A First Draft of the Core Fungal Microbiome of Schedonorus arundinaceus with and without Its Fungal Mutualist Epichloë coenophiala
Abstract
1. Introduction
- What species are present?
- What species were present in all or most plants over time (which we refer to as the draft “core microbiome”)?
- Were these fungal communities influenced by the presence of the E. coenophiala endophyte?
- Were these fungal communities different between plants infected with different strains of the Epichloë endophyte?
- Were these fungal communities correlated with surrogates of plant fitness?
2. Methods
2.1. Field Samples
2.2. DNA Extraction and Sequencing
2.3. Epichloë Concentration
2.4. Bioinformatics
2.5. Statistical Analyses
3. Results
3.1. Taxonomic Variability across Epichloë Treatments and Years
3.2. ASV Co-Occurrences
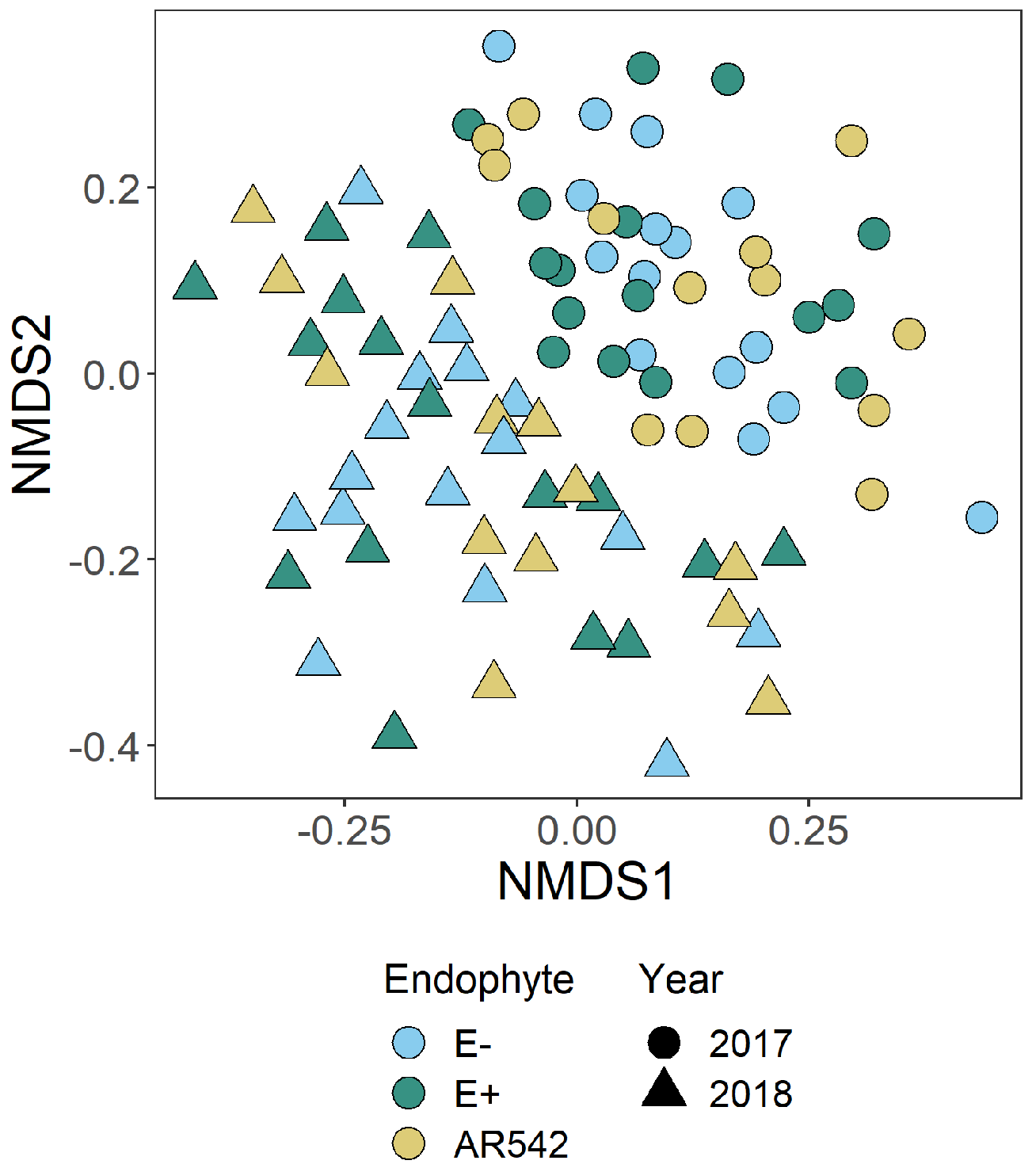
3.3. Fungal Community Structure
3.4. Draft Core Microbiome
3.5. Guilds
3.6. Differential Reads
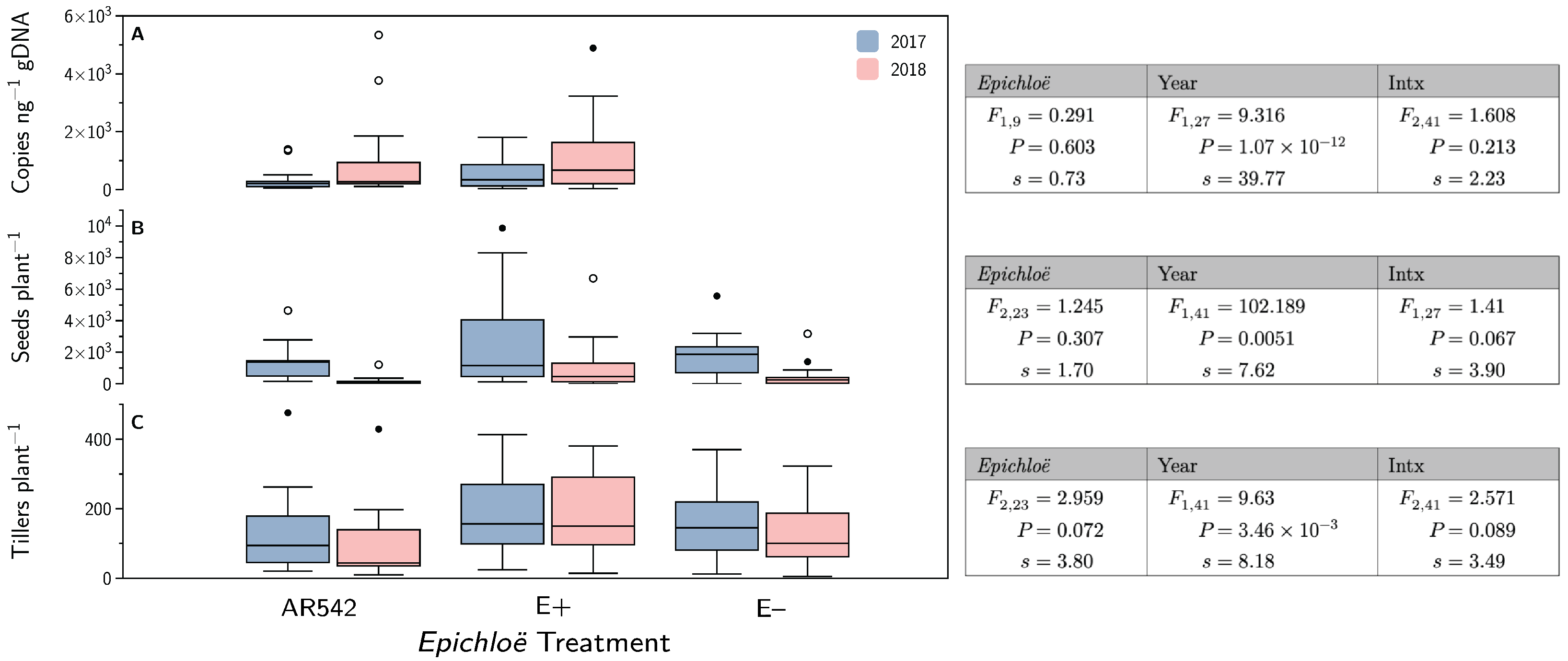
3.7. Epichloë coenophiala Concentrations and Plant Fitness
3.8. Relationship between Core ASVs, Plant Fitness, and Epichloë Concentrations
4. Discussion
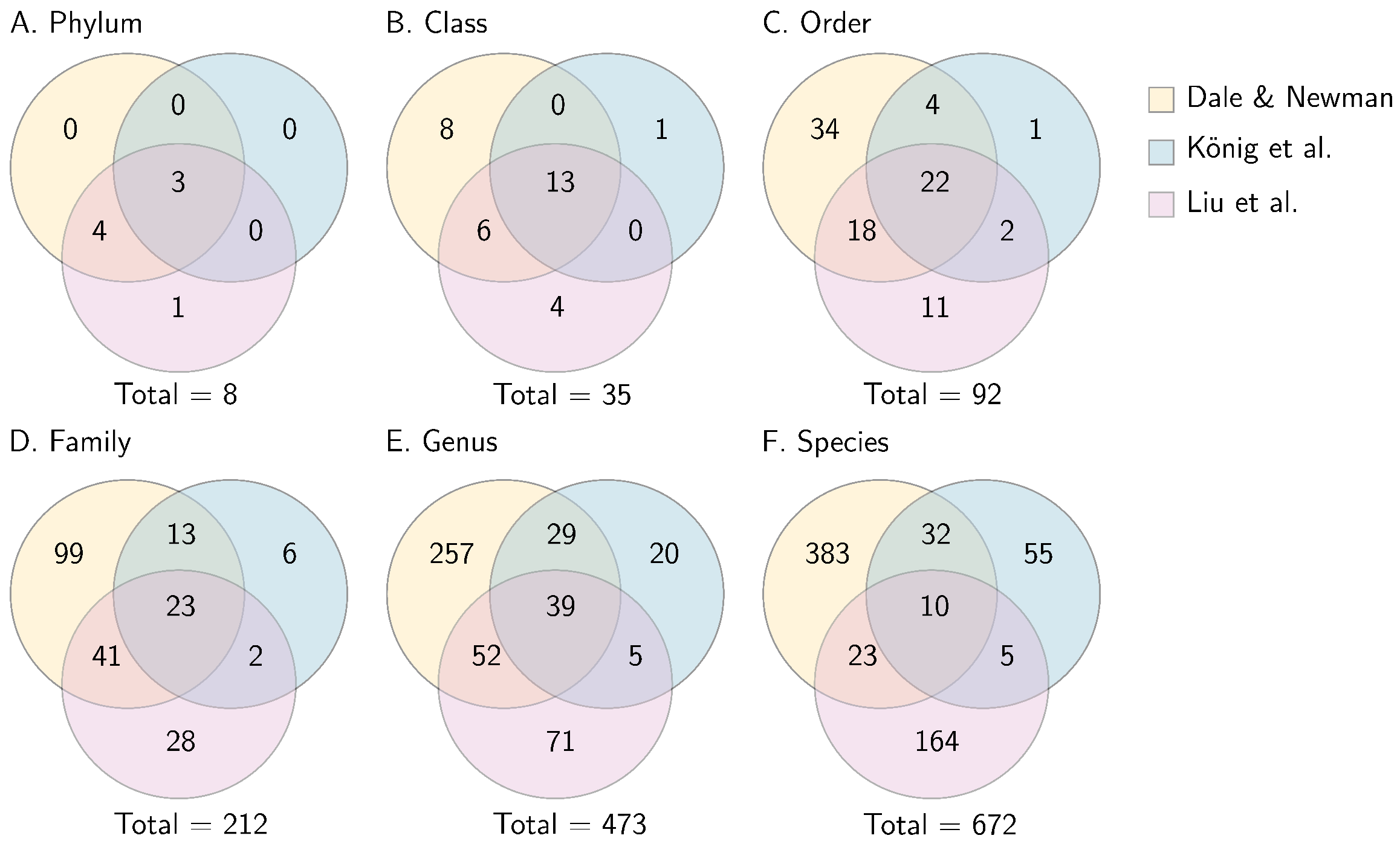
4.1. Taxonomy
4.2. Effect of Epichloë and Comparisons with Previous Research
4.3. Variation over Time
4.4. Plant Fitness
4.5. Limitations
4.6. Conclusions and Future Directions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gibson, D.; Newman, J. Festuca arundinacea Schreber (F. elatior L. ssp. arundinacea (Schreber) Hackel). J. Ecol. 2001, 89, 304–324. [Google Scholar] [CrossRef]
- Leuchtmann, A.; Bacon, C.W.; Schardl, C.L.; White, J.F., Jr.; Tadych, M. Nomenclatural realignment of Neotyphodium species with genus Epichloë. Mycologia 2014, 106, 202–215. [Google Scholar] [CrossRef] [PubMed]
- Newman, J.A.; Gillis, S.; Hager, H.A. Costs, benefits, parasites and mutualists: The use and abuse of the mutualism-parasitism continuum concept for Epichloë fungi. Philos. Theory, Pract. Biol. 2022, 14, 9. [Google Scholar] [CrossRef]
- Rasmussen, S.; Parsons, A.J.; Newman, J.A. Metabolomics analysis of the Lolium perenne–Neotyphodium lolii symbiosis: More than just alkaloids? Phytochem. Rev. 2009, 8, 535–550. [Google Scholar] [CrossRef]
- Bastías, D.A.; Martínez-Ghersa, M.A.; Ballaré, C.L.; Gundel, P.E. Epichloë fungal endophytes and plant defenses: Not just alkaloids. Trends Plant Sci. 2017, 22, 939–948. [Google Scholar] [CrossRef] [PubMed]
- Fernando, K.; Reddy, P.; Spangenberg, G.C.; Rochfort, S.J.; Guthridge, K.M. Metabolic potential of Epichloë endophytes for host grass fungal disease resistance. Microorganisms 2022, 10, 64. [Google Scholar] [CrossRef]
- Schardl, C.L. The Epichloae, symbionts of the grass subfamily Poöidae. Ann. Mo. Bot. Gard. 2010, 97, 646–665. [Google Scholar] [CrossRef]
- Nagabhyru, P.; Dinkins, R.D.; Wood, C.L.; Bacon, C.W.; Schardl, C.L. Tall fescue endophyte effects on tolerance to water-deficit stress. BMC Plant Biol. 2013, 13, 127. [Google Scholar] [CrossRef]
- Pańka, D.; West, C.P.; Guerber, C.A.; Richardson, M.D. Susceptibility of tall fescue to Rhizoctonia zeae infection as affected by endophyte symbiosis. Ann. Appl. Biol. 2013, 163, 257–268. [Google Scholar] [CrossRef]
- Hunt, M.G.; Newman, J.A. Reduced herbivore resistance from a novel grass–endophyte association. J. Appl. Ecol. 2005, 42, 762–769. [Google Scholar] [CrossRef]
- Bourguignon, M.; Nelson, J.A.; Carlisle, E.; Ji, H.; Dinkins, R.D.; Phillips, T.D.; McCulley, R.L. Ecophysiological responses of tall fescue genotypes to fungal endophyte infection, elevated temperature, and precipitation. Crop Sci. 2015, 55, 2895–2909. [Google Scholar] [CrossRef]
- Gwinn, K.; Gavin, A. Relationship between endophyte infestation level of tall fescue seed lots and Rhizoctonia zeae seedling disease. Plant Disease 1992, 76, 911–914. [Google Scholar] [CrossRef]
- Card, S.D.; Bastías, D.A.; Caradus, J.R. Antagonism to plant pathogens by Epichloë fungal endophytes—A review. Plants 2021, 10, 1997. [Google Scholar] [CrossRef] [PubMed]
- Kauppinen, M.; Helander, M.; Anttila, N.; Saloniemi, I.; Saikkonen, K. Epichloë endophyte effects on leaf blotch pathogen (Rhynchosporium sp.) of tall fescue (Schedonorus phoenix) vary among grass origin and environmental conditions. Plant Ecol. Divers. 2018, 11, 625–635. [Google Scholar] [CrossRef]
- Chen, W.; Liu, H.; Wurihan.; Gao, Y.; Card, S.D.; Ren, A. The advantages of endophyte-infected over uninfected tall fescue in the growth and pathogen resistance are counteracted by elevated CO2. Sci. Rep. 2017, 7, 6952. [Google Scholar] [CrossRef]
- Welty, R.E.; Barker, R.E.; Azevedo, M.D. Response of field-grown tall fescue infected by Acremonium coenophialum to Puccinia graminis subsp. graminicola. Plant Dis. 1993, 77, 574–575. [Google Scholar] [CrossRef]
- Rojas, X.; Guo, J.; Leff, J.W.; McNear, D.H.; Fierer, N.; McCulley, R.L. Infection with a shoot-specific fungal endophyte (Epichloë) alters tall fescue soil microbial communities. Microb. Ecol. 2016, 72, 197–206. [Google Scholar] [CrossRef]
- Mahmud, K.; Lee, K.; Hill, N.S.; Mergoum, A.; Missaoui, A. Influence of tall fescue Epichloë endophytes on rhizosphere soil microbiome. Microorganisms 2021, 9, 1843. [Google Scholar] [CrossRef]
- Panke-Buisse, K.; Cheng, L.; Gan, H.; Wickings, K.; Petrovic, M.; Kao-Kniffin, J. Root fungal endophytes and microbial extracellular enzyme activities show patterned responses in tall fescues under drought conditions. Agronomy 2020, 10, 1076. [Google Scholar] [CrossRef]
- Mack, K.M.L.; Rudgers, J.A. Balancing multiple mutualists: Asymmetric interactions among plants, arbuscular mycorrhizal fungi, and fungal endophytes. Oikos 2008, 117, 310–320. [Google Scholar] [CrossRef]
- Omacini, M.; Semmartin, M.; Pérez, L.I.; Gundel, P.E. Grass–endophyte symbiosis: A neglected aboveground interaction with multiple belowground consequences. Appl. Soil Ecol. 2012, 61, 273–279. [Google Scholar] [CrossRef]
- Kalosa-Kenyon, E.; Slaughter, L.C.; Rudgers, J.A.; McCulley, R.L. Asexual Epichloë endophytes do not consistently alter arbuscular mycorrhizal fungi colonization in three grasses. Am. Midl. Nat. 2018, 179, 157–165. [Google Scholar] [CrossRef]
- Slaughter, L.C.; Nelson, J.A.; Carlisle, E.; Bourguignon, M.; Dinkins, R.D.; Phillips, T.D.; McCulley, R.L. Climate change and Epichloë coenophiala association modify belowground fungal symbioses of tall fescue host. Fungal Ecol. 2018, 31, 37–46. [Google Scholar] [CrossRef]
- Slaughter, L.C.; McCulley, R.L. Aboveground Epichloë coenophiala-grass associations do not affect belowground fungal symbionts or associated plant, soil parameters. Microb. Ecol. 2016, 682–691. [Google Scholar] [CrossRef] [PubMed]
- Slaughter, L.C.; Nelson, J.A.; Carlisle, A.E.; Bourguignon, M.; Dinkins, R.D.; Phillips, T.D.; McCulley, R.L. Tall fescue and Epichloë coenophiala genetics influence root-associated soil fungi in a temperate grassland. Front. Microbiol. 2019, 10, 2380. [Google Scholar] [CrossRef]
- Nissinen, R.; Helander, M.; Kumar, M.; Saikkonen, K. Heritable Epichloë symbiosis shapes fungal but not bacterial communities of plant leaves. Sci. Rep. 2019, 9, 5253. [Google Scholar] [CrossRef]
- König, J.; Guerreiro, M.A.; Peršoh, D.; Begerow, D.; Krauss, J. Knowing your neighbourhood—The effects of Epichloë endophytes on foliar fungal assemblages in perennial ryegrass in dependence of season and land-use intensity. PeerJ 2018, 6, e4660. [Google Scholar] [CrossRef]
- Liu, B.; Ju, Y.; Xia, C.; Zhong, R.; Christensen, M.J.; Zhang, X.; Nan, Z. The effect of Epichloë endophyte on phyllosphere microbes and leaf metabolites in Achnatherum inebrians. iScience 2022, 25, 104144. [Google Scholar] [CrossRef]
- Zahn, G.; Amend, A.S. Foliar fungi alter reproductive timing and allocation in Arabidopsis under normal and water-stressed conditions. Fungal Ecol. 2019, 41, 101–106. [Google Scholar] [CrossRef]
- Busby, P.E.; Peay, K.G.; Newcombe, G. Common foliar fungi of Populus trichocarpa modify Melampsora rust disease severity. New Phytol. 2016, 209, 1681–1692. [Google Scholar] [CrossRef]
- Christian, N.; Herre, E.A.; Clay, K. Foliar endophytic fungi alter patterns of nitrogen uptake and distribution in Theobroma cacao. New Phytol. 2019, 222, 1573–1583. [Google Scholar] [CrossRef] [PubMed]
- Bouton, J.; Gates, R.; Hill, G.; Owsley, M.; Wood, D. Registration of ‘Georgia 5’ tall fescue. Crop Sci. 1993, 33, 1405. [Google Scholar] [CrossRef]
- Bellemain, E.; Carlsen, T.; Brochmann, C.; Coissac, E.; Taberlet, P.; Kauserud, H. ITS as an environmental DNA barcode for fungi: An in silico approach reveals potential PCR biases. BMC Microbiol. 2010, 10, 189. [Google Scholar] [CrossRef] [PubMed]
- Ihrmark, K.; Bödeker, I.T.; Cruz-Martinez, K.; Friberg, H.; Kubartova, A.; Schenck, J.; Strid, Y.; Stenlid, J.; Brandström-Durling, M.; Clemmensen, K.E.; et al. New primers to amplify the fungal ITS2 region - evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol. 2012, 82, 666–677. [Google Scholar] [CrossRef] [PubMed]
- Ryan, G.D.; Rasmussen, S.; Xue, H.; Parsons, A.J.; Newman, J.A. Metabolite analysis of the effects of elevated CO2 and nitrogen fertilization on the association between tall fescue (Schedonorus arundinaceus) and its fungal symbiont Neotyphodium coenophialum. Plant Cell Environ. 2014, 37, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package; R Package Version 2.5-7; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Martinez Arbizu, P. pairwiseAdonis: Pairwise Multilevel Comparison Using Adonis. R Package Version 0.4. 2020. Available online: https://github.com/pmartinezarbizu/pairwiseAdonis (accessed on 29 August 2022).
- McMurdie, P.J.; Holmes, S. Waste not, want not: Why rarefying microbiome data is inadmissible. PLoS Comput. Biol. 2014, 10, e1003531. [Google Scholar] [CrossRef]
- Willis, A.D. Rarefaction, alpha diversity, and statistics. Front. Microbiol. 2019, 10, 2407. [Google Scholar] [CrossRef] [PubMed]
- Weiss, S.; Xu, Z.Z.; Peddada, S.; Amir, A.; Bittinger, K.; Gonzalez, A.; Lozupone, C.; Zaneveld, J.R.; Vázquez-Baeza, Y.; Birmingham, A.; et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 2017, 5, 1–18. [Google Scholar] [CrossRef] [PubMed]
- McKnight, D.T.; Huerlimann, R.; Bower, D.S.; Schwarzkopf, L.; Alford, R.A.; Zenger, K.R. Methods for normalizing microbiome data: An ecological perspective. Methods Ecol. Evol. 2019, 10, 389–400. [Google Scholar] [CrossRef]
- Lin, H.; Peddada, S.D. Analysis of compositions of microbiomes with bias correction. Nat. Commun. 2020, 11, 3514. [Google Scholar] [CrossRef] [PubMed]
- Wasserstein, R.L.; Schirm, A.L.; Lazar, N.A. Moving to a world beyond “p < 0.05”. Am. Stat. 2019, 73, 1–19. [Google Scholar]
- Greenland, S. Valid p-values behave exactly as they should: Some misleading criticisms of p-values and their resolution with s-values. Am. Stat. 2019, 73, 106–114. [Google Scholar] [CrossRef]
- Põlme, S.; Abarenkov, K.; Henrik Nilsson, R.; Lindahl, B.D.; Clemmensen, K.E.; Kauserud, H.; Nguyen, N.; Kjøller, R.; Bates, S.T.; Baldrian, P.; et al. FungalTraits: A user-friendly traits database of fungi and fungus-like stramenopiles. Fungal Divers. 2020, 105, 1–16. [Google Scholar] [CrossRef]
- CABI. Invasive Species Compendium. 2022. Available online: www.cabi.org/isc (accessed on 6 August 2022).
- Bonos, S.A.; Clarke, B.B.; Meyer, W.A. Breeding for disease resistance in the major cool-season turfgrasses. Annu. Rev. Phytopathol. 2006, 44, 213–234. [Google Scholar] [CrossRef]
- Heineck, G.C.; Qiu, Y.; Ehlke, N.J.; Watkins, E. The fungal endophyte Epichloë festucae var. lolii plays a limited role in mediating crown rust severity in perennial ryegrass. Crop Sci. 2020, 60, 1090–1104. [Google Scholar] [CrossRef]
- Reddy, M.N.; Faeth, S.H. Damping-off of Festuca arizonica caused by Fusarium. Am. J. Plant Sci. 2010, 1, 104–105. [Google Scholar] [CrossRef]
- Blaalid, R.; Kumar, S.; Nilsson, R.H.; Abarenkov, K.; Kirk, P.M.; Kauserud, H. ITS1 versus ITS2 as DNA metabarcodes for fungi. Mol. Ecol. Resour. 2013, 13, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Fernando, K.; Reddy, P.; Vassiliadis, S.; Spangenberg, G.C.; Rochfort, S.J.; Guthridge, K.M. The known antimammalian and insecticidal alkaloids are not responsible for the antifungal activity of Epichloë endophytes. Plants 2021, 10, 2486. [Google Scholar] [CrossRef] [PubMed]
- Zabalgogeazcoa, I.; Gundel, P.E.; Helander, M.; Saikkonen, K. Non-systemic fungal endophytes in Festuca rubra plants infected by Epichloë festucae in subarctic habitats. Fungal Divers. 2013, 60, 25–32. [Google Scholar] [CrossRef]
- Siegel, M.R.; Latch, G.C.M. Expression of antifungal activity in agar culture by isolates of grass endophytes. Mycologia 1991, 83, 529–537. [Google Scholar] [CrossRef]
- Dastogeer, K.M.; Tumpa, F.H.; Sultana, A.; Akter, M.A.; Chakraborty, A. Plant microbiome–an account of the factors that shape community composition and diversity. Curr. Plant Biol. 2020, 23, 100161. [Google Scholar] [CrossRef]
- Vacher, C.; Hampe, A.; Porté, A.; Sauer, U.; Compant, S.; Morris, C.E. The phyllosphere: Microbial jungle at the plant-climate interface. Annu. Rev. Ecol. Evol. Syst. 2016, 47, 1–24. [Google Scholar] [CrossRef]
- Rice, J.S.; Pinkerton, B.W.; Stringer, W.C.; Undersander, D.J. Seed production in tall fescue as affected by fungal endophyte. Crop Sci. 1990, 30, 1303–1305. [Google Scholar] [CrossRef]
- Wang, Z.; Li, C.; White, J. Effects of Epichloë endophyte infection on growth, physiological properties and seed germination of wild barley under saline conditions. J. Agron. Crop Sci. 2020, 206, 43–51. [Google Scholar] [CrossRef]
- Zarean, M.; Sabzalian, M.R.; Mirlohi, A.; Davoudi, M.; Ataii, E. Epichloë endophyte and plant genotype determine seed production through self-pollination in tall fescue. Euphytica 2017, 213, 250. [Google Scholar] [CrossRef]
- Torkian, M.; Sabzalian, M.R.; Ehtemam, M.H. A simultaneous effect of selfing and Epichloë endophyte on forage, seed yield and turf characteristics of perennial ryegrass (Lolium perenne L.). Grass Forage Sci. 2019, 74, 559–570. [Google Scholar] [CrossRef]
- Adams, A.E.; Kazenel, M.R.; Rudgers, J.A. Does a foliar endophyte improve plant fitness under flooding? Plant Ecol. 2017, 218, 711–723. [Google Scholar] [CrossRef]
- Saari, S.; Helander, M.; Faeth, S.H.; Saikkonen, K. The effects of endophytes on seed production and seed predation of tall fescue and meadow fescue. Microb. Ecol. 2010, 60, 928–934. [Google Scholar] [CrossRef] [PubMed]
- Gundel, P.E.; Biganzoli, F.; Freitas, P.P.; Landesmann, J.B.; Martínez-Ghersa, M.A.; Ghersa, C.M. Plant damage, seed production and persistence of the fungal endophyte Epichloë occultans in Lolium multiflorum plants under an herbivore lepidopteran attack and ozone pollution. Ecol. Austral 2020, 30, 321–330. [Google Scholar] [CrossRef]
- Lücking, R.; Aime, M.C.; Robbertse, B.; Miller, A.N.; Ariyawansa, H.A.; Aoki, T.; Cardinali, G.; Crous, P.W.; Druzhinina, I.S.; Geiser, D.M.; et al. Unambiguous identification of fungi: Where do we stand and how accurate and precise is fungal DNA barcoding? IMA Fungus 2020, 11, 14. [Google Scholar] [CrossRef] [PubMed]



| Phylum (P), Class (C), Order(O) | Family | Genus | Species | ASVs Raw | ASVs Rarefied | Reads Raw | Reads Rarefied |
|---|---|---|---|---|---|---|---|
| P: Ascomycota | 0.680 | 0.670 | 0.896 | 0.895 | |||
| C: Dothideomycetes | 0.418 | 0.411 | 0.662 | 0.649 | |||
| O: Capnodiales | 0.037 | 0.039 | 0.047 | 0.041 | |||
| Cladosporiaceae | Cladosporium | cladosporioides | 0.017 | 0.018 | |||
| Neodevriesiaceae | Neodevriesia | poagena | 0.004 | 0.005 | |||
| Teratosphaeriaceae | 0.011 | ||||||
| O: Pleosporales | 0.352 | 0.343 | 0.633 | 0.618 | |||
| Cucurbitariaceae | 0.059 | 0.062 | |||||
| Cucurbitariaceae | Pyrenochaetopsis | 0.059 | 0.062 | ||||
| Pyrenochaetopsis | leptospora | 0.036 | 0.038 | ||||
| Coniothyriaceae | Coniothyrium | crepinianum | 0.003 | ||||
| Didymellaceae | 0.017 | 0.020 | 0.060 | 0.074 | |||
| Neoascochyta | 0.049 | 0.062 | |||||
| Neoascochyta | tardicrescens | 0.049 | 0.060 | ||||
| Didymosphaeriaceae | 0.012 | 0.062 | 0.061 | ||||
| Paraphaeosphaeria | 0.062 | 0.061 | |||||
| Paraphaeosphaeria | rosae | 0.014 | 0.012 | ||||
| Lentitheciaceae | 0.021 | 0.020 | 0.030 | 0.034 | |||
| Keissleriella | 0.009 | 0.010 | |||||
| Keissleriella | quadriseptata | 0.004 | |||||
| Phaeosphaeriaceae | 0.204 | 0.188 | 0.307 | 0.275 | |||
| Neosetophoma | samararum | 0.004 | |||||
| Parastagonospora | 0.0253 | 0.244 | 0.0396 | 0.0299 | |||
| Phaeosphaeria | 0.017 | 0.019 | |||||
| Piniphoma | wesendahlina | 0.019 | 0.020 | ||||
| Septoriella | 0.044 | 0.032 | 0.091 | 0.085 | |||
| Septoriella | phragmitis | 0.042 | 0.031 | 0.092 | 0.087 | ||
| Pleosporaceae | 0.015 | 0.017 | 0.028 | 0.028 | |||
| Alternaria | 0.009 | 0.011 | 0.028 | 0.028 | |||
| Alternaria | infectoria | 0.005 | 0.006 | ||||
| C: Eurotiomycetes | 0.028 | 0.029 | 0.023 | 0.023 | |||
| O: Chaetothyriales | 0.027 | 0.028 | 0.023 | 0.023 | |||
| C: Lecanoromycetes | 0.016 | 0.018 | 0.064 | 0.063 | |||
| O: Ostropales | 0.013 | 0.063 | 0.063 | ||||
| Gomphillaceae | 0.043 | 0.042 | |||||
| Corticifraga | 0.043 | 0.041 | |||||
| Corticifraga | peltigerae | 0.004 | 0.043 | 0.041 | |||
| Stictidaceae | Neofitzroyomyces | 0.020 | 0.021 | ||||
| Neofitzroyomyces | nerii | 0.005 | 0.006 | 0.020 | 0.021 | ||
| C: Leotiomycetes | 0.057 | 0.064 | 0.093 | 0.108 | |||
| O: Helotiales | 0.053 | 0.061 | 0.093 | 0.108 | |||
| Helotiaceae | 0.053 | 0.061 | 0.093 | 0.108 | |||
| Articulospora | 0.012 | 0.014 | 0.020 | ||||
| Articulospora | proliferata | 0.012 | 0.014 | 0.020 | 0.020 | ||
| Hyaloscyphaceae | 0.012 | 0.015 | |||||
| C: Saccharomycetes | 0.012 | ||||||
| O: Saccharomycetales | 0.012 | ||||||
| Metschnikowiaceae | Metschnikowia | 0.011 | |||||
| C: Sordariomycetes | 0.072 | 0.075 | 0.046 | 0.042 | |||
| O: Glomerellales | 0.020 | 0.017 | |||||
| O: Hypocreales | 0.029 | 0.030 | 0.014 | 0.016 | |||
| O: Xylariales | 0.007 | ||||||
| P: Basidiomycota | 0.254 | 0.272 | 0.100 | 0.100 | |||
| C: Agaricomycetes | 0.112 | 0.119 | 0.059 | 0.055 | |||
| O: Agaricales | 0.036 | 0.038 | |||||
| Psathyrellaceae | 0.011 | 0.012 | |||||
| O: Cantharellales | 0.012 | 0.005 | |||||
| O: Corticiales | 0.032 | 0.039 | 0.047 | 0.041 | |||
| Corticiaceae | 0.032 | 0.038 | 0.047 | 0.041 | |||
| Laetisaria | 0.011 | ||||||
| Laetisaria | lichenicola | 0.009 | 0.011 | ||||
| Limonomyces | 0.014 | 0.017 | 0.032 | 0.032 | |||
| C: Cystobasidiomycetes | 0.022 | 0.023 | 0.003 | 0.003 | |||
| C: Microbotryomycetes | 0.012 | 0.013 | 0.002 | 0.002 | |||
| C: Tremellomycetes | 0.065 | 0.077 | 0.034 | 0.038 | |||
| O: Tremellales | 0.049 | 0.056 | 0.028 | 0.032 | |||
| Bulleribasidiaceae | 0.030 | 0.034 | 0.025 | 0.029 | |||
| Dioszegia | 0.010 | 0.012 | |||||
| Vishniacozyma | 0.014 | 0.016 | 0.022 | ||||
| Vishniacozyma | dimennae | 0.004 | 0.004 | ||||
| Vishniacozyma | tephrensis | 0.010 | 0.012 | ||||
| Vishniacozyma | victoriae | 0.004 | 0.004 | ||||
| P: Chytridiomycota | 0.013 | 0.014 | 0.003 | 0.003 | |||
| C: Spizellomycetes | 0.013 | 0.003 | 0.003 | ||||
| O: Spizellomycetales | 0.013 | ||||||
| P: Mortierellomycota | 0.002 | 0.001 | 0.000 | 0.000 | |||
| P: Mucoromycota | 0.000 | 0.000 | 0.000 | 0.000 | |||
| P: Olpidiomycota | 0.000 | 0.000 | 0.000 | ||||
| P: Rozellomycota | 0.000 | 0.000 | 0.000 |
| Epichloë Treatment | 2017 Only | 2018 Only | Both Years | Subtotal |
|---|---|---|---|---|
| E+ only | 264 | 315 | 33 | 612 |
| E− only | 272 | 264 | 45 | 581 |
| AR542 only | 232 | 191 | 24 | 447 |
| E+, E− only | 42 | 15 | 15 | 72 |
| E+, AR542 only | 27 | 20 | 6 | 53 |
| E−, AR542 only | 32 | 11 | 9 | 52 |
| E+, E−, AR542 | 30 | 16 | 302 | 348 |
| Subtotal | 899 | 832 | 434 | 2165 |
| A. 2017 | |||||||
|---|---|---|---|---|---|---|---|
| Source | df | SS | MS | F | P | s | |
| Epichloë | 2 | 0.34 | 0.17 | 1.04 | 0.04 | 0.3873 | 1.37 |
| Row | 16 | 3.15 | 0.20 | 1.20 | 0.41 | 0.0144 | 6.12 |
| Column | 2 | 0.45 | 0.236 | 1.37 | 0.06 | 0.0349 | 4.84 |
| Residuals | 23 | 3.78 | 0.16 | 0.49 | |||
| Total | 43 | 7.72 | 1.00 | ||||
| B. 2018 | |||||||
| Source | df | SS | MS | F | P | s | |
| Epichloë | 2 | 0.39 | 0.19 | 1.42 | 0.05 | 0.0453 | 4.46 |
| Row | 16 | 3.03 | 0.19 | 1.39 | 0.42 | 0.0007 | 10.48 |
| Column | 2 | 0.48 | 0.24 | 1.76 | 0.07 | 0.0061 | 7.35 |
| Residuals | 23 | 3.26 | 0.14 | 0.46 | |||
| Total | 43 | 7.16 | 1.00 | ||||
| 2017 Plants | 2018 Plants | Proportion | Epichloë | Year | Intx | Consistent Correlations | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Taxonomic Identity | ASV | AR542 | E+ | E− | AR542 | E+ | E− | Reads | p-Value | p-Value | p-Value | Tiller # | Seed # | [Epichloë] | Guild | |
| plants | family: Phaeosphaeriaceae | ASV1 | 13 | 15 | 16 | 13 | 16 | 16 | 0.095 | 3.04 × 10 | 1.11 × 10 | 9.64 × 10 | ||||
| family: Phaeosphaeriaceae | ASV2 | 13 | 15 | 16 | 13 | 16 | 16 | 0.082 | 4.55 × 10 | 9.80 × 10 | 7.37 × 10 | |||||
| family: Didymosphaeriaceae | ASV3 | 13 | 15 | 16 | 13 | 16 | 16 | 0.057 | 8.27 × 10 | 1.06 × 10 | 3.89 × 10 | |||||
| order: Pleosporales | ASV4 | 13 | 15 | 16 | 13 | 16 | 16 | 0.057 | 5.31 × 10 | 2.39 × 10 | 2.33 × 10 | |||||
| Neoascochyta tardicrescens | ASV5 | 13 | 15 | 16 | 13 | 16 | 16 | 0.049 | 6.80 × 10 | 1.94 × 10 | 9.22 × 10 | |||||
| Corticifraga peltigerae | ASV6 | 13 | 15 | 16 | 13 | 16 | 16 | 0.039 | 2.07 × 10 | 9.99 × 10 | 5.81 × 10 | |||||
| Pyrenochaetopsis leptospora | ASV7 | 13 | 15 | 16 | 13 | 16 | 16 | 0.036 | 7.71 × 10 | 4.86 × 10 | 7.61 × 10 | |||||
| family: Lentitheciaceae | ASV9 | 13 | 15 | 16 | 13 | 16 | 16 | 0.027 | 5.00 × 10 | 7.65 × 10 | 6.05 × 10 | |||||
| Volucrispora graminea | ASV10 | 13 | 15 | 16 | 13 | 16 | 16 | 0.026 | 7.01 × 10 | 4.09 × 10 | 9.54 × 10 | |||||
| genus: Pyrenochaetopsis | ASV11 | 13 | 15 | 16 | 13 | 16 | 16 | 0.024 | 8.45 × 10 | 2.51 × 10 | 8.04 × 10 | |||||
| Cladosporium cladosporioides | ASV15 | 13 | 15 | 16 | 13 | 16 | 16 | 0.018 | 6.21 × 10 | 7.86 × 10 | 5.40 × 10 | E,PP,WS | ||||
| order: Chaetothyriales | ASV19 | 13 | 15 | 16 | 13 | 16 | 16 | 0.014 | 4.20 × 10 | 5.92 × 10 | 6.93 × 10 | |||||
| Epicoccum nigrum | ASV32 | 13 | 15 | 16 | 13 | 16 | 16 | 0.006 | 4.58 × 10 | 4.15 × 10 | 7.52 × 10 | E,PP | ||||
| plant | genus: Colletotrichum | ASV12 | 13 | 14 | 16 | 13 | 16 | 16 | 0.015 | 8.58 × 10 | 6.27 × 10 | 6.65 × 10 | E,PP | |||
| genus: Parastagonospora | ASV18 | 13 | 15 | 15 | 13 | 15 | 16 | 0.012 | 5.54 × 10 | 5.56 × 10 | 3.88 × 10 | |||||
| Alternaria rosae | ASV21 | 13 | 15 | 15 | 13 | 16 | 16 | 0.011 | 4.85 × 10 | 2.25 × 10 | 6.46 × 10 | AP,E,PP,WS | ||||
| Alternaria alternata | ASV27 | 13 | 15 | 15 | 13 | 16 | 16 | 0.008 | 4.00 × 10 | 4.64 × 10 | 8.69 × 10 | AP,E,PP,WS | ||||
| Vishniacozyma tephrensis | ASV29 | 13 | 15 | 16 | 12 | 16 | 16 | 0.008 | 7.33 × 10 | 2.00 × 10 | 7.25 × 10 | |||||
| genus: Parastagonospora | ASV28 | 13 | 14 | 15 | 13 | 16 | 16 | 0.007 | 6.26 × 10 | 1.18 × 10 | 4.15 × 10 | |||||
| Vishniacozyma tephrensis | ASV61 | 13 | 15 | 16 | 12 | 16 | 15 | 0.003 | ||||||||
| family: Tubeufiaceae | ASV76 | 13 | 15 | 15 | 13 | 16 | 15 | 0.002 | 8.09 × 10 | 2.61 × 10 | 6.42 × 10 | |||||
| plants | Vishniacozyma victoriae | ASV50 | 13 | 15 | 16 | 11 | 16 | 15 | 0.004 | 3.37 × 10 | 8.44 × 10 | 3.29 × 10 | ||||
| Phialophora livistonae | ASV62 | 11 | 15 | 15 | 12 | 16 | 15 | 0.003 | 3.88 × 10 | 4.85 × 10 | 2.92 × 10 | |||||
| Devriesia pseudoamericana | ASV67 | 12 | 15 | 14 | 11 | 16 | 15 | 0.002 | ||||||||
| order: Pleosporales | ASV68 | 11 | 13 | 15 | 12 | 15 | 14 | 0.002 | 2.93 × 10 | 4.81 × 10 | 9.01 × 10 | |||||
| Epicoccum nigrum | ASV95 | 11 | 14 | 16 | 12 | 14 | 15 | 0.001 | 9.93 × 10 | 1.95 × 10 | 3.61 × 10 | E,PP | ||||
| plants | family: Phaeosphaeriaceae | ASV17 | 13 | 14 | 15 | 11 | 13 | 13 | 0.011 | 2.90 × 10 | 3.51 × 10 | 9.22 × 10 | ||||
| Articulospora proliferata | ASV23 | 12 | 15 | 15 | 13 | 13 | 14 | 0.010 | 1.78 × 10 | 1.61 × 10 | 4.30 × 10 | |||||
| Alternaria infectoria | ASV47 | 10 | 15 | 13 | 12 | 13 | 15 | 0.004 | AP,E,PP,WS | |||||||
| genus: Cyphellophora | ASV53 | 10 | 15 | 15 | 10 | 13 | 15 | 0.003 | AP | |||||||
| Paraophiobolus plantaginis | ASV144 | 10 | 12 | 13 | 10 | 15 | 14 | 0.001 | ||||||||
| plants | Piniphoma wesendahlina | ASV13 | 10 | 11 | 12 | 12 | 12 | 13 | 0.020 | |||||||
| genus: Cryptocoryneum | ASV48 | 9 | 11 | 13 | 12 | 12 | 15 | 0.004 | ||||||||
| Alternaria infectoria | ASV60 | 11 | 13 | 12 | 12 | 16 | 16 | 0.003 | pp | |||||||
| Vishniacozyma dimennae | ASV84 | 13 | 15 | 16 | 9 | 13 | 14 | 0.002 | 3.47 × 10 | 1.03 × 10 | 2.61 × 10 | |||||
| genus: Acremonium | ASV93 | 12 | 13 | 13 | 13 | 12 | 14 | 0.001 | ||||||||
| plants | Neofitzroyomyces nerii | ASV30 | 10 | 10 | 13 | 9 | 12 | 13 | 0.008 | |||||||
| Fusarium langsethiae | ASV56 | 9 | 14 | 11 | 10 | 15 | 16 | 0.004 | AP,E,PP,WS | |||||||
| genus: Parastagonospora | ASV55 | 13 | 12 | 11 | 12 | 14 | 13 | 0.003 | gpp | |||||||
| family: Lentitheciaceae | ASV78 | 12 | 14 | 15 | 10 | 11 | 11 | 0.003 | ||||||||
| Articulospora proliferata | ASV72 | 9 | 15 | 12 | 9 | 11 | 14 | 0.002 | ||||||||
| Paraphaeosphaeria michotii | ASV90 | 8 | 14 | 11 | 13 | 16 | 15 | 0.002 | pp | |||||||
| Chrysozyma griseoflava | ASV110 | 11 | 13 | 15 | 11 | 13 | 11 | 0.001 | ||||||||
| sum: | 0.691 | |||||||||||||||
| Different across Epichloë Treatment | Different across Years | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ASV | Taxonomy | E− | E+ | AR542 | 2017 | 2018 | |||||
| Reads | Plants | Reads | Plants | Reads | Plants | Reads | Plants | Reads | Plants | ||
| ASV1 | Family: Phaeosphaeriaceae | 19,623 | 44 | 82,628 | 45 | ||||||
| ASV2 | Septoriella phragmitis | 30,098 | 44 | 57,643 | 45 | ||||||
| ASV12 | Genus: Colletotrichum | 1589 | 43 | 14,207 | 45 | ||||||
| ASV15 | Cladosporium cladosporioides | 5562 | 44 | 13,512 | 45 | ||||||
| ASV17 | Family: Phaeosphaeriaceae | 9803 | 43 | 2097 | 39 | ||||||
| ASV21 | Alternaria rosae | 2777 | 43 | 9361 | 45 | ||||||
| ASV29 | Vishniacozyma tephrensis | 6888 | 44 | 1502 | 44 | ||||||
| ASV41 | Order: Pleosporales | 304 | 17 | 3072 | 36 | ||||||
| ASV46 | Genus: Dioszegia | 3839 | 44 | 177 | 31 | ||||||
| ASV50 | Vishniacozyma victoriae | 3179 | 44 | 1290 | 42 | ||||||
| ASV60 | Alternaria infectoria | 1096 | 36 | 2431 | 44 | ||||||
| ASV61 | Vishniacozyma tephrensis | 2667 | 44 | 1075 | 43 | ||||||
| ASV70 | Colletotrichum eleusines | 152 | 15 | 942 | 30 | ||||||
| ASV78 | Family: Lentitheciaceae | 2263 | 41 | 432 | 34 | ||||||
| ASV84 | Vishniacozyma dimennae | 1591 | 44 | 402 | 37 | ||||||
| ASV88 | Order: Pleosporales | 1157 | 42 | 348 | 32 | ||||||
| ASV90 | Paraphaeosphaeria michotii | 581 | 34 | 1047 | 44 | ||||||
| ASV98 | Ramularia collo-cygni | 392 | 30 | 805 | 43 | ||||||
| ASV108 | Alfaria ossiformis | 223 | 17 | 988 | 34 | ||||||
| ASV114 | Cystofilobasidium macerans | 1340 | 42 | 9 | 6 | ||||||
| ASV125 | Coniothyrium crepinianum | 31 | 3 | 834 | 18 | ||||||
| ASV137 | Dioszegia rishiriensis | 930 | 40 | 74 | 19 | ||||||
| ASV142 | Genus: Cryptococcus | 572 | 39 | 116 | 19 | ||||||
| ASV145 | Taphrina tormentillae | 747 | 33 | 94 | 19 | ||||||
| ASV149 | Dioszegia rishiriensis | 686 | 40 | 132 | 19 | ||||||
| ASV158 | Vishniacozyma victoriae | 669 | 26 | 42 | 6 | ||||||
| ASV160 | Order: Hypocreales | 110 | 27 | 548 | 38 | ||||||
| ASV169 | Order: Erythrobasidiales | 490 | 34 | 121 | 21 | ||||||
| ASV178 | Genus: Dioszegia | 902 | 38 | 17 | 5 | ||||||
| ASV195 | Filobasidium magnum | 520 | 32 | 89 | 17 | ||||||
| ASV196 | Zymoseptoria verkleyi | 54 | 18 | 342 | 32 | ||||||
| ASV197 | Class: Cystobasidiomycetes | 385 | 34 | 84 | 12 | ||||||
| ASV227 | Genus: Limonomyces | 213 | 7 | 0 | 0 | 135 | 6 | ||||
| ASV234 | Filobasidium stepposum | 514 | 28 | 18 | 10 | ||||||
| ASV237 | Papiliotrema frias | 485 | 32 | 32 | 12 | ||||||
| ASV249 | Class: Lecanoromycetes | 100 | 6 | 77 | 5 | 0 | 0 | ||||
| ASV260 | Genus: Filobasidium | 282 | 29 | 63 | 12 | ||||||
| ASV272 | Kingdom: Fungi | 168 | 28 | 34 | 13 | ||||||
| ASV273 | Filobasidium oeirense | 245 | 26 | 9 | 4 | ||||||
| ASV281 | Genus: Phaeosphaeria | 49 | 10 | 193 | 28 | ||||||
| ASV283 | Protomyces inouyei | 324 | 24 | 0 | 0 | ||||||
| ASV288 | Order: Holtermanniales | 283 | 28 | 38 | 6 | ||||||
| ASV289 | Dioszegia rishiriensis | 151 | 18 | 18 | 4 | ||||||
| ASV298 | Genus: Parastagonospora | 29 | 4 | 181 | 21 | ||||||
| ASV301 | Filobasidium wieringae | 259 | 29 | 15 | 5 | ||||||
| ASV318 | Sporobolomyces roseus | 179 | 28 | 1 | 2 | ||||||
| ASV323 | Order: Microstromatales | 142 | 22 | 30 | 2 | ||||||
| ASV367 | Order: Pleosporales | 18 | 3 | 117 | 20 | ||||||
| ASV387 | Genus: Phaeosphaeria | 21 | 4 | 94 | 17 | ||||||
| ASV533 | Family: Extremaceae | 33 | 8 | 6 | 3 | 0 | 0 | ||||
| ASV552 | Apenidiella strumelloidea | 3 | 2 | 40 | 13 | ||||||
| ASV837 | Phylum: Ascomycota | 6 | 6 | 7 | 4 | 0 | 0 | ||||
| ASV875 | Phylum: Ascomycota | 13 | 8 | 0 | 0 | 0 | 3 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dale, J.C.M.; Newman, J.A. A First Draft of the Core Fungal Microbiome of Schedonorus arundinaceus with and without Its Fungal Mutualist Epichloë coenophiala. J. Fungi 2022, 8, 1026. https://doi.org/10.3390/jof8101026
Dale JCM, Newman JA. A First Draft of the Core Fungal Microbiome of Schedonorus arundinaceus with and without Its Fungal Mutualist Epichloë coenophiala. Journal of Fungi. 2022; 8(10):1026. https://doi.org/10.3390/jof8101026
Chicago/Turabian StyleDale, Jenna C. M., and Jonathan A. Newman. 2022. "A First Draft of the Core Fungal Microbiome of Schedonorus arundinaceus with and without Its Fungal Mutualist Epichloë coenophiala" Journal of Fungi 8, no. 10: 1026. https://doi.org/10.3390/jof8101026
APA StyleDale, J. C. M., & Newman, J. A. (2022). A First Draft of the Core Fungal Microbiome of Schedonorus arundinaceus with and without Its Fungal Mutualist Epichloë coenophiala. Journal of Fungi, 8(10), 1026. https://doi.org/10.3390/jof8101026








