The Combination of Iron and Copper Increases Pathogenicity and Induces Proteins Related to the Main Virulence Factors in Clinical Isolates of Cryptococcus neoformans var. grubii
Abstract
1. Introduction
2. Materials and Methods
2.1. Microorganisms
2.2. Growth Medium
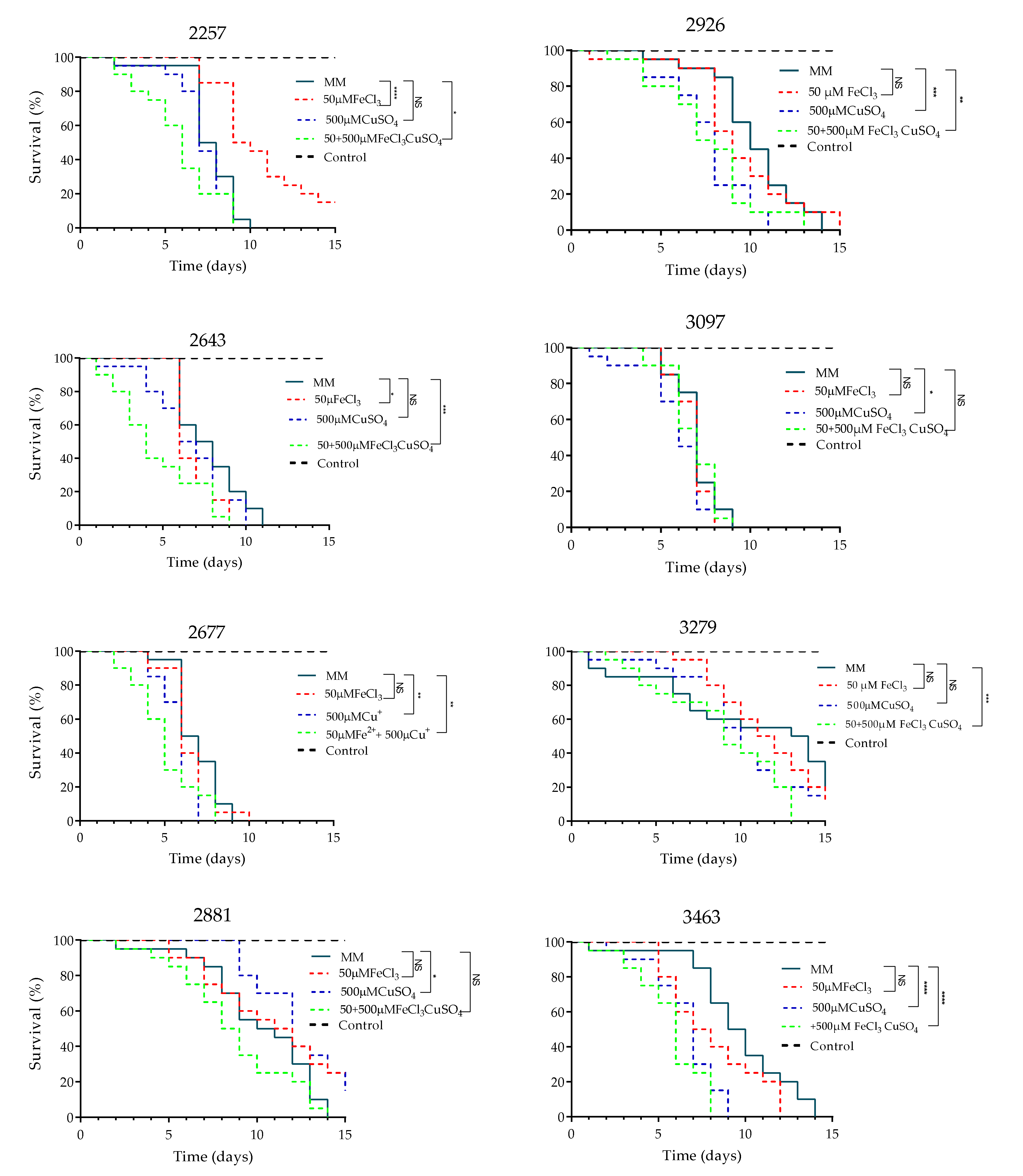
2.3. Effect of Iron and Copper on the Pathogenicity of C. neoformans in G. mellonella
2.4. Capsule Induction
2.5. Melanin Production
2.6. Cytoplasmic Protein Extraction
2.7. Protein Preparation for Mass Spectrometry and Label-Free Protein Quantification
2.8. Statistical Analysis
2.9. Biological Data Bases and Bioinformatic Resources
3. Results
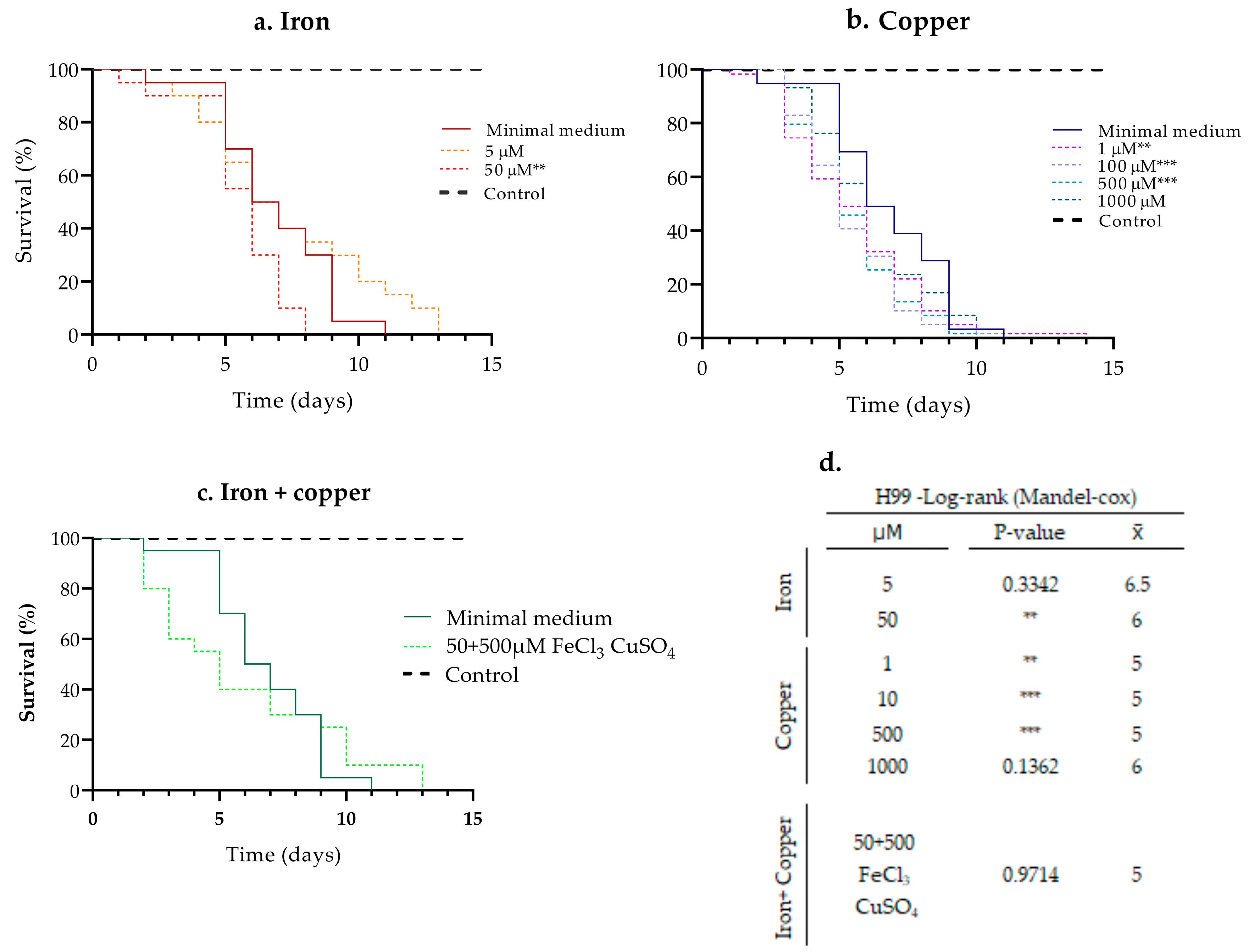
3.1. Evaluation of the Pathogenic Capacity of the H99 Strain in Different Pre-Incubation Concentrations of Iron and Copper
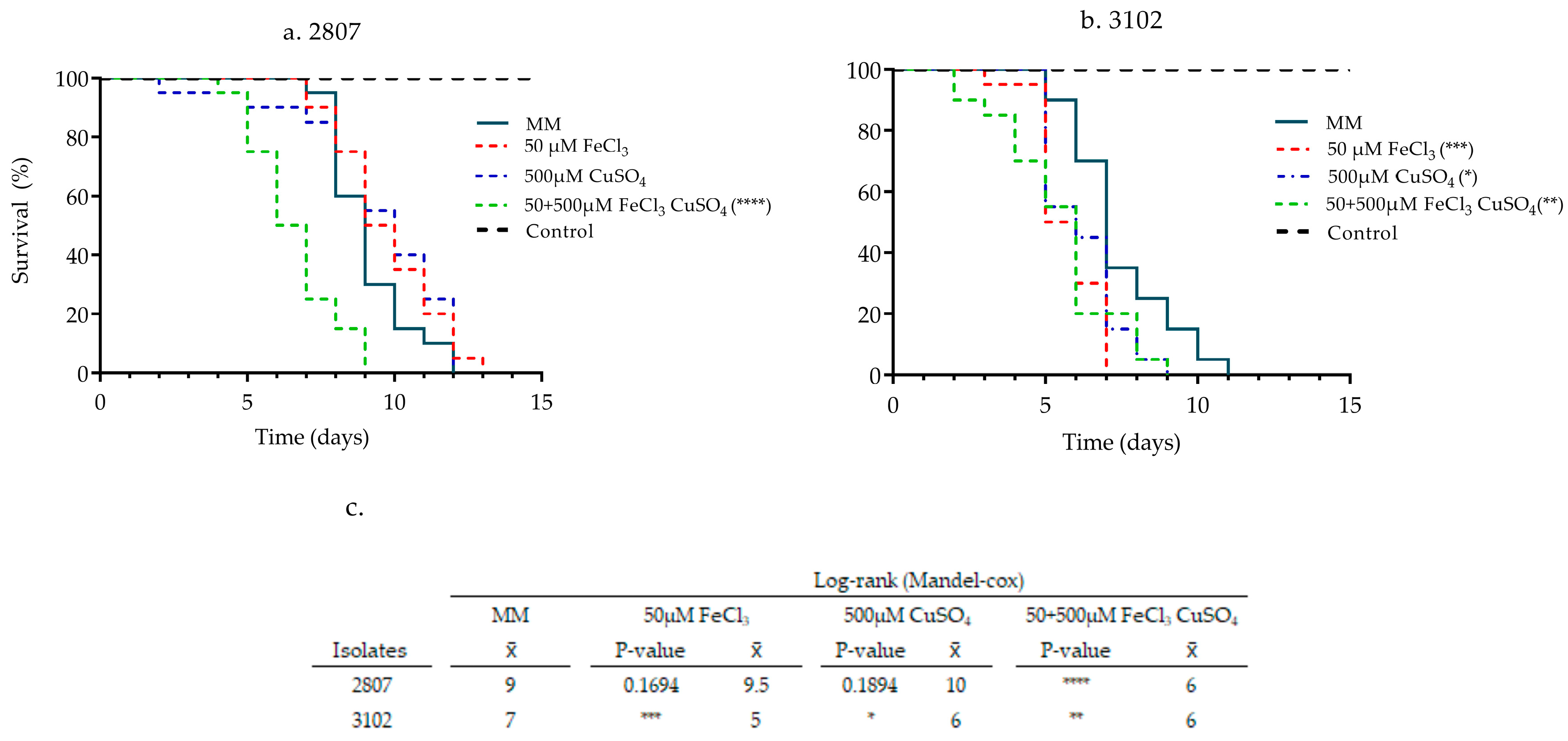
3.2. Effect of Pre-Incubation with Iron and Copper on the Pathogenicity of Clinical Isolates
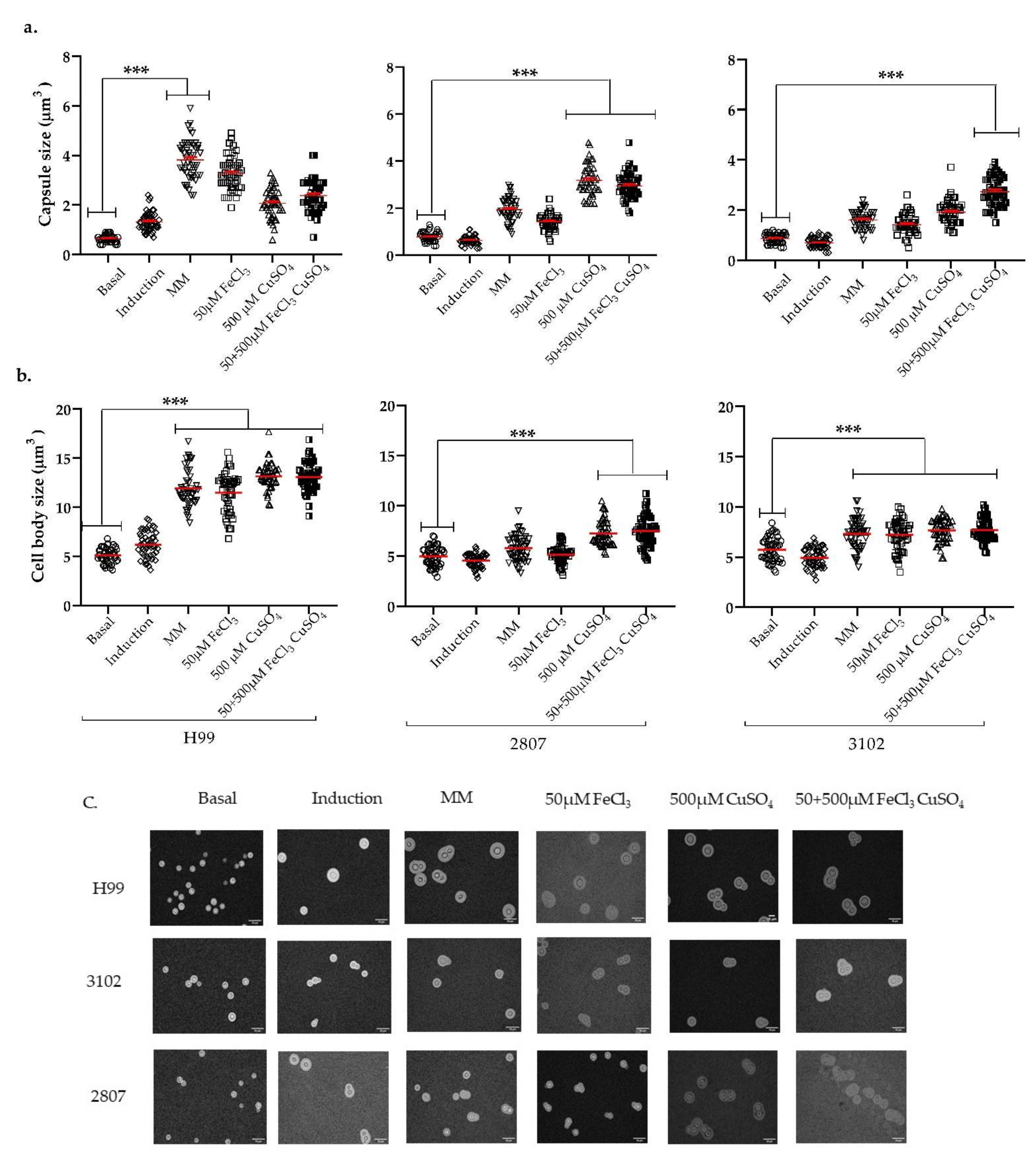
3.3. Effect of Iron, Copper, and Their Combination on Capsular Size
3.4. Effect of Iron, Copper, and Their Combination on Melanin Production
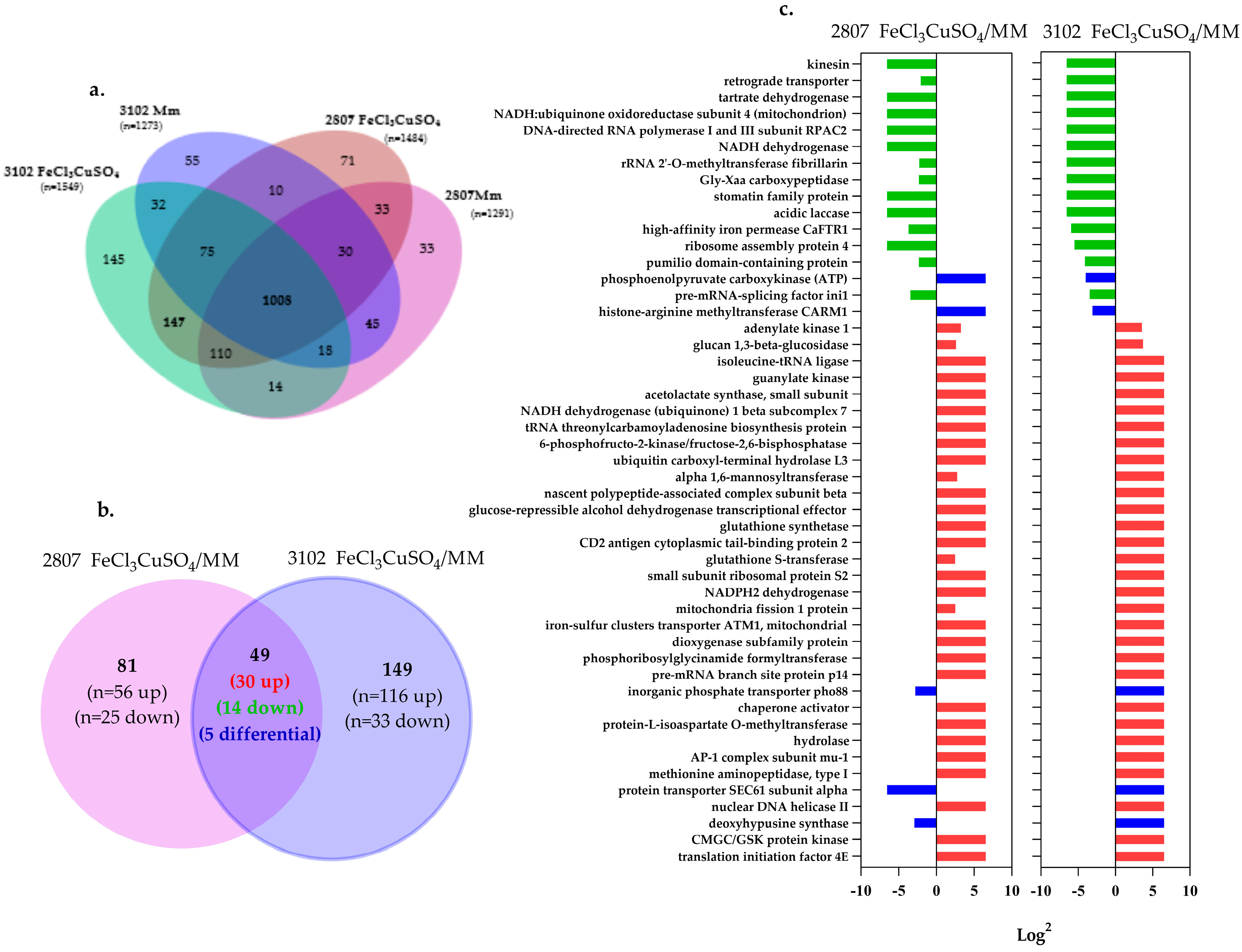
3.5. Proteomic Analysis of the Effect of Iron and Copper
3.6. Individual Analysis of Proteomic Changes Induced by Iron and Copper in Each Clinical Isolate
3.7. Proteins Related to the Capsule, Melanin, and Other Virulence Factors in Response to the Stimulus-Induced by Iron and Copper Combination
3.8. Effect of Iron and Copper Combination on the Pathogenicity of Eight Clinical Isolates of C. neoformans var. grubii in the G. mellonella Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Crawford, A.; Wilson, D. Essential metals at the host-pathogen interface: Nutritional immunity and micronutrient assimilation by human fungal pathogens. FEMS Yeast Res. 2015, 15, fov071. [Google Scholar] [CrossRef]
- Zaragoza, O. Basic principles of the virulence of Cryptococcus. Virulence 2019, 10, 490–501. [Google Scholar] [CrossRef] [PubMed]
- Potrykus, J.; Ballou, E.R.; Childers, D.S.; Brown, A.J.P. Conflicting Interests in the Pathogen-Host Tug of War: Fungal Micronutrient Scavenging Versus Mammalian Nutritional Immunity. PLoS Pathog. 2014, 10, e1003910. [Google Scholar] [CrossRef] [PubMed]
- Ene, I.V.; Brunke, S.; Brown, A.J.P.; Hube, B. Metabolism in fungal pathogenesis. Cold Spring Harb. Perspect. Med. 2014, 4, a019695. [Google Scholar] [CrossRef]
- Askwith, C.; Kaplan, J. Iron and copper transport in yeast and its relevance to human disease. Trends Biochem. Sci. 1998, 23, 135–138. [Google Scholar] [CrossRef]
- Bahn, Y.; Jung, K. Stress Signaling Pathways for the Pathogenicity of Cryptococcus. ASM 2013, 12, 1564–1577. [Google Scholar] [CrossRef] [PubMed]
- Charlier, C.; Chretien, F.; Baudrimont, M.; Mordelet, E.; Lortholary, O.; Dromer, F. Capsule structure changes associated with Cryptococcus neoformans crossing of the blood brain barrier. Am. J. Pathol. 2005, 166, 421–432. [Google Scholar] [CrossRef]
- Bose, I.; Reese, A.J.; Ory, J.J.; Janbon, G.; Doering, T.L. A Yeast under Cover: The Capsule of Cryptococcus neoformans. Eukaryot. Cell 2003, 2, 655–663. [Google Scholar] [CrossRef]
- Helsel, M.E.; White, E.J.; Razvi, S.Z.A.; Alies, B.; Franz, K.J. Chemical and functional properties of metal chelators that mobilize copper to elicit fungal killing of Cryptococcus neoformans. Metallomics 2017, 9, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Balhara, M.; Chaudhary, R.; Ruhil, S.; Singh, B.; Dahiya, N.; Parmar, V.S.; Jaiwal, P.K.; Chhillar, A.K. Siderophores; iron scavengers: The novel & promising targets for pathogen specific antifungal therapy. Expert Opin. Ther. Targets 2016, 20, 1477–1489. [Google Scholar] [CrossRef]
- Walton, F.J.; Idnurm, A.; Heitman, J. Novel gene functions required for melanization of the human pathogen Cryptococcus neoformans. Mol. Microbiol. 2005, 57, 1381–1396. [Google Scholar] [CrossRef] [PubMed]
- Haas, H.; Eisendle, M.; Turgeon, B.G. Siderophores in Fungal Physiology and Virulence. Annu. Rev. Phytopathol. 2008, 46, 149–187. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L. Iron and siderophores in fungal?host interactions. Mycol. Res. 2008, 112, 170–183. [Google Scholar] [CrossRef] [PubMed]
- Jung, W.H.; Sham, A.; White, R.; Kronstad, J.W. Iron Regulation of the Major Virulence Factors in the AIDS-Associated Pathogen Cryptococcus neoformans. PLoS Biol. 2006, 4, e410. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Festa, R.A.; Chen, Y.L.; Espart, A.; Palacios, Ò.; Espin, J. Cryptococcus Neoformans Copper Detoxificacion Machinery Is Critical For Fungal Virulence. Cell Host Microbe 2013, 13, 265–276. [Google Scholar] [CrossRef]
- Ding, C.; Festa, R.A.; Sun, T.; Wang, Z. Iron and copper as virulence modulators in human fungal pathogens. Mol. Microbiol. 2014, 93, 10–23. [Google Scholar] [CrossRef] [PubMed]
- O’ Meara, T.R.; Xu, W.; Selving, K.M.; O’meara, M.J.; Mitchell, A.P.; Alspaught, J. The Cryptococcus Neoformans Rim101 Transcription Factor Directly Regulates Genes Required for Adaptation to the Host. Mol. Cell. Biol. 2014, 34, 673–684. [Google Scholar] [CrossRef]
- Kronstad, J.; Saikia, S.; Nielson, E.D.; Kretschmer, M.; Jung, W.; Hu, G.; Geddes, J.M.H.; Griffiths, E.J.; Choi, J.; Cadieux, B.; et al. Adaptation of Cryptococcus neoformans to mammalian hosts: Integrated regulation of metabolism and virulence. Eukaryot. Cell 2012, 11, 109–118. [Google Scholar] [CrossRef]
- Rajasingham, R.; Smith, R.M.; Park, B.J.; Jarvis, J.N.; Govender, N.P.; Chiller, T.M.; Denning, D.W.; Loyse, A. Global burden of disease of HIV-associated cryptococcal meningitis: An updated analysis. Lancet Infect. Dis. 2017, 17, 873–881. [Google Scholar] [CrossRef]
- De Freitas, J.; Wintz, H.; Kim, J.H.; Poynton, H.; Fox, T.; Vulpe, C. Yeast, a model organism for iron and copper metabolism studies. BioMetals 2003, 16, 185–197. [Google Scholar] [CrossRef]
- Hood MI, S.E. Nutritional immunity: Transition metals at the pathogen-host interface. Nat. Rev. Micro. Biol. 2012, 10, 525–537. [Google Scholar] [CrossRef]
- Christin, C.; Bischoff, R.; Horvatovich, P. Data processing pipelines for comprehensive profiling of proteomics samples by label-free LC–MS for biomarker discovery. Talanta 2011, 83, 1209–1224. [Google Scholar] [CrossRef][Green Version]
- Vartivarian, S.E.; Anaissie, E.J.; Cowart, R.E.; Sprigg, H.A.; Tingler, M.J.; Jacobson, E.S. Regulation of Cryptococcal Capsular Polysaccharide by Iron. J. Infect. Dis. 1993, 167, 186–190. [Google Scholar] [CrossRef]
- Mylonakis, E.; Moreno, R.; El Khoury, J.B.; Idnurm, A.; Heitman, J.; Calderwood, S.B.; Ausubel, F.M.; Diener, A. Galleria mellonella as a Model System To Study Cryptococcus neoformans Pathogenesis. Infect. Immun. 2005, 73, 3842–3850. [Google Scholar] [CrossRef] [PubMed]
- Zaragoza, O.; Casadevall, A. Experimental modulation of capsule size in Cryptococcus neoformans. Biol. Proced. Online 2004, 6, 10–15. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676. [Google Scholar] [CrossRef]
- Selvan, L.D.N.; Kaviyil, J.E.; Nirujogi, R.S.; Muthusamy, B.; Puttamallesh, V.N.; Subbannayya, T.; Syed, N.; Radhakrishnan, A.; Kelkar, D.S.; Ahmad, S.; et al. Proteogenomic analysis of pathogenic yeast Cryptococcus neoformans using high resolution mass spectrometry. Clin. Proteom. 2014, 11, 5. [Google Scholar] [CrossRef][Green Version]
- Ceballos-Garzon, A.; Monteoliva, L.; Gil, C.; Alvarez-Moreno, C.; Vega-Vela, N.E.; Engelthaler, D.M.; Bowers, J.; Le Pape, P.; Parra-Giraldo, C.M. Genotypic, proteomic, and phenotypic approaches to decipher the response to caspofungin and calcineurin inhibitors in clinical isolates of echinocandin-resistant Candida glabrata. J. Antimicrob. Chemother. 2021, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Jung, W.H.; Sham, A.; Lian, T.; Singh, A.; Kosman, D.J.; Kronstad, J.W. Iron Source Preference and Regulation of Iron Uptake in Cryptococcus neoformans. PLOS Pathog. 2008, 4, e45. [Google Scholar] [CrossRef] [PubMed]
- Jung, W.H.; Kronstad, J.W. Iron influences the abundance of the iron regulatory protein Cir1 in the fungal pathogen Cryptococcus neoformans. FEBS Lett. 2011, 585, 3342–3347. [Google Scholar] [CrossRef]
- Waterman, S.; Hacham, M.; Hu, G.; Zhu, X.; Park, Y.D.; Shin, S.; Panepinto, J.; Valyi-Nagy, T.; Beam, C.; Husain, S.; et al. Role of a CUF1/CTR4 copper regulatory axis in the virulence of Cryptococcus neoformans. J. Clin. Investig. 2007, 117, 794–802. [Google Scholar] [CrossRef]
- Mauch, R.M.; Cunha, V.D.; Dias, A.L. The copper interference with the melanogenesis OF Cryptococcus neoformans. Rev. Inst. Med. Trop. Sao Paulo 2013, 55, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Santamarina, S.; Festa, R.A.; Smith, A.D.; Yu, C.H.; Probst, C.; Ding, C.; Homer, C.M.; Yin, J.; Noonan, J.P.; Madhani, H.; et al. Genome-wide analysis of the regulation of Cu metabolism in Cryptococcus neoformans. Mol Microbiol. 2018, 5, 473–494. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Yin, J.; Tovar, E.M.M.; Fitzpatrick, D.A.; Higgins, D.G.; Thiele, D.J. The Cu regulon of the human fungal pathogen Cryptococcus neoformans H99: Cuf1 activates distinct genes in response to both Cu excess and deficiency. Mol. Microbiol. 2011, 81, 1560–1576. [Google Scholar] [CrossRef]
- Norwood, S.J.; Shaw, D.J.; Cowieson, N.P.; Owen, D.J.; Teasdale, R.; Collins, B.M. Assembly and Solution Structure of the Core Retromer Protein Complex. Traffic 2011, 12, 56–71. [Google Scholar] [CrossRef]
- Nothwehr, S.F.; Ha, S.-A.; Bruinsma, P. Sorting of Yeast Membrane Proteins into an Endosome-to-Golgi Pathway Involves Direct Interaction of Their Cytosolic Domains with Vps35p. J. Cell Biol. 2000, 151, 297–310. [Google Scholar] [CrossRef]
- Park, J.-N.; Lee, D.-J.; Kwon, O.; Oh, D.-B.; Bahn, Y.-S.; Kang, H.A. Unraveling Unique Structure and Biosynthesis Pathway of N-Linked Glycans in Human Fungal Pathogen Cryptococcus neoformans by Glycomics Analysis. J. Biol. Chem. 2012, 287, 19501–19515. [Google Scholar] [CrossRef] [PubMed]
- Thak, E.J.; Lee, S.-B.; Xu-Vanpala, S.; Lee, D.-J.; Chung, S.-Y.; Bahn, Y.-S.; Oh, D.-B.; Shinohara, M.L.; Kang, H.A. Core N-Glycan Structures Are Critical for the Pathogenicity of Cryptococcus neoformans by Modulating Host Cell Death. MBio 2020, 11, e00711–e00720. [Google Scholar] [CrossRef] [PubMed]
- Havel, V.E.; Wool, N.K.; Ayad, D.; Downey, K.M.; Wilson, C.F.; Larsen, P.; Djordjevic, J.T.; Panepinto, J.C. Ccr4 promotes resolution of the endoplasmic reticulum stress response during host temperature adaptation in Cryptococcus neoformans. Eukaryot. Cell 2010, 10, 895–901. [Google Scholar] [CrossRef]
- Panepinto, J.C.; Heinz, E.; Traven, A. The cellular roles of Ccr4-NOT in model and pathogenic fungi—Implications for fungal virulence. Front. Genet. 2013, 4, 302. [Google Scholar] [CrossRef]
- Grütter, C.; Simard, J.R.; Mayer-Wrangowski, S.C.; Schreier, P.H.; Pérez-Martín, J.; Richters, A.; Getlik, M.; Gutbrod, O.; Braun, C.A.; Beck, M.E.; et al. argeting GSK3 from Ustilago maydis: Type-II Kinase Inhibitors as Potential Antifungals. ACS Chem. Biol. 2012, 7, 1257–1267. [Google Scholar] [CrossRef]
- Haynes, B.C.; Skowyra, M.L.; Spencer, S.J.; Gish, S.R.; Williams, M.; Held, E.P.; Brent, M.R.; Doering, T.L. Toward an Integrated Model of Capsule Regulation in Cryptococcus neoformans. PLoS Pathog 2011, 7, e1002411. [Google Scholar] [CrossRef] [PubMed]
- Kauppinen, R. Porphyrias. Lancet 2005, 365, 241–252. [Google Scholar] [CrossRef]
- Lee, I.R.; Chow, E.W.; Morrow, C.A.; Djordjevic, J.T.; Fraser, J.A. Nitrogen Metabolite Repression of Metabolism and Virulence in the Human Fungal Pathogen Cryptococcus neoformans. Genetics 2011, 188, 309–323. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Santamarina, S.; Uzarska, M.A.; Festa, R.A.; Lill, R.; Thiele, D.J. Cryptococcus neoformans Iron-Sulfur Protein Biogenesis Machinery Is a Novel Layer of Protection against Cu Stress. MBio 2017, 8, e01742-17. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.W.; Lee, Y.; Huh, E.Y.; Lee, S.C.; Lim, S.; Bahn, Y.S. Rad53- and Chk1-Dependent DNA Damage Response Pathways Cooperatively Promote Fungal Pathogenesis and Modulate Antifungal Drug Susceptibility. mBio 2019, 10, e01726-18. [Google Scholar] [CrossRef]
- Sahu, M.S.; Patra, S.; Kumar, K.; Kaur, R. SUMOylation in Human Pathogenic Fungi: Role in Physiology and Virulence. J. Fungi 2020, 6, 32. [Google Scholar] [CrossRef]
- Zamudio-Arroyo, J.M.; Peña-Rangel, M.T.; Riesgo-Escovar, J.R. La ubiquitinación: Un sistema de regulación dinámico de los organismos. Rev. Esp. Cienc. Quím. Biol. 2012, 15, 135–141. [Google Scholar]
- Broomfield, S.; Chow, B.L.; Xiao, W. MMS2, encoding a ubiquitin-conjugating-enzyme-like protein, is a member of the yeast error-free postreplication repair pathway. Proc. Natl. Acad. Sci. USA 1998, 95, 5678–5683. [Google Scholar] [CrossRef] [PubMed]
- O’Meara, T.R.; Hay, C.; Price, M.S.; Giles, S.; Alspaugh, J.A. Cryptococcus neoformans Histone Acetyltransferase Gcn5 Regulates Fungal Adaptation to the Host. Eukaryot. Cell 2010, 9, 1193–1202. [Google Scholar] [CrossRef]
- Thiagalingam, S.; Cheng, K.-H.; Lee, H.J.; Mineva, N.; Thiagalingam, A.; Ponte, J.F. Histone Deacetylases: Unique Players in Shaping the Epigenetic Histone Code. Ann. N. Y. Acad. Sci. 2003, 983, 84–100. [Google Scholar] [CrossRef]
- Lee, K.-T.; So, Y.-S.; Yang, D.-H.; Jung, K.-W.; Choi, J.; Lee, D.-G.; Kwon, H.; Jang, J.; Wang, L.L.; Cha, S.; et al. Systematic functional analysis of kinases in the fungal pathogen Cryptococcus neoformans. Nat. Commun. 2016, 7, 12766. [Google Scholar] [CrossRef] [PubMed]
- Roelants, F.M.; Torrance, P.D.; Bezman, N.; Thorner, J. Pkh1 and Pkh2 Differentially Phosphorylate and Activate Ypk1 and Ykr2 and Define Protein Kinase Modules Required for Maintenance of Cell Wall Integrity. Mol. Biol. Cell 2002, 13, 3005–3028. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.-T.; Byun, H.-J.; Jung, K.-W.; Hong, J.; Cheong, E.; Bahn, Y.-S. Distinct and Redundant Roles of Protein Tyrosine Phosphatases Ptp1 and Ptp2 in Governing the Differentiation and Pathogenicity of Cryptococcus neoformans. Eukaryot Cell 2014, 13, 796–812. [Google Scholar] [CrossRef]
- Kraus, P.R.; Fox, D.S.; Cox, G.M.; Heitman, J. The Cryptococcus neoformans MAP kinase Mpk1 regulates cell integrity in response to antifungal drugs and loss of calcineurin function. Mol. Microbiol. 2003, 48, 1377–1387. [Google Scholar] [CrossRef]
- Cabib, E.; Roh, D.-H.; Schmidt, M.; Crotti, L.B.; Varma, A. The Yeast Cell Wall and Septum as Paradigms of Cell Growth and Morphogenesis. J. Biol. Chem. 2001, 276, 19679–19682. [Google Scholar] [CrossRef]
- Sanguinetti, M.; Posteraro, B.; La Sorda, M.; Torelli, R.; Fiori, B.; Santangelo, R.; Delogu, G.; Fadda, G. Role of AFR1, an ABC Transporter-Encoding Gene, in the In Vivo Response to Fluconazole and Virulence of Cryptococcus neoformans. Infect. Immun. 2006, 74, 1352–1359. [Google Scholar] [CrossRef] [PubMed]
- Orsi, C.F.; Colombari, B.; Ardizzoni, A.; Peppoloni, S.; Neglia, R.; Posteraro, B.; Morace, G.; Fadda, G.; Blasi, E. The ABC transporter-encoding gene AFR1 affects the resistance of Cryptococcus neoformans to microglia-mediated antifungal activity by delaying phagosomal maturation. FEMS Yeast Res. 2009, 9, 301–310. [Google Scholar] [CrossRef]
- Vélez, N.; Escandón, P. Report on novel environmental niches for Cryptococcus neoformans and Cryptococcus gattii in Colombia: Tabebuia guayacan and Roystonea regia. Med. Mycol. 2017, 55. [Google Scholar] [CrossRef]
- McFadden, D.C.; Casadevall, A. Capsule and melanin synthesis in Cryptococcus neoformans. Med. Mycol. 2001, 39 (Suppl. S1), 19–30. [Google Scholar] [CrossRef][Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vélez, N.; Monteoliva, L.; Sánchez-Quitian, Z.-A.; Amador-García, A.; García-Rodas, R.; Ceballos-Garzón, A.; Gil, C.; Escandón, P.; Zaragoza, Ó.; Parra-Giraldo, C.-M. The Combination of Iron and Copper Increases Pathogenicity and Induces Proteins Related to the Main Virulence Factors in Clinical Isolates of Cryptococcus neoformans var. grubii. J. Fungi 2022, 8, 57. https://doi.org/10.3390/jof8010057
Vélez N, Monteoliva L, Sánchez-Quitian Z-A, Amador-García A, García-Rodas R, Ceballos-Garzón A, Gil C, Escandón P, Zaragoza Ó, Parra-Giraldo C-M. The Combination of Iron and Copper Increases Pathogenicity and Induces Proteins Related to the Main Virulence Factors in Clinical Isolates of Cryptococcus neoformans var. grubii. Journal of Fungi. 2022; 8(1):57. https://doi.org/10.3390/jof8010057
Chicago/Turabian StyleVélez, Nórida, Lucía Monteoliva, Zilpa-Adriana Sánchez-Quitian, Ahinara Amador-García, Rocío García-Rodas, Andrés Ceballos-Garzón, Concha Gil, Patricia Escandón, Óscar Zaragoza, and Claudia-Marcela Parra-Giraldo. 2022. "The Combination of Iron and Copper Increases Pathogenicity and Induces Proteins Related to the Main Virulence Factors in Clinical Isolates of Cryptococcus neoformans var. grubii" Journal of Fungi 8, no. 1: 57. https://doi.org/10.3390/jof8010057
APA StyleVélez, N., Monteoliva, L., Sánchez-Quitian, Z.-A., Amador-García, A., García-Rodas, R., Ceballos-Garzón, A., Gil, C., Escandón, P., Zaragoza, Ó., & Parra-Giraldo, C.-M. (2022). The Combination of Iron and Copper Increases Pathogenicity and Induces Proteins Related to the Main Virulence Factors in Clinical Isolates of Cryptococcus neoformans var. grubii. Journal of Fungi, 8(1), 57. https://doi.org/10.3390/jof8010057







