | ON533684 | ON533691 | This study |
| A. jiangxiensis | JAUCC 3008T | MZ871501 | MZ871502 | MZ855767 | MZ855768 | Peng et al. (2022) |
| A. lignicola | MFLU 15-1172T | KU221018 | MZ868774 | MZ892980 | MZ892986 | Yang et al. (2015) |
| Distoseptispora adscendens | HKUCC 10820 | DQ408561 | – | – | DQ435092 | Shenoy et al. (2006) |
| D. amniculi | MFLU 21-0138T | MZ868761 | MZ868770 | – | MZ892982 | Yang et al. (2021) |
| D. appendiculata | MFLUCC 18-0259T | MN163023 | MN163009 | MN174866 | – | Luo et al. (2019) |
| D. aqualignicola | KUNCC 21-10729T | ON400845 | OK341186 | OP413480 | OP413474 | Zhang et al. (2022) |
| D. aquamyces | KUNCC 21-10731T | OK341199 | OK341187 | OP413482 | OP413476 | Zhang et al. (2022) |
| D. aquatica | MFLUCC 15-0374T | KU376268 | MF077552 | – | – | Su et al. (2016) |
| D. aquatica | MFLUCC 18-0646 | MK849793 | MK828648 | – | – | Luo et al. (2019) |
| D. aquisubtropica | GZCC 22-0075T | ON527941 | ON527933 | ON533677 | ON533685 | This study |
| D. atroviridis | HKAS 112616T | MZ868763 | MZ868772 | MZ892978 | MZ892984 | Yang et al. (2021) |
| D. bambusae | MFLUCC 20-0091T | MT232718 | MT232713 | MT232880 | MT232881 | Sun et al. (2020) |
| D. bambusae | MFLUCC 14-0583 | MT232717 | MT232712 | – | MT232882 | Sun et al. (2020) |
| D. bambusicola | GZCC 21-0667T | MZ474872 | MZ474873 | – | – | |
| D. bangkokensis | MFLU 21-0110T | MZ518206 | MZ518205 | – | – | Shen et al. (2021) |
| D. cangshanensis | MFLUCC 16-0970T | MG979761 | MG979754 | MG988419 | – | Luo et al. (2018) |
| D. caricis | CPC 36498T | MN567632 | MN562124 | – | MN556805 | Crous et al. (2019) |
| D. caricis | CPC 36442 | – | MN562125 | – | MN556806 | Crous et al. (2019) |
| D. chinensis | GZCC 21-0665T | MZ474867 | MZ474871 | MZ501609 | – | Hyde et al. (2021) |
| D. clematidis | MFLUCC 17-2145T | MT214617 | MT310661 | – | MT394721 | Phukhamsakda et al. (2020) |
| D. crassispora | KUMCC 21-10726T | OK341196 | OK310698 | OP413479 | OP413473 | Zhang et al. (2022) |
| D. curvularia | KUMCC 21-10725T | OK341195 | OK310697 | OP413478 | OP413472 | Zhang et al. (2022) |
| D. cylindricospora | HKAS 115796T | OK513523 | OK491122 | OK524220 | – | Phukhamsakda et al. (2022) |
| D. dehongensis | KUMCC 18-0090T | MK079662 | MK085061 | MK087659 | – | Hyde et al. (2019) |
| D. effusa | GZCC 19-0532T | MZ227224 | MW133916 | – | – | Yang et al. (2021) |
| D. eusptata | MFUCC 20-0154T | MW081544 | MW081539 | – | MW151860 | Li et al. (2021) |
| D. eusptata | MFLU 20-0568 | MW081545 | MW081540 | MW084994 | MW084996 | Li et al. (2021) |
| D. fasciculata | KUMCC 19-0081T | MW287775 | MW286501 | MW396656 | – | Dong et al. (2021) |
| D. fluminicola | MFLUCC 15-0417T | KU376270 | MF077553 | – | – | Su et al. (2016) |
| D. fusiformis | HKAS 112617T | MZ868764 | MZ868773 | MZ892979 | MZ892985 | Yang et al. (2021) |
| D. guizhouensis | GZCC 21-0666T | MZ474869 | MZ474868 | MZ501610 | MZ501611 | Hyde et al. (2021) |
| D. guttulata | MFLUCC 16-0183T | MF077554 | MF077543 | MF135651 | – | Yang et al. (2018) |
| D. guttulata | DLUCC B43 | MN163016 | MN163011 | – | – | Luo et al. (2019) |
| D. hyalina | MFLU 21-0137T | MZ868760 | MZ868769 | MZ892976 | MZ892981 | Yang et al. (2021) |
| D. hydei | MFLUCC 20-0481T | MT742830 | MT734661 | – | MT767128 | Monkai et al. (2020) |
| D. lancangjiangensis | KUN-HKAS 112712T | MW879522 | MW723055 | – | MW882260 | Shen et al. (2021) |
| D. leonensis | HKUCC 10822 | DQ408566 | – | – | DQ435089 | Shenoy et al. (2006) |
| D. lignicola | MFLUCC 18-0198T | MK849797 | MK828651 | – | – | Luo et al. (2019) |
| D. longispora | HFJAU 0705T | MH555357 | MH555359 | – | – | Yang et al. (2021) |
| D. martinii | CGMCC 318651T | KX033566 | KU999975 | – | – | Xia et al. (2017) |
| D. meilingensis | JAUCC 4727T | OK562396 | OK562390 | OK562408 | – | Zhai et al. (2022) |
| D. multiseptata | MFLUCC 16-1044 | MF077555 | MF077544 | MF135652 | MF135644 | Yang et al. (2018) |
| D. multiseptata | MFLUCC 15-0609T | KX710140 | KX710145 | MF135659 | – | Hyde et al. (2016) |
| D. neorostrata | MFLUCC 18-0376T | MN163017 | MN163008 | – | – | Luo et al. (2019) |
| D. nonrostrata | KUNCC 21-10730T | OK341198 | OK310699 | OP413481 | OP413475 | Zhang et al. (2022) |
| D. obclavata | MFLUCC 18-0329T | MN163010 | MN163012 | – | – | Luo et al. (2019) |
| D. obpyriformis | MFLUCC 17-01694T | MG979764 | – | MG988422 | MG988415 | Luo et al. (2018) |
| D. obpyriformis | DLUCC 0867 | MG979765 | MG979757 | MG988423 | MG988416 | Luo et al. (2018) |
| D. pachyconidia | KUMCC 21-10724T | OK341194 | OK310696 | OP413477 | OP413471 | Zhang et al. (2022) |
| D. pachyconidia | GZCC 22-0074 | ON527942 | ON527934 | ON533678 | ON533686 | This study |
| D. palmarum | MFLUCC 18-1446T | MK079663 | MK085062 | MK087660 | MK087670 | Hyde et al. (2019) |
| D. phangngaensis | MFLUCC 16-0857T | MF077556 | MF077545 | MF135653 | – | Yang et al. (2018) |
| D. rayongensis | MFLUCC 18-0415T | MH457137 | MH457172 | MH463253 | MH463255 | Hyde et al. (2020) |
| D. rayongensis | MFLUCC 18-0417 | MH457138 | MH457173 | MH463254 | MH463256 | Hyde et al. (2020) |
| D. rostrata | MFLUCC 16-0969T | MG979766 | MG979758 | MG988424 | MG988417 | Luo et al. (2018) |
| D. rostrata | DLUCC 0885 | MG979767 | MG979759 | MG988425 | – | Luo et al. (2018) |
| D. saprophytica | MFLUCC 18-1238T | MW287780 | MW286506 | MW396651 | MW504069 | Dong et al. (2021) |
| D. septata | GZCC 22-0078T | ON527947 | ON527939 | ON533683 | ON533690 | This study |
| D. songkhlaensis | MFLUCC 18-1234T | MW287755 | MW286482 | MW396642 | – | Dong et al. (2021) |
| D. submersa | MFLUCC 16-0946T | MG979768 | MG979760 | MG988426 | MG988418 | Luo et al. (2018) |
| D. suoluoensis | MFLUCC 17-0224T | MF077557 | MF077546 | MF135654 | – | Yang et al. (2018) |
| D. suoluoensis | MFLUCC 17-1305 | MF077558 | MF077547 | – | – | Yang et al. (2018) |
| D. tectonae | MFLUCC 12-0291T | KX751713 | KX751711 | KX751710 | KX751708 | Hyde et al. (2016) |
| D. tectonae | MFLU 20-0262 | MT232719 | MT232714 | – | – | Sun et al. (2020) |
| D. tectonae | MFLUCC 15-0981 | MW287763 | MW286489 | MW396641 | – | Dong et al. (2021) |
| D. tectonigena | MFLUCC 12-0292T | KX751714 | KX751712 | – | KX751709 | Hyde et al. (2016) |
| D. thailandica | MFLUCC 16-0270T | MH260292 | MH275060 | MH412767 | – | Tibpromma et al. (2018) |
| D. thysanolaenae | KUMCC 18-0182T | MK064091 | MK045851 | MK086031 | – | Phukhamsak et al. (2019) |
| D. tropica | GZCC 22-0076T | ON527943 | ON527935 | ON533679 | ON533687 | This study |
| D. verrucosa | HKAS 112652T | MZ868762 | MZ868771 | MZ892977 | MZ892983 | Yang et al. (2021) |
| D. wuzhishanensis | GZCC 22-0077T | ON527946 | ON527938 | ON533682 | – | This study |
| D. xishuangbannaensis | KUMCC 17-0290T | MH260293 | MH275061 | MH412768 | MH412754 | Tibpromma et al. (2018) |
| D. xishuangbannaensis | GZCC 22-0079 | ON527944 | ON527936 | ON533680 | ON533688 | This study |
| D. yongxiuensis | JAUCC 4725T | OK562394 | OK562388 | OK562406 | – | Zhai et al. (2022) |
| D. yunjushanensis | JAUCC 4723T | OK562398 | OK562392 | OK562410 | – | Zhai et al. (2022) |
| D. yunnanensis | MFLUCC 20-0153T | MW081546 | MW081541 | MW084995 | MW151861 | Li et al. (2021) |
| Myrmecridium aquaticum | MFLUCC 15-0366T | MK849804 | – | – | – | Luo et al. (2019) |
| M. aquaticum | S 1158 | MK849803 | MK828656 | MN194061 | MN124540 | Luo et al. (2019) |
| M. banksiae | CBS 132536T | JX069855 | JX069871 | – | – | Crous et al. (2012) |
| M. montsegurinum | JF 13180T | KT991664 | KT991674 | – | KT991654 | Réblová et al. (2016) |
| M. schulzeri | CBS 100.54 | EU041826 | EU041769 | – | – | Arzanlou et al. (2007) |
| Pseudostanjehughesia aquitropica | MFLUCC 16-0569T | MF077559 | MF077548 | MF135655 | – | Yang et al. (2018) |
| Ps. lignicola | MFLUCC 15-0352T | MK849787 | MK828643 | MN194047 | MN124534 | Luo et al. (2019) |
| Sporidesmium bambusicola | HKUCC 3578T | DQ408562 | – | – | – | Shenoy et al. (2006) |
| S. dulongense | MFLUCC 17-0116T | MH795817 | MH795812 | MH801191 | MH801190 | Luo et al. (2019) |
| S. lageniforme | DLUCC 0880T | MK849782 | MK828640 | MN194044 | MN124533 | Luo et al. (2019) |
| S. pyriformatum | MFLUCC 15-0620T | KX710141 | KX710146 | MF135662 | MF135649 | Hyde et al. (2016) |
| S. thailandense | MFLUCC 15-0617 | MF077561 | MF077550 | MF135657 | – | Yang et al. (2018) |
| S. thailandense | MFLUCC 15-0964T | MF374370 | MF374361 | MF370957 | MF370955 | Zhang et al. (2017) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, J.; Zhang, J.-Y.; Xiao, X.-J.; Xiao, Y.-P.; Tang, X.; Boonmee, S.; Kang, J.-C.; Lu, Y.-Z. Multi-Gene Phylogenetic Analyses Revealed Five New Species and Two New Records of Distoseptisporales from China. J. Fungi 2022, 8, 1202. https://doi.org/10.3390/jof8111202
Ma J, Zhang J-Y, Xiao X-J, Xiao Y-P, Tang X, Boonmee S, Kang J-C, Lu Y-Z. Multi-Gene Phylogenetic Analyses Revealed Five New Species and Two New Records of Distoseptisporales from China. Journal of Fungi. 2022; 8(11):1202. https://doi.org/10.3390/jof8111202
Chicago/Turabian StyleMa, Jian, Jing-Yi Zhang, Xing-Juan Xiao, Yuan-Pin Xiao, Xia Tang, Saranyaphat Boonmee, Ji-Chuan Kang, and Yong-Zhong Lu. 2022. "Multi-Gene Phylogenetic Analyses Revealed Five New Species and Two New Records of Distoseptisporales from China" Journal of Fungi 8, no. 11: 1202. https://doi.org/10.3390/jof8111202
APA StyleMa, J., Zhang, J.-Y., Xiao, X.-J., Xiao, Y.-P., Tang, X., Boonmee, S., Kang, J.-C., & Lu, Y.-Z. (2022). Multi-Gene Phylogenetic Analyses Revealed Five New Species and Two New Records of Distoseptisporales from China. Journal of Fungi, 8(11), 1202. https://doi.org/10.3390/jof8111202







