Beauveria bassiana Xylanase: Characterization and Wastepaper Deinking Potential of a Novel Glycosyl Hydrolase from an Endophytic Fungal Entomopathogen
Abstract
:1. Introduction
2. Methodology
2.1. Chemicals and Reagents
2.2. Microorganism
2.3. Enzyme Production
2.4. Enzyme Assay
2.5. Protein Estimation
2.6. Enzyme Purification
2.6.1. Ammonium Sulphate Precipitation
2.6.2. Ion-Exchange Chromatography
2.6.3. Size Exclusion Chromatography
2.7. Characterization of Enzymes
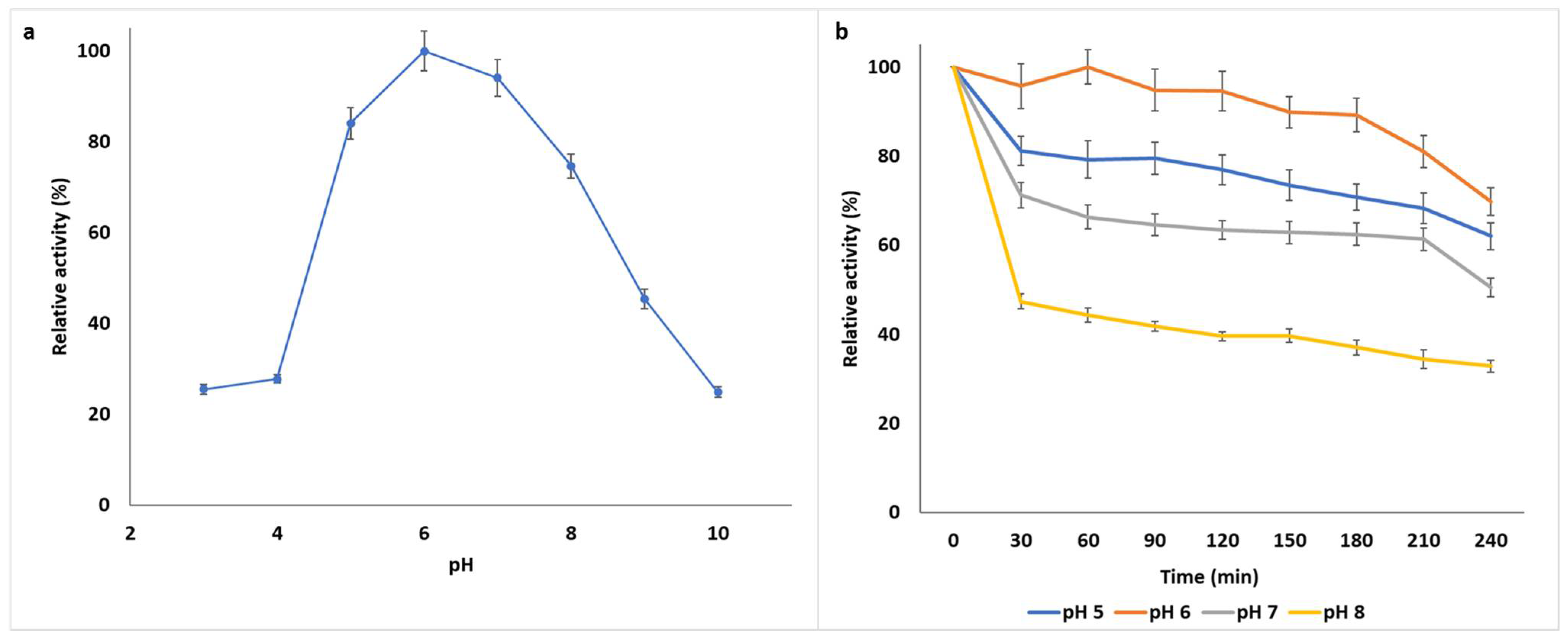
2.7.1. pH Optima and pH Stability
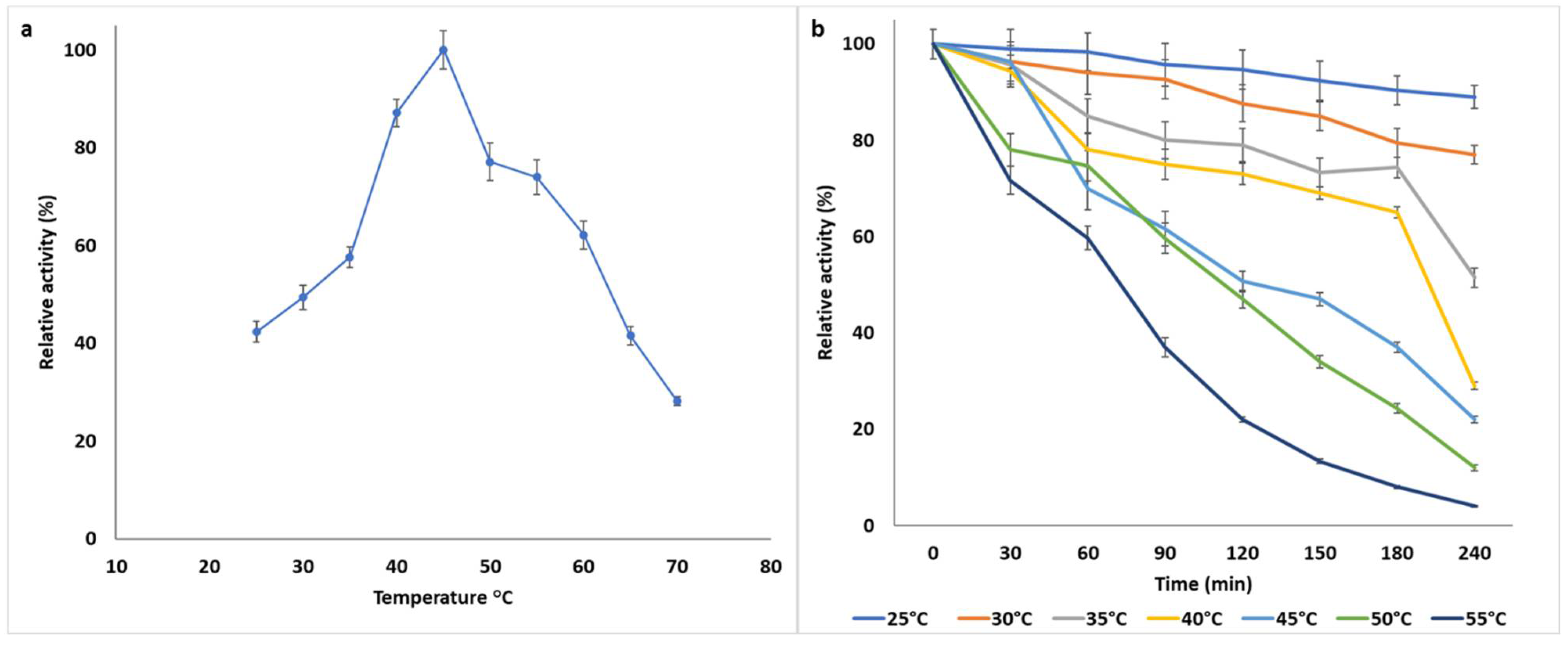
2.7.2. Temperature Optima and Thermostability
2.7.3. Effect of Metal Ions and Salt Concentration on Enzyme Activity
2.7.4. Effect of Various Additives on Enzyme Activity
2.7.5. Effects of Different Solvents on Enzyme Activity
2.8. Kinetic Study and Substrate Specificity
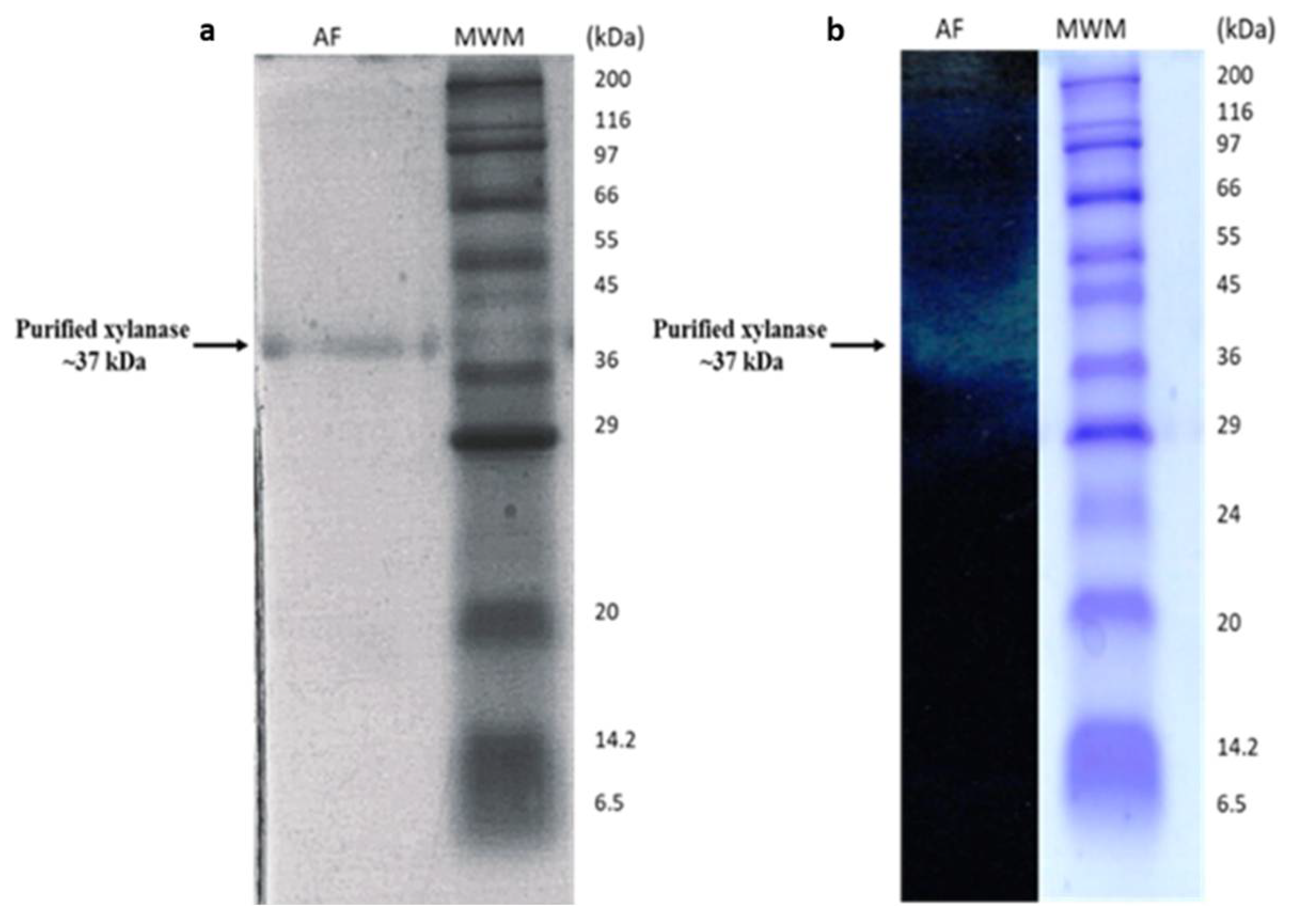
2.9. Analysis of Protein Pattern by SDS-PAGE
2.10. Zymogram Analysis of Xylanase
2.11. Deinking of Wastepaper
2.11.1. Optimization of Deinking Process
2.11.2. Release of Ink
2.11.3. Scanning Electron Microscopy
2.11.4. FTIR Analysis
2.12. Statistical Analysis
3. Results and Discussion
3.1. Purification and Characterization of B. bassiana SAN01 Xylanase
3.1.1. Purification of B. bassiana SAN01 Xylanase
3.1.2. Biochemical Characterization of B. bassiana SAN01 Xylanase
3.1.3. Kinetic Analysis of B. bassiana SAN01 Xylanase
3.1.4. Substrate Specificity of B. bassiana SAN01 Xylanase
3.2. Deinking of Printed Paper by B. bassiana SAN01 Xylanase
3.2.1. Optimization of Deinking Process
3.2.2. Validation of the Experimental Model
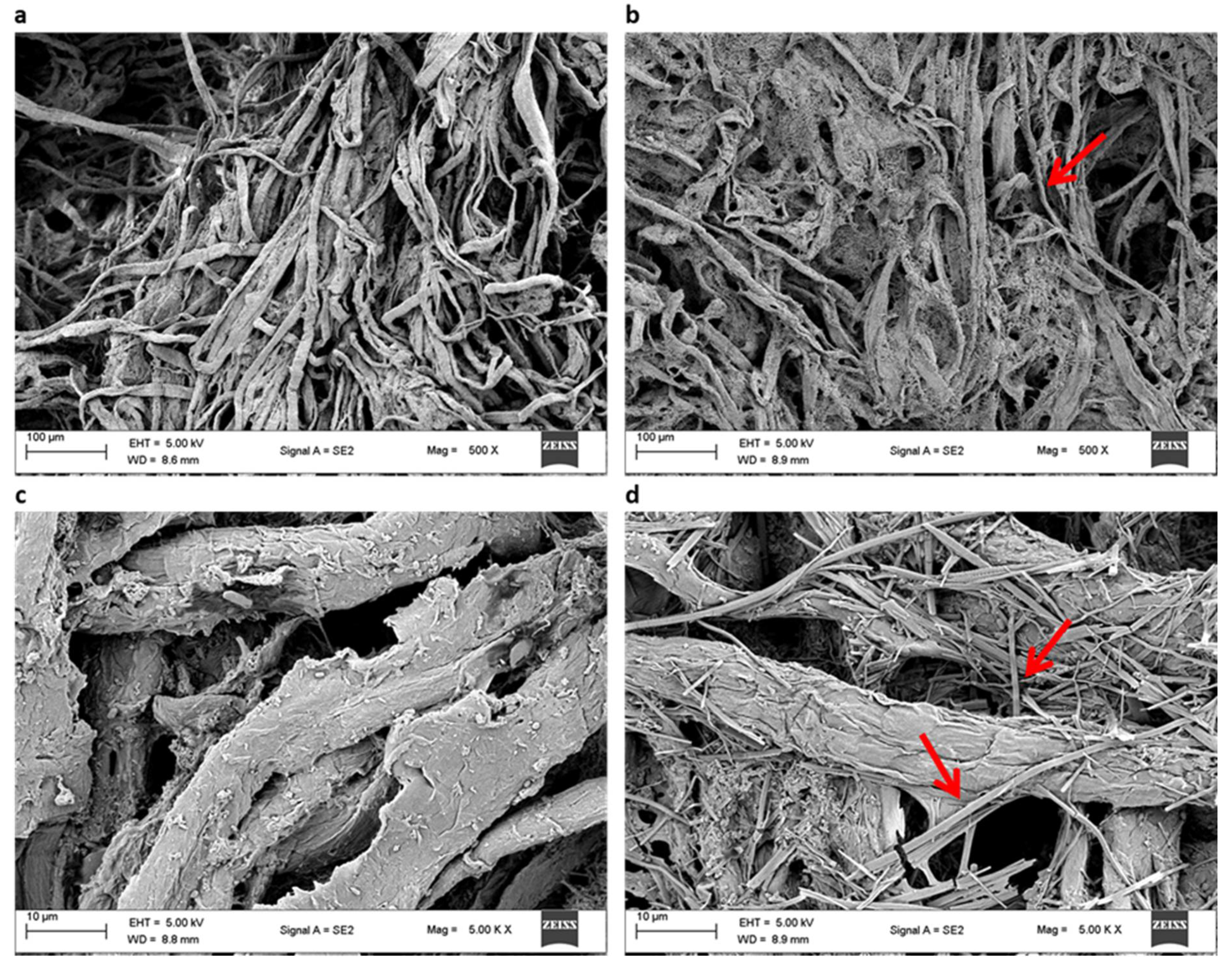
3.2.3. Scanning Electron Microscopy
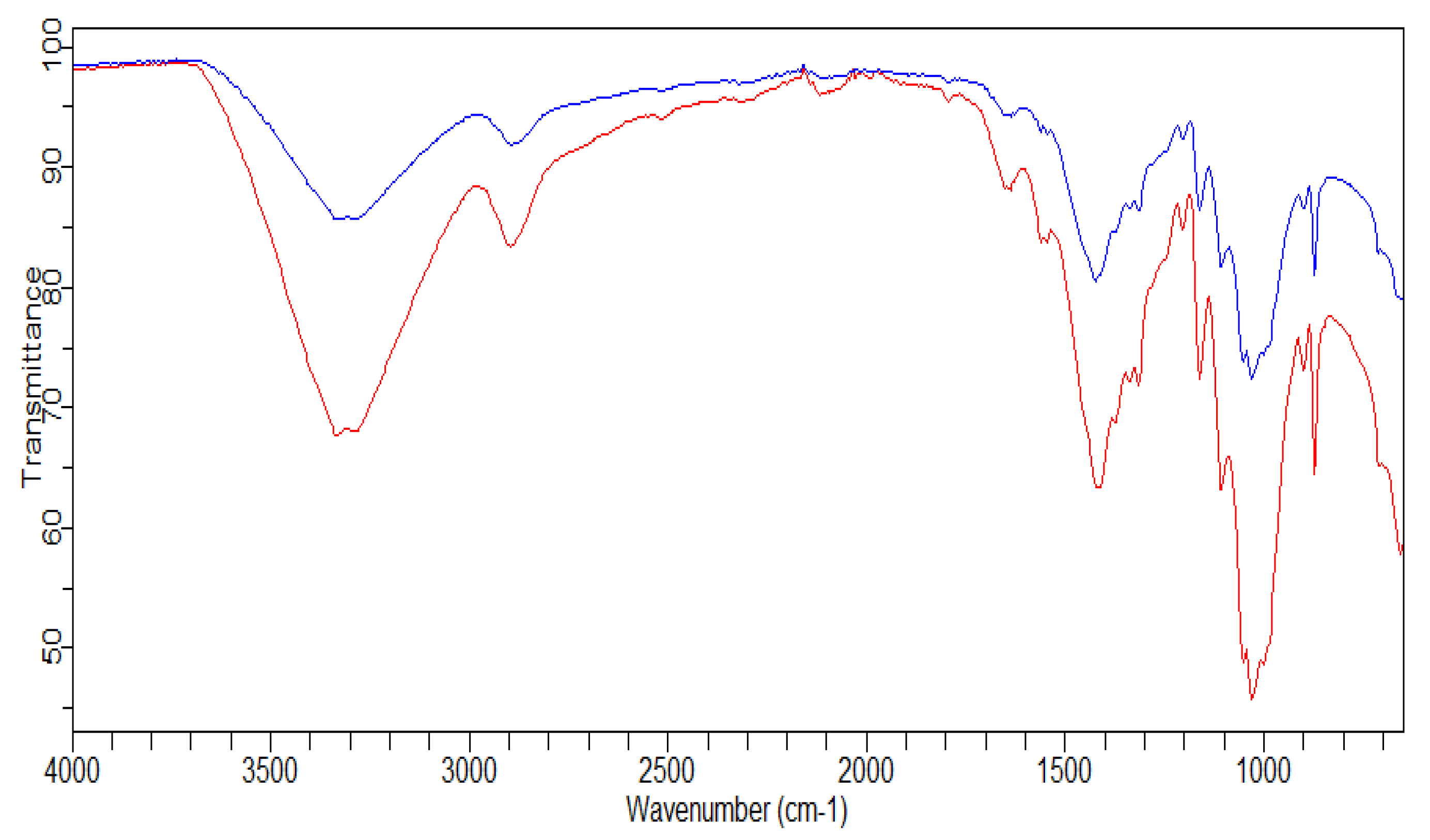
3.2.4. FTIR Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Ethics Approval
Consent to Participate
References
- Litwin, A.; Nowak, M.; Różalska, S. Entomopathogenic fungi: Unconventional applications. Rev. Environ. Sci. Bio. Technol. 2020, 19, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, P.; Gonzalez, D.; Rodríguez Giordano, S. Endophytic microorganisms: A source of potentially useful biocatalysts. J. Mol. Catal. B Enzym. 2016, 133, S569–S581. [Google Scholar] [CrossRef]
- Borgi, I.; Gargouri, A. A novel high molecular weight thermo-acidoactive β-glucosidase from Beauveria bassiana. Appl. Biochem. Microbiol. 2016, 52, 602–607. [Google Scholar] [CrossRef]
- Ryali, S.L.; Shankar, S.; More, S.V.; Khandelwal, H.B.; Narasimhan, C.B.K.; Palanivel, S.; Balaram, P. Fungal Strain Beauveria sp. MTCC 5184 and a Process for the Preparation of Enzymes Therefrom. U.S. Patent US10544473B2, 28 January 2020. [Google Scholar]
- Amobonye, A.; Bhagwat, P.; Pandey, A.; Singh, S.; Pillai, S. Biotechnological potential of Beauveria bassiana as a source of novel biocatalysts and metabolites. Crit. Rev. Biotechnol. 2020, 40, 1019–1034. [Google Scholar] [CrossRef]
- Zibaee, A.; Sadeghi-Sefidmazgi, A.; Fazeli-Dinan, M. Properties of a lipase produced by Beauveria bassiana: Purification and biochemical studies. Biocontrol Sci. Technol. 2011, 21, 317–331. [Google Scholar] [CrossRef]
- Zibaee, A.; Ramzi, S. Cuticle-degrading proteases of entomopathogenic fungi: From biochemistry to biological performance. Arch. Phytopathol. Plant Prot. 2018, 51, 779–794. [Google Scholar] [CrossRef]
- Bhagwat, P.; Amobonye, A.; Singh, S.; Pillai, S. A comparative analysis of GH18 chitinases and their isoforms from Beauveria bassiana: An in-silico approach. Process Biochem. 2020, 100, 207–216. [Google Scholar] [CrossRef]
- Petlamul, W.; Buakaew, S. Optimisation and stabilisation of cellulase and xylanase production by Beauveria bassiana. Environ. Asia 2019, 12, 11–19. [Google Scholar] [CrossRef]
- Chen, Z.; Zaky, A.A.; Liu, Y.; Chen, Y.; Liu, L.; Li, S.; Jia, Y. Purification and characterization of a new xylanase with excellent stability from Aspergillus flavus and its application in hydrolyzing pretreated corncobs. Protein Expr. Purif. 2019, 154, 91–97. [Google Scholar] [CrossRef]
- Malgas, S.; Mafa, M.S.; Pletschke, B.I. The effects of xylanase synergistic interactions during lignocellulose degradation and their significance for industry. In Industrial Applications of Glycoside Hydrolases; Springer: Cham, Switzerland, 2020; pp. 229–246. [Google Scholar] [CrossRef]
- Bajpai, P. Chapter 6—Purification of xylanases. In Xylanolytic Enzymes; Bajpai, P., Ed.; Academic Press: Amsterdam, The Netherlands, 2014; pp. 53–61. [Google Scholar] [CrossRef]
- Fan, Y.; Zhang, Y.; Yang, X.; Pei, X.; Guo, S.; Pei, Y. Expression of a Beauveria bassiana chitinase (Bbchit1) in Escherichia coli and Pichia pastoris. Protein Expr. Purif. 2007, 56, 93–99. [Google Scholar] [CrossRef]
- Amobonye, A.; Bhagwat, P.; Singh, S.; Pillai, S. Enhanced xylanase and endoglucanase production from Beauveria bassiana SAN01, an entomopathogenic fungal endophyte. Fungal Biol. 2021, 125, 39–48. [Google Scholar] [CrossRef]
- Bailey, M.J.; Biely, P.; Poutanen, K. Interlaboratory testing of methods for assay of xylanase activity. J. Biotechnol. 1992, 23, 257–270. [Google Scholar] [CrossRef]
- Hartree, E. Determination of protein: A modification of the Lowry method. Anal. Biochem. 1972, 48, 422–427. [Google Scholar] [CrossRef]
- Guillaume, A.; Thorigné, A.; Carré, Y.; Vinh, J.; Levavasseur, L. Contribution of proteases and cellulases produced by solid-state fermentation to the improvement of corn ethanol production. Bioresour. Bioprocess. 2019, 6, 7. [Google Scholar] [CrossRef]
- Kohli, I.; Joshi, N.C.; Varma, A. Production, purification and applications of raw starch degrading and calcium-independent α-amylase from soil rich in extremophile. Int. J. Biol. Macromol. 2020, 162, 873–881. [Google Scholar] [CrossRef] [PubMed]
- Melnichuk, N.; Braia, M.J.; Anselmi, P.A.; Meini, M.-R.; Romanini, D. Valorization of two agroindustrial wastes to produce alpha-amylase enzyme from Aspergillus oryzae by solid-state fermentation. Waste Manag. 2020, 106, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Gui, Y.; Li, J.; Zhang, X.; Zilda, D.S. Identification and characterization of a novel alkali- and high temperature-tolerant lipase (Lip4346) from a macroalgae-associated bacterial strain. J. Ocean Univ. China 2021, 20, 181–188. [Google Scholar] [CrossRef]
- Ozdemir, S.; Fincan, S.A.; Karakaya, A.; Enez, B. A novel raw starch hydrolyzing thermostable α-amylase produced by newly isolated Bacillus mojavensis SO-10: Purification, characterization and usage in starch industries. Braz. Arch. Biol. Technol. 2018, 61, 1–16. [Google Scholar] [CrossRef]
- Mhiri, S.; Bouanane-Darenfed, A.; Jemli, S.; Neifar, S.; Ameri, R.; Mezghani, M.; Bouacem, K.; Jaouadi, B.; Bejar, S. A thermophilic and thermostable xylanase from Caldicoprobacter algeriensis: Recombinant expression, characterization and application in paper biobleaching. Int. J. Biol. Macromol. 2020, 164, 808–817. [Google Scholar] [CrossRef]
- Zhu, T.; Li, R.; Sun, J.; Cui, Y.; Wu, B. Characterization and efficient production of a thermostable, halostable and organic solvent-stable cellulase from an oil reservoir. Int. J. Biol. Macromol. 2020, 159, 622–629. [Google Scholar] [CrossRef]
- Ahmed, S.A.; Abdella, M.A.; El-Sherbiny, G.M.; Ibrahim, A.M.; El-Shamy, A.R.; Atalla, S.M.; Hassan, M.E. Catalytic, kinetic and thermal properties of free andimmobilized Bacillus subtilis MK1 α-amylase on chitosan-magnetic nanoparticles. Biotechnol. Rep. 2020, 26, e00443. [Google Scholar] [CrossRef]
- Marangoni, A.G. Enzyme Kinetics: A Modern Approach; John Wiley & Sons: Hoboken, NJ, USA, 2003. [Google Scholar]
- Laemmli, U. Slab gel electrophoresis: SDS–PAGE with discontinuous buffers. Nature 1979, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.Y.; Wang, K.T.; Wu, Y.S.; Yeh, S.P.; Wu, T.M. Secretory expression and characterization of xylanase isolated from Bacillus subtilis E20 increase the utilization of plant ingredients in tilapia feed. Aquac. Res. 2019, 50, 2240–2250. [Google Scholar] [CrossRef]
- Saxena, D.; Sabikhi, L.; Chakraborty, S.K.; Singh, D. Process optimization for enzyme aided clarification of watermelon juice. J. Food Sci. Technol. 2014, 51, 2490–2498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cerreti, M.; Liburdi, K.; Benucci, I.; Spinelli, S.E.; Lombardelli, C.; Esti, M. Optimization of pectinase and protease clarification treatment of pomegranate juice. LWT Food Sci. Technol. 2017, 82, 58–65. [Google Scholar] [CrossRef]
- Thomas, L.; Ushasree, M.V.; Pandey, A. An alkali-thermostable xylanase from Bacillus pumilus functionally expressed in Kluyveromyces lactis and evaluation of its deinking efficiency. Bioresour. Technol. 2014, 165, 309–313. [Google Scholar] [CrossRef] [PubMed]
- Chutani, P.; Sharma, K.K. Biochemical evaluation of xylanases from various filamentous fungi and their application for the deinking of ozone treated newspaper pulp. Carbohydr. Polym. 2015, 127, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Borges, M.S.; Barbosa, R.S.; Rambo, M.K.D.; Rambo, M.C.D.; Scapin, E. Evaluation of residual biomass produced in Cerrado Tocantinense as potential raw biomass for biorefinery. Biomass Convers. Biorefinery 2020, 1–12. [Google Scholar] [CrossRef]
- Bhardwaj, N.; Verma, V.K.; Chaturvedi, V.; Verma, P. Cloning, expression and characterization of a thermo-alkali-stable xylanase from Aspergillus oryzae LC1 in Escherichia coli BL21 (DE3). Protein Expr. Purif. 2020, 168, 105551. [Google Scholar] [CrossRef]
- Podestá, M.V.; Morilla, E.A.; Allasia, M.B.; Woitovich Valetti, N.; Tubio, G.; Boggione, M.J. An Eco-friendly Method of Purification for xylanase from Aspergillus niger by polyelectrolyte precipitation. J. Polym. Environ. 2019, 27, 2895–2905. [Google Scholar] [CrossRef]
- Hassan, N.; Rafiq, M.; Rehman, M.; Sajjad, W.; Hasan, F.; Abdullah, S. Fungi in acidic fire: A potential source of industrially important enzymes. Fungal Biol. Rev. 2019, 33, 58–71. [Google Scholar] [CrossRef]
- Somboon, C.; Boonrung, S.; Katekaew, S.; Ekprasert, J.; Aimi, T.; Boonlue, S. Purification and characterization of low molecular weight alkali stable xylanase from Neosartorya spinosa UZ-2-11. Mycoscience 2020, 61, 128–135. [Google Scholar] [CrossRef]
- Knob, A.; Carmona, E.C. Purification and characterization of two extracellular xylanases from Penicillium sclerotiorum: A novel acidophilic xylanase. Appl. Biochem. Biotechnol. 2010, 162, 429–443. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zheng, X.; Pilgaard, B.; Holck, J.; Muschiol, J.; Li, S.; Lange, L. Identification and characterization of GH11 xylanase and GH43 xylosidase from the chytridiomycetous fungus, Rhizophlyctis rosea. Appl. Microbiol. Biotechnol. 2019, 103, 777–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marques, G.L.; dos Santos Reis, N.; Silva, T.P.; Ferreira, M.L.O.; Aguiar-Oliveira, E.; de Oliveira, J.R.; Franco, M. Production and characterisation of xylanase and endoglucanases produced by Penicillium roqueforti ATCC 10110 through the solid-state fermentation of rice husk residue. Waste Biomass Valorization 2018, 9, 2061–2069. [Google Scholar] [CrossRef]
- Adiguzel, G.; Faiz, O.; Sisecioglu, M.; Sari, B.; Baltaci, O.; Akbulut, S.; Genc, B.; Adiguzel, A. A novel endo-β-1,4-xylanase from Pediococcus acidilactici GC25; purification, characterization and application in clarification of fruit juices. Int. J. Biol. Macromol. 2019, 129, 571–578. [Google Scholar] [CrossRef]
- Terrone, C.C.; Freitas, C.d.; Terrasan, C.R.F.; Almeida, A.F.d.; Carmona, E.C. Agroindustrial biomass for xylanase production by Penicillium chrysogenum: Purification, biochemical properties and hydrolysis of hemicelluloses. Electron. J. Biotechnol. 2018, 33, 39–45. [Google Scholar] [CrossRef]
- Michelin, M.; Silva, T.M.; Jorge, J.A.; Polizeli, M.d.L.T.M. Purification and biochemical properties of multiple xylanases from Aspergillus ochraceus tolerant to Hg2+ ion and a wide range of pH. Appl. Biochem. Biotechnol. 2014, 174, 206–220. [Google Scholar] [CrossRef]
- Palavesam, A. Investigation on lignocellulosic saccharification and characterization of haloalkaline solvent tolerant endo-1,4 β-d-xylanase from Halomonas meridiana APCMST-KS4. Biocatal. Agric. Biotechnol. 2015, 4, 761–766. [Google Scholar] [CrossRef]
- Lin, S.L.; Aniza, R.; Lee, Y.Y.; Wang, C.L. Reduction of traditional pollutants and polychlorinated dibenzo-p-dioxins and dibenzofurans emitted from a diesel engine generator equipped with a catalytic ceramic fiber filter system. Clean Technol. Environ. Policy 2018, 20, 1297–1309. [Google Scholar] [CrossRef]
- Souza, L.O.; de Brito, A.R.; Bonomo, R.C.F.; Santana, N.B.; Almeida Antunes Ferraz, J.L.d.; Aguiar-Oliveira, E.; Araújo Fernandes, A.G.d.; Ferreira, M.L.O.; de Oliveira, J.R.; Franco, M. Comparison of the biochemical properties between the xylanases of Thermomyces lanuginosus (Sigma®) and excreted by Penicillium roqueforti ATCC 10110 during the solid state fermentation of sugarcane bagasse. Biocatal. Agric. Biotechnol. 2018, 16, 277–284. [Google Scholar] [CrossRef]
- de Cassia Pereira, J.; Giese, E.C.; de Souza Moretti, M.M.; dos Santos Gomes, A.C.; Perrone, O.M.; Boscolo, M.; da Silva, R.; Gomes, E.; Martins, D.A.B. Effect of metal ions, chemical agents and organic compounds on lignocellulolytic enzymes activities. Enzym. Inhib. Act. 2017, 29, 139–164. [Google Scholar] [CrossRef] [Green Version]
- Adigüzel, A.O.; Tunçer, M. Production, characterization and application of a xylanase from Streptomyces sp. AOA40 in fruit juice and bakery industries. Food Biotechnol. 2016, 30, 189–218. [Google Scholar] [CrossRef]
- Hamid, A.; Aftab, M.N. Cloning, purification, and characterization of recombinant thermostable β-xylanase Tnap_0700 from Thermotoga naphthophila. Appl. Biochem. Biotechnol. 2019, 189, 1274–1290. [Google Scholar] [CrossRef] [PubMed]
- Adhyaru, D.N.; Bhatt, N.S.; Modi, H.A. Enhanced production of cellulase-free, thermo-alkali-solvent-stable xylanase from Bacillus altitudinis DHN8, its characterization and application in sorghum straw saccharification. Biocatal. Agric. Biotechnol. 2014, 3, 182–190. [Google Scholar] [CrossRef]
- Gaur, R.; Tiwari, S.; Rai, P.; Srivastava, V. Isolation, Production and characterization of thermotolerant xylanase from solvent tolerant Bacillus vallismortis. Int. J. Polym. Sci. 2015, 2015, 986324. [Google Scholar] [CrossRef] [Green Version]
- Gupta, M.N. Enzyme function in organic solvents. Eur. J. Biochem. 1992, 203, 25–32. [Google Scholar] [CrossRef]
- Li, Z.; Chen, S.; Wang, Y.; Liu, Z.; Xia, Z.; Zhang, M.; Luo, X.; Song, Y.; Zhao, J.; Zhang, T. Biochemical characterization of a novel halo/organic-solvents/final-products tolerant GH39 xylosidase from saline soil and its synergic action with xylanase. Int. J. Biol. Macromol. 2020, 164, 184–192. [Google Scholar] [CrossRef]
- Uday, U.S.P.; Majumdar, R.; Tiwari, O.N.; Mishra, U.; Mondal, A.; Bandyopadhyay, T.K.; Bhunia, B. Isolation, screening and characterization of a novel extracellular xylanase from Aspergillus niger (KP874102.1) and its application in orange peel hydrolysis. Int. J. Biol. Macromol. 2017, 105, 401–409. [Google Scholar] [CrossRef]
- Sudan, R.; Bajaj, B.K. Production and biochemical characterization of xylanase from an alkalitolerant novel species Aspergillus niveus RS2. World J. Microbiol. Biotechnol. 2007, 23, 491–500. [Google Scholar] [CrossRef]
- Fu, L.H.; Jiang, N.; Li, C.X.; Luo, X.M.; Zhao, S.; Feng, J.X. Purification and characterization of an endo-xylanase from Trichoderma sp., with xylobiose as the main product from xylan hydrolysis. World J. Microbiol. Biotechnol. 2019, 35, 171. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; Garg, S.; Capalash, N.; Gupta, N.; Sharma, P. Production of thermo-alkali-stable laccase and xylanase by co-culturing of Bacillus sp. and B. halodurans for biobleaching of kraft pulp and deinking of waste paper. Bioprocess Biosyst. Eng. 2015, 38, 947–956. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Satyanarayana, T. Production of endoxylanase with enhanced thermostability by a novel polyextremophilic Bacillus halodurans TSEV1 and its applicability in waste paper deinking. Process Biochem. 2014, 49, 386–394. [Google Scholar] [CrossRef]
- Saini, S.; Chutani, P.; Kumar, P.; Sharma, K.K. Development of an eco-friendly deinking process for the production of bioethanol using diverse hazardous paper wastes. Renew. Energy 2020, 146, 2362–2373. [Google Scholar] [CrossRef]
- Przybysz Buzała, K.; Przybysz, P.; Kalinowska, H.; Derkowska, M. Effect of cellulases and xylanases on refining process and kraft pulp properties. PLoS ONE 2016, 11, e0161575. [Google Scholar] [CrossRef] [PubMed]
- Sridevi, A.; Sandhya, A.; Ramanjaneyulu, G.; Narasimha, G.; Devi, P.S. Biocatalytic activity of Aspergillus niger xylanase in paper pulp biobleaching. 3 Biotech 2016, 6, 165. [Google Scholar] [CrossRef] [Green Version]
- Kumar, N.V.; Rani, M.E.; Gunaseeli, R.; Kannan, N. Paper pulp modification and deinking efficiency of cellulase-xylanase complex from Escherichia coli SD5. Int. J. Biol. Macromol. 2018, 111, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Desai, D.I.; Iyer, B.D. Biodeinking of old newspaper pulp using a cellulase-free xylanase preparation of Aspergillus niger DX-23. Biocatal. Agric. Biotechnol. 2016, 5, 78–85. [Google Scholar] [CrossRef]
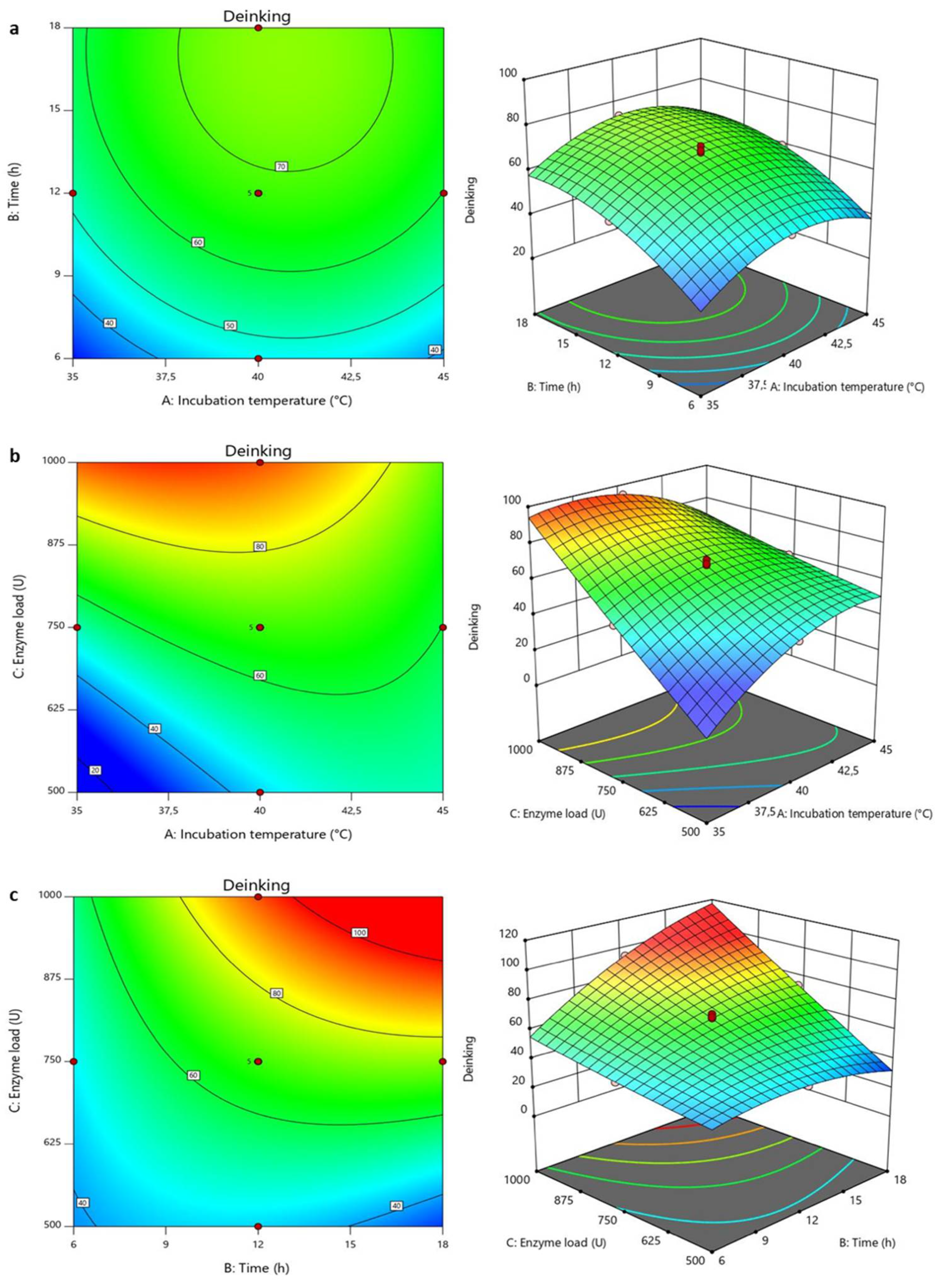
| Independent Variables | Units | Levels | ||||
|---|---|---|---|---|---|---|
| −1.68 | −1 | 0 | +1 | +1.68 | ||
| Temperature (A) | °C | 35 | 36.47 | 40 | 43.54 | 45 |
| Time (B) | h | 6 | 7.76 | 12 | 16.25 | 18 |
| Enzyme load (C) | U·mL−1 | 500 | 573.23 | 750 | 926.78 | 1000 |
| Purification Step | Total Activity (U) b | Total Protein (mg) c | Specific Activity (U·mg−1) | Yield (%) | Purification Fold |
|---|---|---|---|---|---|
| Crude enzyme a | 106,100 | 1452 | 73.07 | 100 | 1 |
| Ammonium sulphate precipitation d | 31272 | 281 | 111.29 | 29.47 | 1.52 |
| Ion exchangechromatography | 5047 | 28.5 | 177.1 | 4.75 | 2.42 |
| Ultrafiltration | 4016 | 14.7 | 273.2 | 3.78 | 3.74 |
| Size exclusion chromatography | 2971.6 | 8.2 | 362.39 | 2.8 | 4.96 |
| Metal Ions | Relative Activity (%) a | |
|---|---|---|
| 1 mM | 10 mM | |
| Control | 100 | 100 |
| Ag2+ | 64.31 ± 3.18 | 30.83 ± 1.44 |
| Ba2+ | 94.27 ± 3.13 | 107.92 ± 6.51 |
| Co2+ | 137.44 ± 7.93 | 144.49 ± 7.92 |
| Cu2+ | 99.55 ± 4.71 | 77.53 ± 3.04 |
| Fe2+ | 92.95 ± 4.19 | 89.86 ± 5.84 |
| Fe3+ | 61.67 ± 3.02 | 77.09 ± 2.51 |
| Hg2+ | 92.95 ± 4.49 | 71.80 ± 3.16 |
| Mg2+ | 111.013 ± 5.16 | 114.97 ± 5.97 |
| Na+ | 85.02 ± 4.02 | 76.65 ± 3.82 |
| Zn2+ | 101.76 ± 5.15 | 95.15 ± 4.18 |
| Additives | Relative Activity (%) a | |
|---|---|---|
| 1 mM | 10 mM | |
| Control | 100 | 100 |
| BME | 123.34 ± 6.02 | 149.33 ± 6.27 |
| DTT | 109.69 ± 5.63 | 121.14 ± 5.74 |
| EDTA | 189.42 ± 7.33 | 125.55 ± 6.61 |
| PMSF | 55.67 ± 2.68 | 50.47 ± 3.06 |
| SDS | 65.19 ± 2.79 | 56.38 ± 2.52 |
| Tween 20 | 103.08 ± 4.37 | 88.15 ± 4.26 |
| Triton X-100 | 90.30 ± 4.37 | 89.86 ± 3.84 |
| Organic Solvent | Relative Activity (%) a |
|---|---|
| Control * | 100 |
| Acetone | 82.7 ± 4.06 |
| Benzene | 81.39 ± 3.88 |
| Butanol | 77.20 ± 3.02 |
| Chloroform | 86.97 ± 4.42 |
| Ethanol | 110.69 ± 5.74 |
| Hexane | 104.65 ± 3.63 |
| Isopropanol | 120 ± 6.18 |
| Methanol | 67.44 ± 1.86 |
| Toluene | 87.44 ± 3.86 |
| Substrate | Relative Activity |
|---|---|
| Arabinoxylan | 76.28 ± 3.36 |
| Avicel | 0 |
| Beechwood xylan | 100 |
| CMC | 17.12 ± 0.56 |
| Oatspelt xylan | 87.7 ± 4.16 |
| Pectin | 0 |
| Soluble starch | 0 |
| Level | Deinking (%) | Reducing Sugar (mg/g) | |||||
|---|---|---|---|---|---|---|---|
| Coded (A) | Coded (B) | Coded (C) | Actual | Predicted | Actual | Predicted | |
| 1 | +1.68 | 0 | 0 | 59.68 | 59.84 | 167.70 | 166.64 |
| 2 | −1 | −1 | −1 | 28.27 | 28.11 | 121.50 | 122.56 |
| 3 | 0 | −1.68 | 0 | 45.83 | 45.99 | 174.17 | 173.11 |
| 4 | 0 | 0 | 0 | 66.90 | 68.00 | 198.67 | 194.28 |
| 5 | +1 | +1 | −1 | 52.08 | 51.92 | 168.62 | 169.68 |
| 6 | 0 | 0 | −1.68 | 43.33 | 43.49 | 140.28 | 139.22 |
| 7 | 0 | 0 | 0 | 71.16 | 68.00 | 190.45 | 194.28 |
| 8 | 0 | 0 | 0 | 68.27 | 68.00 | 187.67 | 194.28 |
| 9 | −1 | +1 | −1 | 100.00 | 99.84 | 196.74 | 197.80 |
| 10 | 0 | 0 | 0 | 69.04 | 68.00 | 197.74 | 194.28 |
| 11 | 0 | +1.68 | 0 | 73.45 | 73.61 | 202.00 | 200.94 |
| 12 | 0 | 0 | 0 | 64.97 | 68.00 | 194.77 | 194.28 |
| 13 | 0 | 0 | +1.68 | 94.32 | 94.48 | 212.56 | 211.50 |
| 14 | −1.68 | 0 | 0 | 51.66 | 51.82 | 159.78 | 158.72 |
| 15 | +1 | −1 | +1 | 53.51 | 53.35 | 170.47 | 171.53 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amobonye, A.; Bhagwat, P.; Singh, S.; Pillai, S. Beauveria bassiana Xylanase: Characterization and Wastepaper Deinking Potential of a Novel Glycosyl Hydrolase from an Endophytic Fungal Entomopathogen. J. Fungi 2021, 7, 668. https://doi.org/10.3390/jof7080668
Amobonye A, Bhagwat P, Singh S, Pillai S. Beauveria bassiana Xylanase: Characterization and Wastepaper Deinking Potential of a Novel Glycosyl Hydrolase from an Endophytic Fungal Entomopathogen. Journal of Fungi. 2021; 7(8):668. https://doi.org/10.3390/jof7080668
Chicago/Turabian StyleAmobonye, Ayodeji, Prashant Bhagwat, Suren Singh, and Santhosh Pillai. 2021. "Beauveria bassiana Xylanase: Characterization and Wastepaper Deinking Potential of a Novel Glycosyl Hydrolase from an Endophytic Fungal Entomopathogen" Journal of Fungi 7, no. 8: 668. https://doi.org/10.3390/jof7080668
APA StyleAmobonye, A., Bhagwat, P., Singh, S., & Pillai, S. (2021). Beauveria bassiana Xylanase: Characterization and Wastepaper Deinking Potential of a Novel Glycosyl Hydrolase from an Endophytic Fungal Entomopathogen. Journal of Fungi, 7(8), 668. https://doi.org/10.3390/jof7080668







