Abstract
Aspergillus flavus (A. flavus) is a ubiquitous and opportunistic fungal pathogen that causes invasive and non-invasive aspergillosis in humans and animals. This fungus is also capable of infecting a large number of agriculture crops (e.g., peanuts, maze, cotton seeds, rice, etc.), causing economic losses and posing serious food-safety concerns when these crops are contaminated with aflatoxins, the most potent naturally occurring carcinogens. In particular, A. flavus and aflatoxins are intensely studied, and they continue to receive considerable attention due to their detrimental effects on humans, animals, and crops. Although several studies have been published focusing on the biosynthesis of the aforementioned secondary metabolites, some of the molecular mechanisms (e.g., posttranslational modifications, transcription factors, transcriptome, proteomics, metabolomics and transcriptome, etc.) involved in the fungal development and aflatoxin biosynthesis in A. flavus are still not fully understood. In this study, a review of the recently published studies on the function of the genes and the molecular mechanisms involved in development of A. flavus and the production of its secondary metabolites is presented. It is hoped that the information provided in this review will help readers to develop effective strategies to reduce A. flavus infection and aflatoxin production.
1. Introduction
Aspergillus flavus (A. flavus) is a common saprophytic fungus. Classically, A. flavus belongs to the kingdom of Fungi, phyllum of Ascomycota, and order of Eurotiales in the Eurotiomycetes class. It also belongs to the family of Trichocomaceae and the genus Aspergillus in species flavus []. This fungus is known to be an opportunistic pathogen, and it is capable of infecting a number of seed crops (e.g., peanuts, maze, cotton seeds, rice, etc.) and living organisms including animals and humans [,]. In general, A. flavus produces highly toxic secondary metabolites named aflatoxins, and the consumption of highly aflatoxin contaminated food can cause death or chronic diseases particularly in immune-suppressed patients. The discovery of aflatoxins in 1961 gave rise to modern mycotoxicology and stirred up scientific interest []. Studies on aflatoxins led to a golden age of mycotoxin research during which numerous other new mycotoxins, such as fumigillin, aspertoxin, cyclopiazonic acid, aflatrem, ochratoxin, and patulin, were also discovered [,]. Among all polyketides and mycotoxins produced by fungal species, aflatoxins are known as the major mycotoxins, including aflatoxin B1 (AFB1), B2 (AFB2), G1 (AFG1), and G2 (AFG2). Additional aflatoxins, such as M1 (AFM1) and M2 (AFM2), which are the metabolites from AFB1 and AFB2, respectively, are generally found in cow milk in areas of high aflatoxin exposure. Subsequently, humans may be exposed to these aflatoxins through milk and milk products, including breast milk, especially in areas where the lowest quality grain is used for animal feed. Of all the aflatoxins identified so far, AFB1 has been found to the most toxic and foremost potent carcinogen natural chemical compound produced by Aspergillus fungi (e.g., A. flavus, A. nomius, A. parasiticus, etc.) [,].
Aflatoxin biosynthesis is a process that involves multiple enzyme reactions with several bioconversion steps. In recent years, worldwide efforts were made to better understand the biochemistry, genetics, and regulation of aflatoxin biosynthesis [,]. In the last decade, for example, significant progress has been made in discovering the genes and their enzymes that are involved in each step of the aflatoxin biosynthetic pathway [,], further confirming that the aflatoxin biosynthesis pathway is one of the most studied fungal secondary metabolite pathways in A. flavus [,]. Although the aflatoxin biosynthesis pathway in A. flavus has been extensively studied, its molecular mechanisms are still not well known. As a result, this review focuses on the recently discovered functions and molecular mechanisms of genes involved in fungal development (e.g., production of conidiophores, conidia, sclerotia, mycelia growth, etc.) and biosynthesis of aflatoxins in A. flavus. In doing so, updated information about A. flavus genomics and its conserved transcription factors involved in morphogenesis and aflatoxins production is provided. Then, an overview on posttranslational modifications (e.g., methylation, acetylation, phosphorylation, sumoylation, succinylation, etc.), signal pathways (e.g., HOG, Slt2, etc.), transcriptome, and metabolomics involved in morphogenesis and aflatoxin production in A. flavus is also provided. To keep our discussion concise, a special focus is directed to the studies that have been published in the last 10 years.
2. A. flavus Genomics and Aflatoxins Biosynthesis Genes
The term genomics denotes the process of revealing the whole genetic content of any kind of organism through the bioinformatics analysis of sequences of the genomic DNA. Additionally, the fungal genomic information allow scientist to analyze the revolutionary relationship between different species []. Interestingly, A. flavus genome is already known, and its whole genome microarray is also available []. The availability of the genome sequence of A. flavus has enabled significant efforts in the understanding of the regulation of aflatoxin biosynthesis and the establishment of new control strategies []. Indeed, the availability of A. flavus genomic data marked the beginning of a new research phase not only in fungal biology but also in other fields, such as medical mycology, pathogenicity, agricultural ecology, aflatoxins biosynthesis, and evolution. The genome microarrays can be used to identify genes responsible for the different mycotoxins’ (typically aflatoxins) biosynthesis and fungal infections in living organisms [,]. Several studies [,] indicated that genome of A. flavus NRRL 3357 consists of eight chromosomes and has an estimated size of approximately 36.8 Mega base pairs (Mb) with about 12,197 predicted number of genes. Recently, a chromosome-level assembly of the A. flavus genome was completed []. In 2006, Bhatnagar et al. [] reported that genome of A. flavus is compressed with several duplicates of genes or less copied sequence. In fact, A. flavus consists of at least 56 secondary metabolic gene clusters containing about 12,000 predicted functional genes that could regulate the production of abundant secondary metabolites, including aflatoxins [].
Until now, about 30 genes have already been identified as being integral components of aflatoxin biosynthetic gene cluster in A. flavus, and all of them are clustered in a 70 kb DNA region on the chromosome 3, named the toxin-producing gene cluster [,]. All these genes contain the same regulatory trend, and their expression levels can directly affect the level of aflatoxin biosynthesis []. The aflatoxin pathway genes are mainly symbolized by a code of three-letter “afl” followed by a capital letter from “A” to “Y” characterizing each individual gene potentially involved or confirmed to be involved in the aflatoxin biosynthesis process. Therefore, different aflatoxin biosynthesis pathway genes were named starting from aflA to aflY. The aflatoxin biosynthesis pathway also involves sterigmatocystin (ST) synthesis genes, including aflA to aflQ, which start from the first conversion of fatty acid to the final product, and some other genes whose involvement in this pathway is still unclear to this date [,]. In particular, AFB1 has been found to be originated from a polyketide precursor, and it has been generally established to be synthesized following a path that was previously described by Caceres et al. []. However, it has been suggested that other aflatoxins, including AFB2, AFG2, and AFG1, could be metabolically linked by a direct interconversion method [,]. To this end, many enzymes do actively contribute to the conversion of the precursors at different stages involved in the aflatoxin biosynthesis pathway [].
Consistent with the aforementioned, aflatoxins biosynthesis requires an intricate regulatory mechanism that is composed of several specific regulatory genes in the pathway. In particular, both aflR and aflS have been reported to be major aflatoxin transcriptional regulator genes [,]. A number of studies conducted by our research group on the genes functions [,,,,] indicated that several other structure genes are equally related to the aflatoxin production in A. flavus. For example, after the deletion of an antioxidant catalase cta1, the expression levels of aflQ, aflC, and aflD were down-regulated, showing that cta1 plays an important role in aflatoxin biosynthesis in A. flavus []. The nucleoside diphosphate kinase (Aflndk) was also found to play an essential role in sclerotia production and conidia development in A. flavus []. Specifically, all Aflndk mutant strains displayed visible effects on sporulation, including the atrophy of spore heads and stalks, while the amount of conidia and sclerotia production decreased. In [], we found that the deletion of hexA gene significantly reduced the production of AFB1, conidia, and conidiophores, while the deletion of sakA significantly decreased the mycelia growth and increased both sclerotia and AFB1 production []. In [], we also found that the deletion of pex5 caused defects in sclerotial formation, sporulation, stress response, carbon metabolism, crop infection, and aflatoxin biosynthesis.
3. A. flavus Transcription Factors Involved in Morphogenesis and Aflatoxin Production
Transcription factors (TFs) are known as sequence-specific DNA-binding factors controlling transcription rate of genetic information from DNA to mRNA through binding to specific DNA sequences []. A recent study [] found that TFs are the key regulatory proteins in living organisms. In fungal species, TFs include either proteins that are encoded by cluster-associated (pathway-specific) regulatory genes or broad-domain regulatory proteins, which can affect the expression of several genes involving in biosynthetic procedures []. In this case, TFs are considered the final connection between the target genes and the signal flow that play important roles in the signal transduction pathways. On the basis of conserved DNA-binding domain, TFs are normally classified into different categories and structures [,]. Up to now, several studies have identified the roles of different TFs in A. flavus. Cary et al. [] reported the function of the Homeobox (hbx1) transcriptional factors in A. flavus. They found that disruption of hbx1 gene leads to loss of production of sclerotia, conidiophores, and conidia as well as aflatrem, cyclopiazonic acid, AFB1, and AFB2. They further observed significant down-regulation in the expression levels of three aflatoxins biosynthetic genes (aflM, aflD, aflC) and one regulatory gene (aflR) in A. flavus. A latter study [] confirmed that hbx1 gene can affect the expression of several genes in the aflatoxins biosynthesis pathway and the expression levels of transcription factor genes involved in A. flavus development, including conidiophores biogenesis genes (flbA, flbC, flbD, and flbE) and conidiation regulatory pathway genes (brlA and wetA). A research conducted by Hu et al. [] found the plant homeodomain (PHD) transcription factor Rum1 to be a regulator of morphogenesis and aflatoxin biosynthesis in A. flavus. Another study discovered that basic leucine zipper (bZIP) transcription factor AflRsmA could regulates sclerotium formation, oxidative stress response, and AFB1 biosynthesis in A. flavus []. It has also been revealed that the Skn7 transcription factor influences aflatoxin production, pathogenicity, morphological development, and stress response in A. flavus []. In addition, in A. flavus, APSES transcription factors AfStuA and AfRafA were shown to play important roles in pathogenicity, fungal development, and mycotoxin synthesis []. Cary et al. [] examined the function of transcription factors NsdD and NsdC on the development and aflatoxin production in A. flavus, and it was found that NsdD and NsdC affect the transcription of genes that are essential for sclerotial morphogenesis and aflatoxin biosynthesis in A. flavus. These TFs play important roles in morphogenesis and aflatoxin production in A. flavus.
4. Aflatoxins Production at the Transcriptome Level in A. flavus
The term transcriptome is derived from the two words transcript and genome, and it is linked with the process of transcript formation during transcription. Two biological techniques are generally used in the study of transcriptome, including RNA sequencing (RNA-seq) (i.e., sequence-based approach) and DNA microarray (i.e., a hybridization-based technique). However, compared to the outdated DNA microarray, RNA-seq is the most preferred and dominant transcriptomics method because it generally detects higher percentages of differentially expressed genes, especially the genes with low expression (i.e., can quantify expression across a larger dynamic range (>105 for RNA-Seq vs. 103 for DNA microarray)) [,,]. A list of the key advantages of RNA-Seq compared with DNA microarray method is provided in [], and interested readers are directed to this specific study for more information.
Consistent with the aforementioned, several studies were conducted to understand the effect of environmental conditions on the development of A. flavus and its aflatoxin production at the transcriptome level. In a study conducted by Zhang et al. [], the RNA-seq approach was used to delineate A. flavus transcriptome and specific data about differentially expressed genes under different water activities (0.99 and 0.93 aw). It was found that 0.93 aw significantly decreased conidiation and aflatoxins biosynthesis in A. flavus compared to 0.99 aw. Among the 33 genes identified in the transcriptome of A. flavus, the expression of 16 genes related to the production of aflatoxins were up-regulated more than twofold under 0.99 aw compared to 0.93 aw. In the same study, the expression of 11 genes related to the development of A. flavus was increased after 0.99 aw treatment, suggesting that A. flavus undergoes a substantial transcriptome response during aw variation.
In Yu et al. [], the authors applied the RNA-seq approach to profile the A. flavus transcriptome under various temperature conditions and identified a number of differentially expressed genes under aflatoxin conducive and non-conducive conditions. More specifically, some of these identified genes (e.g., aflT, aflC, aflNa, aflx, sugR, glcA, hxtA, nadA, etc.) were found to be involved in aflatoxin biosynthesis and adjacent sugar utilization, while others (e.g., maoA, dmaT, pks-nrps, etc.) were involved in cyclopiazonic acid transportation and aflatoxin biosynthesis. In a similar study, the RNA-seq method was also used to explore the transcriptome of A. flavus in response to the treatment with 5-Azacytidine (5-AC) []. 5-AC is a derivative of the nucleoside cytidine that is used in epigenetics as an inactivator of DNA methyltransferase. When A. flavus was treated with 5-AC, 240 genes were significantly expressed, and these included the genes that are involved in A. flavus development (e.g., veA, flbA, stuA, abaA, brlA, rodA, wetA, nsdD, rodB, etc.) and aflatoxin biosynthesis (e.g., aflQ, aflX, aflI, aflLa, aflG, etc.). They also include the genes that are commonly involved in proteolytic functions and catalytic activities (e.g., AFLA_000860, AFLA_004460, AFLA_008370, AFLA_011740, AFLA_121360, etc.). The latter regulate the modification of some essential proteins participating in the formation of aflatoxins in aflatoxisomes (i.e., vesicles that generate aflatoxins) []. A study conducted by Yao et al. [] examined the regulatory mechanism of aflatoxins using comparative transcriptomics in A. flavus and identified 18 novel (i.e., AFLA_084720, NosA, AFLA_019420, AFLA_081920, AFLA_108250, AFLA_108260, AFLA_131810, AFLA_019430, AFLA_138920, AFLA_132980, AFLA_039650, AFLA_108810, AFLA_065960, AFLA_013540, and four others genes whose identities were not provided) and 12 known (i.e, meaB, rtfA, atfB, gprP, pdeH, pdeL, laeB, ppoC, hamE, hamF, hamG, and hamH) regulatory genes that are essential for aflatoxin biosynthesis. In their study, the authors found that the deletion of an LaeA-like methyltransferase (Lael1) from A. flavus resulted in a significant increase in AFB1 production. They also found that the inactivation of both AfStuA and AfRafA in A. flavus significantly decreased the level of AFB1 production. Bai et al. [] showed 135 differentially expressed miRNA-like RNAs under different temperature and aw.
In summary, transcriptomics helps in the determination of the transcription of the genes and the quantification of changes in the expression levels of every transcript during the cell development and aflatoxin production under different environmental conditions. To this end, it is important to determine the transcriptome in molecular biology (i.e., it captures a snapshot in time of the total transcripts present in a cell) to guarantee an accurate determination of the cells’ molecular constituents and, subsequently, the interpretation of the functional elements of the genome in A. flavus.
5. Proteomics Analysis
Proteomic analysis (proteomics) is a powerful tool that helps in the identification of the entire complement of proteins (the proteome) and provides a systematic evaluation of protein expression in a biological organism under a particular condition []. A previous study also reported proteomic analysis as the most influential method to identify proteins in complex mixtures, and it is applied in the study of changes in protein expression in the organism under different environmental states []. Therefore, the development of quantitative, highly accurate, and sensitive proteomic techniques has helped in the discovery of novel proteins and the quantification of their expression. For example, by using isobaric tags for relative and absolute quantitation (iTRAQ) technique, multiple samples of proteins can be analyzed simultaneously [,].
Several studies have been conducted by our research group to investigate the responses of A. flavus proteome under different environmental conditions [,]. In Zhang et al. [], the authors used the iTRAQ technique to analyze the proteomic alterations in A. flavus under two different water activity (aw) levels (0.93 and 0.99 aw)). They identified a total of 3566 proteins, of which 837 proteins were differentially expressed in response to the aforementioned aw levels. They found that two of these proteins, namely AFL2G_04330 and KapK, play a critical role in the production of aflatoxin. They indicated that AFL2G_04330 protein plays an essential role in the activation of asexual sporulation and the production of aflatoxins in A. flavus, while KapK down-regulates aflatoxin biosynthesis. Another study by Bai et al. [] investigated changes in translation and relative protein levels in A. flavus in response to different temperature levels (28 °C and 37 °C). They used the iTRAQ method to identify and quantify the proteins. A total of 3886 proteins were identified, of which 2832 proteins were quantified. A further analysis revealed that 12 of these fully quantified proteins (namely aflE, aflW, aflC, aflD, aflO, aflP, aflK, aflM, aflY, aflJ, aflS, and aflH) were highly expressed at 28 °C, which resulted in an increase in the aflatoxins biosynthesis. In proteogenomic analysis, Yang et al. [] used several samples from eight different stress treatments and found that several novel genes were differently expressed in response to these stress conditions. They identified 732 novel protein-coding genes, and some of these conserved proteins (e.g., YdiU domain protein, Sac7, nmrA-like family protein, protein trm-112, Cds1, MpkC, etc.) were found to play important functions in regulating the pathogenicity, survival, growth, stress responses, and aflatoxin biosynthesis (e.g., autophagy-related protein 7, NADPH cytochrome P450, lipase/esterase family protein, CFEM protein domain, O-methylsterigmatocystin oxidoreductase, etc.) in A. flavus.
Consistent with the aforementioned, it is clear that proteome analysis could be an essential approach in delineating some of the unknown molecular mechanisms contributing to the development and aflatoxins biosynthesis in A. flavus. More importantly, the aforementioned iTRAQ technique can be used to reveal changes in gene-product expression during different biological processes, including the fungal development and the secondary metabolite biosynthesis. Indeed, accurate determination of protein expression would undoubtedly contribute to an improved understanding of processes related to the fungal development and secondary metabolite biosynthesis, which could subsequently help in devising control strategies for their reduction and/or elimination.
6. Metabolomics Analysis
In general, metabolomics refers to the scientific identification and quantification of chemical substances involving metabolites, the intermediates, small molecule substrates, and metabolism products of an organism. The term metabolome represents the total number of metabolites within an organism, and these are the final products of cellular processes. The A. flavus metabolome is of substantial interest since A. flavus is capable of producing the potent carcinogenic and toxic metabolites [,]. Apart from aflatoxins and their precursors, A. flavus produces other secondary metabolites, some of which may be toxic (e.g., cyclopiazonic acid, aflatrem, etc.) [], while others are important for some chemical processes (e.g., kojic acid) []. Although a detailed discussion about these additional secondary metabolites is beyond the thematic topic of the present paper, some important points are worth mentioning. For example, studies [,] suggest that cyclopiazonic acid (PCA) does lead to a variety of human diseases, including muscle necrosis, intestinal hemorrhage, and edema oral lesions, while aflatrem causes neurological disorders. In contrast, kojic acid is sometimes used in the food industry and in some health and cosmetic products as a natural preservative []. Additional information regarding these additional secondary metabolites as well as their effects and genes involving in their biosynthesis can be found in the literature [,,,], and interested readers are directed to these specific studies for more information.
In order to determine and/or quantify metabolites, metabolomics uses different complementary analytical methodologies, such as GC-MS, LC-MS/MS, and/or NMR []. In a recent study, Song et al. [] used GC-MS analysis to investigate the differences in metabolic changes between A. flavus A133 and NT strains treated with 5-AC. Among the 1181 identified volatile metabolites, numerous fatty acid-derived volatiles (e.g., Ergosterol, n-Hexadecanoic acid, Normeperidinic acid, 6-Octadecenoic acid, n-Propyl9-octadecenoate, 6,9-hexadecadienoic acid, Trimethylammonioacetate, etc.) were found to act as important intermediates for the biosynthesis of aflatoxins. The above authors also found that these fatty-acid-derived volatiles affect the balance between the production of conidia and the formation of sclerotia in A. flavus. To find out the function of the genes related to secondary metabolite biosynthesis, methods of over-expression or deletion of the target gene followed by metabolite profiling are generally used []. Recently, the effect of rtfA on the A. flavus metabolome was examined, whereby metabolomic analysis demonstrated that A. flavus rtfA could affect the production of numerous secondary metabolites, such as AFB1, leporins, aflatrem, aspirochlorine, aflavinines, and ditryptophenaline, further confirming the rtfA functions as a global positive regulator of secondary metabolites biosynthesis in the Aspergillus species []. Therefore, metabolomics has the potential to find different metabolites influencing biological processes in A. flavus.
7. Post Translation Modifications Influencing Development and Aflatoxin Biosynthesis in A. flavus
Several studies demonstrated the effects of post-translational modifications (PTMs) in different biological processes (e.g., aflatoxins biosynthesis, development, virulence, etc.) in A. flavus [,,]. In this section of the paper, a brief discussion on the influence of different PTMs, such as methylation acetylation, phosphorylation, sumoylation, and succinylation, on development and biosynthesis of aflatoxins in A. flavus is provided.
In a previous study, Lee et al. [] indicated that DNA methylation could play an essential function in the development and production of secondary metabolites in Aspergillus species. Gowher et al. [] found the levels of methylated DNA to be low in A. flavus. A later study carried out in our laboratory found that an arginine methyltransferase gene aflrmtA is involved in the biosynthesis of AFB1 and the morphogenesis and pathogenicity of A. flavus. Deletion of aflrmtA in A. flavus led to a decrease in the production of AFB1 and sclerotia but increased the production of conidia and conidiophores. The deletion of aflrmtA also decreased expression levels of the AFB1 biosynthesis regulatory gene aflR and AFB1 biosynthesis genes aflK and aflC []. In recent studies conducted by our research group [,,], we demonstrated that both the histone and DNA methyltransferases play a significant role in aflatoxin biosynthesis in A. flavus. In particular, the deletion of the dmtA gene increase sclerotial production but decrease both conidiation and aflatoxin biosynthesis []. In separate studies, we investigated the biological role of the histone methyltransferase Aflset1 [] and histone methyltransferase dot1 []. It was found that Aflset1 is involved in the regulation of virulence, fungal morphogenesis, and AFB1 biosynthesis in A. flavus for the former and the regulation of pathogenicity and aflatoxin attributes for the latter. In A. flavus, acetylation has been shown to involve in sclerotia formation, fungal development, pathogenicity, and aflatoxin biosynthesis []. Recently, protein lysine acetylation sites in the A. flavus proteome were identified by using a combination of both affinity enrichment and LC-MS/MS analysis method. A total of 1383 unique protein lysine acetylation sites in 652 acetylated proteins were found to be involved in different cellular processes, including secondary metabolite synthesis, cell growth, and gene expression []. Among the many different PTMs, lysine acetylation controlled by lysine deacetylases and acetyltransferases is a highly evolutionarily and dynamically conserved PTM occurring in eukaryotes and prokaryotes [,]. Genomic analysis revealed that genes encoding acetyltransferases are present in the A. flavus genome []. Lan et al. [] found that deletion of histone acetyltransferase AflgcnE reduced the growth of A. flavus and decreased the hydrophobicity of the cell’s surface. In their study, the authors demonstrated that the AflgcnE mutant strain does not produce aflatoxins and is unable to generate asexual sporulation and sclerotia. They also found that AflgcnE is required for maintain both cell wall integrity and genotoxic stress responses in A. flavus []. In a similar study, Lan et al. [] demonstrated that SinA (a transcriptional regulator) interacts with HosA histone deacetylase to regulate the development and production of AFB1 in A. flavus.
By using MS-based proteomics, Bai et al. [] identified a comprehensive phosphorylation database containing 62,272 non-redundant phosphorylation sites within a total number of 11,222 phosphoproteins from eight fungal species, including A. flavus. An additional research was conducted on a site-specific and global phosphoproteomic analysis of A. flavus, and 598 high-confidence phosphorylation sites were identified in a total of 283 phosphoproteins. These identified phosphoproteins have been found to be involved in different biological processes, including aflatoxins biosynthesis and signal transduction []. A study published by Yang et al. [] indicated that cdc14 is involved in the regulation of fungal development, stress response, pathogenicity, and aflatoxin biosynthesis in A. flavus. They found that cdc14 reduces biosynthesis of aflatoxins by suppressing the expression levels of aflR, aflS, aflD, aflC, aflK, and aflQ genes in the aflatoxin gene cluster. A high-affinity phosphodiesterase, pdeH, has also been shown to play a regulatory function in development and aflatoxin biosynthesis in A. flavus []. Another study found that the MAPK-related tyrosine phosphatases plays essential functions in the regulation of secondary metabolism as well as the development and pathogenicity in A. flavus [].
In a research conducted by Nie et al. [], sumoylation was found to be involved in both toxin attributes and virulence of A. flavus. They revealed that deletion of AfsumO gene leads to a significant decrease in AFB1 and AFB2 production and impairs the expression of the genes related to aflatoxin biosynthesis, including aflP, aflO, aflD, aflC, aflA, aflS, and aflR in A. flavus. Although sumoylation has been extensively studied in higher plants, vertebrates, and yeasts [,,], our literature survey revealed that there is a significantly high paucity of similar studies on A. flavus.
In their study, Ren et al. [] conducted a global analysis of lysine succinylome in A. flavus using high resolution mass spectrometry method, and they identified a total of 985 succinylation sites in 349 succinylated proteins. These succinylated proteins were found to be involved in diverse biological processes, including aflatoxins biosynthesis and pathogenicity. The above authors also found that lysine succinylation sites on the norsolorinic acid reductase NorA (AflE), which is an essential enzyme in aflatoxins biosynthesis pathway, affects the production of both sclerotia and AFB1 production in A. flavus.
In summary, these modifications are crucial mechanisms that help to identify the functions of different proteins involved in a number of biological processes in A. flavus. In fact, all these modifications can regulate the growth, development, and virulence of A. flavus. In this case, they play an important role in promoting epigenetics in fungal development and the production of secondary metabolites. They can also be used for clinical prevention and control of A. flavus. Apart from the above-discussed PTMs (i.e., succinylation, sumoylation, phosphorylation, acetylation, and methylation), there are currently many other modifications documented in the literature, though to a limited extent. These include but are not limited to farnesylation, glycosylation, ubiquitination, palmitoylation, neddylation, and proteolysis [,,]. All the studies reviewed in this section demonstrate that by studying the functions of these PTMs, one can then identify their effects on the development and aflatoxins biosynthesis in A. flavus.
8. Signal Pathways Involved in Morphogenesis and Secondary Metabolism in A. flavus
8.1. HOG Pathway
The high-osmolarity glycerol (HOG) signaling pathway is one of the mitogen activated protein kinases (MAPKs) pathways in eukaryotic cells. This pathway contains a protein kinase known as HogA, which contributes to the regulation of osmotic stress, and it is also able to recognize all steps that involves in the osmoadaptation process [,]. Recently, several studies were conducted by our group on the functions of the genes that are involved in HOG pathway, including Aflste20, pbsB, and sakA/hogA [,,]. It was found that deletion of Aflste20 gene resulted in complete loss of sclerotia, reduced growth, decreased AFB1 production, as well as decreased expression of key structural genes aflD, aflQ, and aflK and regulatory genes aflS and aflR in the aflatoxin biosynthesis cluster in A. flavus []. In a similar study, it was shown that inactivation of pbsB significantly decreased the production of AFB1, conidiation, mycelia growth, and sclerotia production in A. flavus. The expression levels of aflR, aflQ, aflK, aflD, and aflC were also significantly down-regulated in ΔpbsB mutant, showing that pbsB gene increases AFB1 biosynthesis by up-regulating the expression levels of structural and regulatory genes in the aflatoxin gene cluster []. The membrane mucin Msb2 was found to be involved in the regulation of A. flavus morphological development, pathogenicity, secondary metabolism, and stresses adaptation []. A study on the functions of sakA/hogA also revealed that this gene regulates the morphology development, pathogenicity, stress response, and reduces biosynthesis of aflatoxins by suppressing the expression levels of genes in the aflatoxin gene cluster in A. flavus []. All these findings highlight the importance of HOG pathway in the physiological processes in A. flavus. A speculated HOG signaling pathway in A. flavus is shown in Figure 1.
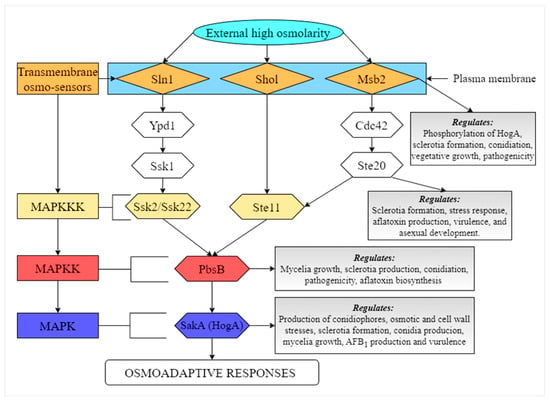
Figure 1.
Illustration of the HOG-MAPK signaling pathway in A. flavus. PbsB integrates signals from two major independent upstream osmosensing mechanisms, which leads to the activation of specific MAPKKKs. Under osmostress, activated PbsB activates the SakA (HogA) MAPK, which induces a set of osmoadaptive responses.
8.2. Slt2 Pathway
A recent study by Zhang et al. [] demonstrated that the MAP kinase Slt2 modulates the fungal development and pathogenicity as well as the aflatoxin biosynthesis and stress response processes in A. flavus. These findings reaffirm the importance of Slt2-MAPK pathway in different physiological processes in A. flavus. Another study [] was been conducted on the functions of MAPKKK Bck1, which is the upstream element of Slt2-MAPK pathway, and it was found that deletion of Bck1 showed more sensitivity to cell wall stress, decreased pathogenicity on peanut seeds, and also caused a significant defect in growth and development in A. flavus. It was also observed that the lack of Bck1 significantly increased the production of AFB1, which indicates that Bck1 decreases AFB1 biosynthesis by down-regulating the expression levels of genes in the aflatoxin gene cluster in A. flavus. A speculated Slt2-MAPK signaling pathway in A. flavus is shown in Figure 2.
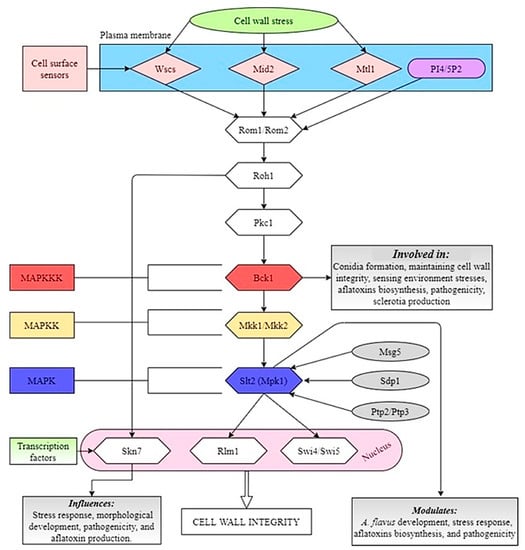
Figure 2.
Illustration of the Slt2-MAPK signaling pathway in A. flavus. Signals are initiated at the plasma membrane (PM) through the cell-surface sensors Wscs, Mid2, and Mtl1. The extracellular domains of these proteins are highly O-mannosylated. Together with PI4, 5P2, which recruits the Rom1/Rom2 guanine nucleotide exchange factors (GEFs) to the plasma membrane, the sensors stimulate nucleotide exchange on Rho1. Two transcription factors, Rlm1 and SBF (Swi4/Swi6), are activated by the pathway. Skn7 may also contribute to the cell wall integrity transcriptional program.
8.3. Other Pathways
Apart from the previously discussed pathways, there are some other signal pathways that have been identified to play important roles in diverse biological processes in eukaryotes, including filamentous growth (Kss1), ascospore formation, mating pheromone (Fus3), and cAMP/PKA pathways [,,]. In A. flavus, additional pathways were been identified and documented in the literature. To this end, different functions of several important components of cAMP/PKA pathway, including GpaB, PdeH, PdeL, AcyA, and Cap, were recently identified by our research group. Liu et al. [] found that G protein alpha, which is a subunit GpaB, is required for fungal development, pathogenicity, and aflatoxin biosynthesis in A. flavus. Yang et al. [] indicated that AcyA regulates the fungal virulence and development as well as the aflatoxin biosynthesis in A. flavus. Additionally, cAMP phosphodiesterases, PdeH and PdeL, have also been reported as being negative regulators of the synthesis of cAMP and the production of AFB1 in A. flavus []. A recent study by Yang et al. [] also indicated that Cap containing multi-domain is engaged in fungal virulence and mycotoxins biosynthesis in A. flavus.
9. Conclusions
In the last decade, significant progress has been made in discovering the different molecules, factors, genes, and enzymes involved in each step of fungal development and aflatoxin biosynthesis in A. flavus. Interestingly, studies are constantly revealing new functions and molecular mechanisms of genes involved in the development of A. flavus, aflatoxin production, and pathogenesis. A full understanding of all these molecular mechanisms and their effects on the different genes encoding proteins in A. flavus is expected. Although there are many genes and molecular mechanisms regulating the fungal development and aflatoxins production in A. flavus, this study only considers the recently published studies (i.e., mainly in the past 10 years) focusing on the functions of the genes/proteins and molecular mechanisms contributing to A. flavus development and aflatoxins biosynthesis. In our future work, we intend to investigate the regulation mechanisms of different pathways on the morphogenesis and the secondary metabolites as well as the crosstalk between these signaling pathways. Additionally, advanced research on omics is highly warranted, as it could bring novel insights into fungal development and aflatoxins biosynthesis in A. flavus. For example, little information is currently known about the function of microRNAs in A. flavus. These small, non-coding RNA are particularly involved in gene silencing in numerous eukaryotic cells, which could play an essential function in the synthesis of fungal secondary metabolites in A. flavus. It is our hope that the information provided in this review may help our readers improve their understanding of gene function, signal transduction, genetic regulation, and other underlying molecular mechanisms in A. flavus, which may help in finding novel therapeutic strategies to control this common pathogen.
Author Contributions
Conceptualization, S.W.; writing—original draft preparation, E.T. and S.W.; writing—review and editing, E.T., R.X. and S.W.; graphic design E.T.; funding acquisition, S.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Natural Science Foundation of China (Grant No. 31972214, 31772105), and Natural Science Foundation of Fujian Province of China (No. 2018J07002).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors acknowledge Walter Nsengiyumva for his input in the proofreading of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Klich, M.A. Aspergillus flavus: The major producer of aflatoxin. Mol. Plant. Pathol. 2007, 8, 713–722. [Google Scholar] [CrossRef]
- Tumukunde, E.; Ma, G.; Li, D.; Yuan, J.; Qin, L.; Wang, S. Current research and prevention of aflatoxins in China. World Mycotoxin J. 2020, 13, 121–138. [Google Scholar] [CrossRef]
- Ahmad, S.; Wang, S.; Wu, W.; Yang, K.; Zhang, Y.; Tumukunde, E.; Wang, S.; Wang, Y. Functional Analysis of Peptidyl-prolyl cis-trans Isomerase from Aspergillus flavus. Int. J. Mol. Sci. 2019, 20, 2206. [Google Scholar] [CrossRef] [Green Version]
- Richard, J.L. Discovery of aflatoxins and significant historical features. Toxin Rev. 2008, 27, 171–201. [Google Scholar] [CrossRef]
- Baker, S.E.; Bennett, J.W. An overview of the genus Aspergillus. Aspergilli: Genom. Med. Asp. Biotechnol. Res. Methods 2007, 2, 3–13. [Google Scholar]
- Heidtmann-Bemvenuti, R.; Mendes, G.; Scaglioni, P.; Badiale-Furlong, E.; Souza-Soares, L. Biochemistry and metabolism of mycotoxins: A review. Afr. J. Food Sci. 2011, 5, 861–869. [Google Scholar] [CrossRef] [Green Version]
- Moore, G.G.; Elliott, J.L.; Singh, R.; Horn, B.W.; Dorner, J.W.; Stone, E.A.; Chulze, S.N.; Barros, G.G.; Naik, M.K.; Wright, G.C. Sexuality generates diversity in the aflatoxin gene cluster: Evidence on a global scale. PLoS Pathog. 2013, 9, e1003574. [Google Scholar] [CrossRef] [Green Version]
- Busby, W., Jr.; Wogan, G. Ochratoxins. In Mycotoxins and N-Nitroso Compounds: Environmental Risks; Shank, R.C., Ed.; CRC Press: Boca Raton, FL, USA, 1981. [Google Scholar]
- Cleveland, T.E.; Yu, J.; Bhatnagar, D.; Chen, Z.Y.; Brown, R.L.; Chang, P.K.; Cary, J.W. Progress in elucidating the molecular basis of the host plant—Aspergillus flavus interaction, a basis for devising strategies to reduce aflatoxin contamination in crops. J. Toxicol. Toxin Rev. 2004, 23, 345–380. [Google Scholar] [CrossRef]
- Bhatnagar, D.; Ehrlich, K.; Cleveland, T. Molecular genetic analysis and regulation of aflatoxin biosynthesis. Appl. Microbiol. Biotechnol. 2003, 61, 83–93. [Google Scholar] [CrossRef]
- Yabe, K.; Nakajima, H. Enzyme reactions and genes in aflatoxin biosynthesis. Appl. Microbiol. Biotechnol. 2004, 64, 745–755. [Google Scholar] [CrossRef]
- Yu, J.; Chang, P.-K.; Ehrlich, K.C.; Cary, J.W.; Bhatnagar, D.; Cleveland, T.E.; Payne, G.A.; Linz, J.E.; Woloshuk, C.P.; Bennett, J.W. Clustered pathway genes in aflatoxin biosynthesis. Appl. Environ. Microbiol. 2004, 70, 1253–1262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tumukunde, E.; Li, D.; Qin, L.; Li, Y.; Shen, J.; Wang, S.; Yuan, J. Osmotic-Adaptation Response of sakA/hogA Gene to Aflatoxin Biosynthesis, Morphology Development and Pathogenicity in Aspergillus flavus. Toxins 2019, 11, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibbons, J.G.; Rokas, A. The function and evolution of the Aspergillus genome. Trends Microbiol. 2013, 21, 14–22. [Google Scholar] [CrossRef] [Green Version]
- Schmidt-Heydt, M.; Abdel-Hadi, A.; Magan, N.; Geisen, R. Complex regulation of the aflatoxin biosynthesis gene cluster of Aspergillus flavus in relation to various combinations of water activity and temperature. Int. J. Food Microbiol. 2009, 135, 231–237. [Google Scholar] [CrossRef] [Green Version]
- Nierman, W.C.; Yu, J.; Fedorova-Abrams, N.D.; Losada, L.; Cleveland, T.E.; Bhatnagar, D.; Bennett, J.W.; Dean, R.; Payne, G.A. Genome sequence of Aspergillus flavus NRRL 3357, a strain that causes aflatoxin contamination of food and feed. Genome Announc. 2015, 3, e00168-15. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Nierman, W.C.; Fedorova, N.D.; Bhatnagar, D.; Cleveland, T.E.; Bennett, J.W. What can the Aspergillus flavus genome offer to mycotoxin research? Mycology 2011, 2, 218–236. [Google Scholar]
- Cleveland, T.E.; Yu, J.; Fedorova, N.; Bhatnagar, D.; Payne, G.A.; Nierman, W.C.; Bennett, J.W. Potential of Aspergillus flavus genomics for applications in biotechnology. Trends Biotechnol. 2009, 27, 151–157. [Google Scholar] [CrossRef]
- Rudramurthy, S.M.; Paul, R.A.; Chakrabarti, A.; Mouton, J.W.; Meis, J.F. Invasive aspergillosis by Aspergillus flavus: Epidemiology, diagnosis, antifungal resistance, and management. J. Fungi 2019, 5, 55. [Google Scholar] [CrossRef] [Green Version]
- Han, X.; Chakrabortti, A.; Zhu, J.; Liang, Z.-X.; Li, J. Sequencing and functional annotation of the whole genome of the filamentous fungus Aspergillus westerdijkiae. BMC Genom. 2016, 17, 633–647. [Google Scholar] [CrossRef] [Green Version]
- Drott, M.T.; Satterlee, T.R.; Skerker, J.M.; Pfannenstiel, B.T.; Glass, N.L.; Keller, N.P.; Milgroom, M.G. The Frequency of Sex: Population Genomics Reveals Differences in Recombination and Population Structure of the Aflatoxin-Producing Fungus Aspergillus flavus. mBio 2020, 11, e00963-20. [Google Scholar] [CrossRef]
- Bhatnagar, D.; Cary, J.W.; Ehrlich, K.; Yu, J.; Cleveland, T.E. Understanding the genetics of regulation of aflatoxin production and Aspergillus flavus development. Mycopathologia 2006, 162, 155. [Google Scholar] [CrossRef] [PubMed]
- Amaike, S.; Keller, N.P. Aspergillus flavus. Annu. Rev. Phytopathol. 2011, 49, 107–133. [Google Scholar] [CrossRef] [PubMed]
- Yu, J. Current understanding on aflatoxin biosynthesis and future perspective in reducing aflatoxin contamination. Toxins 2012, 4, 1024–1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caceres, I.; Khoury, A.A.; Khoury, R.E.; Lorber, S.; Oswald, I.P.; Khoury, A.E.; Atoui, A.; Puel, O.; Bailly, J.-D. Aflatoxin Biosynthesis and Genetic Regulation: A Review. Toxins 2020, 12, 150. [Google Scholar] [CrossRef] [Green Version]
- OBrian, G.; Georgianna, D.; Wilkinson, J.; Yu, J.; Abbas, H.; Bhatnagar, D.; Cleveland, T.; Nierman, W.; Payne, G. The effect of elevated temperature on gene transcription and aflatoxin biosynthesis. Mycologia 2007, 99, 232–239. [Google Scholar] [CrossRef]
- Zhu, Z.; Yang, M.; Bai, Y.; Ge, F.; Wang, S. Antioxidant-related catalase CTA1 regulates development, aflatoxin biosynthesis, and virulence in pathogenic fungus Aspergillus flavus. Environ. Microbiol. 2020, 22, 2792–2810. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, S.; Nie, X.; Yang, K.; Xu, P.; Wang, X.; Liu, M.; Yang, Y.; Chen, Z.; Wang, S. Molecular and structural basis of nucleoside diphosphate kinase–mediated regulation of spore and sclerotia development in the fungus Aspergillus flavus. J. Biol. Chem. 2019, 294, 12415–12431. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Li, D.; Qin, L.; Shen, J.; Guo, X.; Tumukunde, E.; Li, M.; Wang, S. HexA is required for growth, aflatoxin biosynthesis and virulence in Aspergillus flavus. BMC Mol. Biol. 2019, 20, 4. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Geng, L.; Huang, L.; Deng, J.; Fasoyin, O.E.; Yao, G.; Wang, S. Contribution of peroxisomal protein importer AflPex5 to development and pathogenesis in the fungus Aspergillus flavus. Curr. Genet. 2018, 64, 1335–1348. [Google Scholar] [CrossRef]
- Yang, M.; Zhu, Z.; Zhuang, Z.; Bai, Y.; Wang, S.; Ge, F. Proteogenomic characterization of the pathogenic fungus Aspergillus flavus reveals novel genes involved in aflatoxin production. Mol. Cell. Proteom. 2021, 20, 100013. [Google Scholar] [CrossRef]
- Latchman, D.S. Transcription factors: An overview. Int. J. Biochem. Cell Biol. 1997, 29, 1305–1312. [Google Scholar] [CrossRef] [Green Version]
- García-Estrada, C.; Domínguez-Santos, R.; Kosalková, K.; Martín, J.-F. Transcription factors controlling primary and secondary metabolism in filamentous fungi: The β-Lactam paradigm. Fermentation 2018, 4, 47. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zha, W.; Liang, L.; Fasoyin, O.E.; Wu, L.; Wang, S. The bZIP Transcription Factor AflRsmA Regulates Aflatoxin B1 Biosynthesis, Oxidative Stress Response and Sclerotium Formation in Aspergillus flavus. Toxins 2020, 12, 271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cary, J.W.; Harris-Coward, P.; Scharfenstein, L.; Mack, B.M.; Chang, P.-K.; Wei, Q.; Lebar, M.; Carter-Wientjes, C.; Majumdar, R.; Mitra, C. The Aspergillus flavus homeobox gene, hbx1, is required for development and aflatoxin production. Toxins 2017, 9, 315. [Google Scholar] [CrossRef] [Green Version]
- Cary, J.W.; Entwistle, S.; Satterlee, T.; Mack, B.M.; Gilbert, M.K.; Chang, P.K.; Scharfenstein, L.; Yin, Y.; Calvo, A.M. The transcriptional regulator Hbx1 affects the expression of thousands of genes in the aflatoxin-producing fungus Aspergillus flavus. G3 Genes Genomes Genet. 2019, 9, 167–178. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Yang, G.; Zhang, D.; Liu, Y.; Li, Y.; Lin, G.; Guo, Z.; Wang, S.; Zhuang, Z. The PHD Transcription Factor Rum1 Regulates Morphogenesis and Aflatoxin Biosynthesis in Aspergillus flavus. Toxins 2018, 10, 301. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Xu, G.; Geng, L.; Lu, X.; Yang, K.; Yuan, J.; Nie, X.; Zhuang, Z.; Wang, S. The stress response regulator AflSkn7 influences morphological development, stress response, and pathogenicity in the fungus Aspergillus flavus. Toxins 2016, 8, 202. [Google Scholar] [CrossRef] [Green Version]
- Yao, G.; Zhang, F.; Nie, X.; Wang, X.; Yuan, J.; Zhuang, Z.; Wang, S. Essential APSES transcription factors for mycotoxin synthesis, fungal development, and pathogenicity in Aspergillus flavus. Front. Microbiol. 2017, 8, 2277–2292. [Google Scholar] [CrossRef]
- Cary, J.W.; Harris-Coward, P.Y.; Ehrlich, K.C.; Mack, B.M.; Kale, S.P.; Larey, C.; Calvo, A.M. NsdC and NsdD affect Aspergillus flavus morphogenesis and aflatoxin production. Eukaryot. Cell 2012, 11, 1104–1111. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Zhao, S.; Fung-Leung, W.-P.; Bittner, A.; Ngo, K.; Liu, X. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS ONE 2014, 9, e78644. [Google Scholar] [CrossRef]
- Wilhelm, B.T.; Landry, J.-R. RNA-Seq—Quantitative measurement of expression through massively parallel RNA-sequencing. Methods 2009, 48, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Guo, Z.; Zhong, H.; Wang, S.; Yang, W.; Liu, Y.; Wang, S. RNA-Seq-based transcriptome analysis of aflatoxigenic Aspergillus flavus in response to water activity. Toxins 2014, 6, 3187–3207. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Fedorova, N.D.; Montalbano, B.G.; Bhatnagar, D.; Cleveland, T.E.; Bennett, J.W.; Nierman, W.C. Tight control of mycotoxin biosynthesis gene expression in Aspergillus flavus by temperature as revealed by RNA-Seq. FEMS Microbiol. Lett. 2011, 322, 145–149. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.-Q.; Zhao, X.-X.; Zhi, Q.-Q.; Zhao, M.; He, Z.-M. Transcriptomic profiling of Aspergillus flavus in response to 5-azacytidine. Fungal Genet. Biol. 2013, 56, 78–86. [Google Scholar] [CrossRef]
- Yao, G.; Yue, Y.; Fu, Y.; Fang, Z.; Xu, Z.; Ma, G.; Wang, S. Exploration of the regulatory mechanism of secondary metabolism by comparative transcriptomics in Aspergillus flavus. Front. Microbiol. 2018, 9, 1568–1583. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Lan, F.; Yang, W.; Zhang, F.; Yang, K.; Li, Z.; Gao, P.; Wang, S. sRNA profiling in Aspergillus flavus reveals differentially expressed miRNA-like RNAs response to water activity and temperature. Fungal Genet. Biol. 2015, 81, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Szopinska, A.; Degand, H.; Hochstenbach, J.-F.; Nader, J.; Morsomme, P. Rapid response of the yeast plasma membrane proteome to salt stress. Mol. Cell. Proteom. 2011, 10, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medina, M.L. Proteomic Analysis of Differentially Expressed Secreted Proteins from Aspergillus flavus; Department of Chemistry and Biochemistry, Arizona State University: Tempe, AZ, USA; ProQuest Dissertations Publishing: Ann Arbor, MI, USA, 2004. [Google Scholar]
- Zieske, L.R. A perspective on the use of iTRAQ™ reagent technology for protein complex and profiling studies. J. Exp. Bot. 2006, 57, 1501–1508. [Google Scholar] [CrossRef]
- Wang, Y.-H.; Que, F.; Wang, G.-L.; Hao, J.-N.; Li, T.; Xu, Z.-S.; Xiong, A.-S. iTRAQ-based quantitative proteomics and transcriptomics provide insights into the importance of expansins during root development in carrot. Front. Genet. 2019, 10, 247. [Google Scholar] [CrossRef]
- Zhang, F.; Zhong, H.; Han, X.; Guo, Z.; Yang, W.; Liu, Y.; Yang, K.; Zhuang, Z.; Wang, S. Proteomic profile of Aspergillus flavus in response to water activity. Fungal Biol. 2015, 119, 114–124. [Google Scholar] [CrossRef]
- Bai, Y.; Wang, S.; Zhong, H.; Yang, Q.; Zhang, F.; Zhuang, Z.; Yuan, J.; Nie, X.; Wang, S. Integrative analyses reveal transcriptome-proteome correlation in biological pathways and secondary metabolism clusters in A. flavus in response to temperature. Sci. Rep. 2015, 5, 14582–14595. [Google Scholar] [CrossRef] [PubMed]
- Qiu, S.; Zeng, B. Advances in Understanding the Acyl-CoA-Binding Protein in Plants, Mammals, Yeast, and Filamentous Fungi. J. Fungi 2020, 6, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jun, S.-C.; Kim, J.-H.; Han, K.-H. The Conserved MAP Kinase MpkB Regulates Development and Sporulation without Affecting Aflatoxin Biosynthesis in Aspergillus flavus. J. Fungi 2020, 6, 289. [Google Scholar] [CrossRef] [PubMed]
- Han, Z. Aspergillus and Fusarium Toxins: Analysis, Metabolic Profiling, In Vivo Kinetics and Metabolism, and Risk Assessment. Ph.D. Thesis, Ghent University, Ghent, Belgium, 2016. [Google Scholar]
- Terabayashi, Y.; Sano, M.; Yamane, N.; Marui, J.; Tamano, K.; Sagara, J.; Dohmoto, M.; Oda, K.; Ohshima, E.; Tachibana, K. Identification and characterization of genes responsible for biosynthesis of kojic acid, an industrially important compound from Aspergillus oryzae. Fungal Genet. Biol. 2010, 47, 953–961. [Google Scholar] [CrossRef] [PubMed]
- Sheikh-Ali, S.I.; Ahmad, A.; Mohd-Setapar, S.-H.; Zakaria, Z.A.; Abdul-Talib, N.; Khamis, A.K.; Hoque, M.E. The potential hazards of Aspergillus sp. in foods and feeds, and the role of biological treatment: A review. J. Microbiol. 2014, 52, 807–818. [Google Scholar] [CrossRef]
- Cary, J.W.; Gilbert, M.K.; Lebar, M.D.; Majumdar, R.; Calvo, A.M. Aspergillus flavus secondary metabolites: More than just aflatoxins. Food Safety 2018, 6, 7–32. [Google Scholar] [CrossRef] [Green Version]
- Rank, C.; Klejnstrup, M.L.; Petersen, L.M.; Kildgaard, S.; Frisvad, J.C.; Held Gotfredsen, C.; Ostenfeld Larsen, T. Comparative chemistry of Aspergillus oryzae (RIB40) and A. flavus (NRRL 3357). Metabolites 2012, 2, 39–56. [Google Scholar] [CrossRef]
- Duran, R.M.; Cary, J.W.; Calvo, A.M. Production of cyclopiazonic acid, aflatrem, and aflatoxin by Aspergillus flavus is regulated by veA, a gene necessary for sclerotial formation. Appl. Microbiol. Biotechnol. 2007, 73, 1158. [Google Scholar] [CrossRef] [PubMed]
- Roessner, U.; Bowne, J. What is metabolomics all about? Biotechniques 2009, 46, 363–365. [Google Scholar] [CrossRef]
- Song, F.; Geng, Q.; Wang, X.; Gao, X.; He, X.; Zhao, W.; Lan, H.; Tian, J.; Yang, K.; Wang, S. Gas Chromatography–Mass Spectrometry Profiling of Volatile Compounds Reveals Metabolic Changes in a Non-Aflatoxigenic Aspergillus flavus Induced by 5-Azacytidine. Toxins 2020, 12, 57. [Google Scholar] [CrossRef] [Green Version]
- Chang, P.-K.; Scharfenstein, L.L.; Wei, Q.; Bhatnagar, D. Development and refinement of a high-efficiency gene-targeting system for Aspergillus flavus. J. Microbiol. Methods 2010, 81, 240–246. [Google Scholar] [CrossRef]
- Lohmar, J.M.; Puel, O.; Cary, J.W.; Calvo, A.M. The Aspergillus flavus rtfA gene regulates plant and animal pathogenesis and secondary metabolism. Appl. Environ. Microbiol. 2019, 85, e02446-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nie, X.; Yu, S.; Qiu, M.; Wang, X.; Wang, Y.; Bai, Y.; Zhang, F.; Wang, S. Aspergillus flavus SUMO contributes to fungal virulence and toxin attributes. J. Agric. Food Chem. 2016, 64, 6772–6782. [Google Scholar] [CrossRef] [PubMed]
- Lan, H.; Sun, R.; Fan, K.; Yang, K.; Zhang, F.; Nie, X.Y.; Wang, X.; Zhuang, Z.; Wang, S. The Aspergillus flavus histone acetyltransferase AflGcnE regulates morphogenesis, aflatoxin biosynthesis, and pathogenicity. Front. Microbiol. 2016, 7, 1324–1338. [Google Scholar] [CrossRef] [Green Version]
- Ren, S.; Yang, M.; Yue, Y.; Ge, F.; Li, Y.; Guo, X.; Zhang, J.; Zhang, F.; Nie, X.; Wang, S. Lysine succinylation contributes to aflatoxin production and pathogenicity in Aspergillus flavus. Mol. Cell. Proteom. 2018, 17, 457–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, D.W.; Freitag, M.; Selker, E.U.; Aramayo, R. A cytosine methyltransferase homologue is essential for sexual development in Aspergillus nidulans. PLoS ONE 2008, 3, e2531. [Google Scholar] [CrossRef] [Green Version]
- Gowher, H.; Ehrlich, K.C.; Jeltsch, A. DNA from Aspergillus flavus contains 5-methylcytosine. FEMS Microbiol. Lett. 2001, 205, 151–155. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; He, Y.; Li, X.; Fasoyin, O.E.; Hu, Y.; Liu, Y.; Yuan, J.; Zhuang, Z.; Wang, S. Histone Methyltransferase aflrmtA gene is involved in the morphogenesis, mycotoxin biosynthesis, and pathogenicity of Aspergillus flavus. Toxicon 2017, 127, 112–121. [Google Scholar] [CrossRef]
- Yang, K.; Liang, L.; Ran, F.; Liu, Y.; Li, Z.; Lan, H.; Gao, P.; Zhuang, Z.; Zhang, F.; Nie, X. The DmtA methyltransferase contributes to Aspergillus flavus conidiation, sclerotial production, aflatoxin biosynthesis and virulence. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Zhang, M.; Xie, R.; Zhang, F.; Wang, S.; Pan, X.; Wang, S.; Zhuang, Z. The Methyltransferase AflSet1 Is Involved in Fungal Morphogenesis, AFB1 Biosynthesis, and Virulence of Aspergillus flavus. Front. Microbiol. 2020, 11, 234–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, L.; Liu, Y.; Yang, K.; Lin, G.; Xu, Z.; Lan, H.; Wang, X.; Wang, S. The putative histone methyltransferase DOT1 regulates aflatoxin and pathogenicity attributes in Aspergillus flavus. Toxins 2017, 9, 232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, G.; Yue, Y.; Ren, S.; Yang, M.; Zhang, Y.; Cao, X.; Wang, Y.; Zhang, J.; Ge, F.; Wang, S. Lysine acetylation contributes to development, aflatoxin biosynthesis and pathogenicity in Aspergillus flavus. Environ. Microbiol. 2019, 21, 4792–4807. [Google Scholar] [CrossRef]
- Lv, Y. Proteome-wide profiling of protein lysine acetylation in Aspergillus flavus. PLoS ONE 2017, 12, e0178603. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.-W.; Yang, X.-J. Comprehensive lysine acetylomes emerging from bacteria to humans. Trends Biochem. Sci. 2011, 36, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.; O’Connor, C.D. Protein acetylation in prokaryotes. Proteomics 2011, 11, 3012–3022. [Google Scholar] [CrossRef] [PubMed]
- Lan, H.; Wu, L.; Sun, R.; Keller, N.P.; Yang, K.; Ye, L.; He, S.; Zhang, F.; Wang, S. The HosA histone deacetylase regulates aflatoxin biosynthesis through direct regulation of aflatoxin cluster genes. Mol. Plant. Microbe Interact. 2019, 32, 1210–1228. [Google Scholar] [CrossRef]
- Bai, Y.; Chen, B.; Li, M.; Zhou, Y.; Ren, S.; Xu, Q.; Chen, M.; Wang, S. FPD: A comprehensive phosphorylation database in fungi. Fungal Biol. 2017, 121, 869–875. [Google Scholar] [CrossRef]
- Ren, S.; Yang, M.; Li, Y.; Zhang, F.; Chen, Z.; Zhang, J.; Yang, G.; Yue, Y.; Li, S.; Ge, F. Global phosphoproteomic analysis reveals the involvement of phosphorylation in aflatoxins biosynthesis in the pathogenic fungus Aspergillus flavus. Sci. Rep. 2016, 6, 34078–34092. [Google Scholar] [CrossRef]
- Yang, G.; Hu, Y.; Wang, S.; Fasoyin, O.E.; Yue, Y.; Qiu, Y.; Wang, X. The Aspergillus flavus Phosphatase CDC14 Regulates Development, Aflatoxin Biosynthesis and Pathogenicity. Front. Cell. Infect. Microbiol. 2018, 8, 141–156. [Google Scholar] [CrossRef]
- Yang, K.; Liu, Y.; Liang, L.; Li, Z.; Qin, Q.; Nie, X.; Wang, S. The high-affinity phosphodiesterase PdeH regulates development and aflatoxin biosynthesis in Aspergillus flavus. Fungal Genet. Biol. 2017, 101, 7–19. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Cao, X.; Ma, G.; Qin, L.; Wu, Y.; Lin, J.; Ye, P.; Yuan, J.; Wang, S. MAPK pathway-related tyrosine phosphatases regulate development, secondary metabolism and pathogenicity in fungus Aspergillus flavus. Environ. Microbiol. 2020, 22, 5232–5247. [Google Scholar] [CrossRef] [PubMed]
- Harting, R.; Bayram, Ö.; Laubinger, K.; Valerius, O.; Braus, G.H. Interplay of the fungal sumoylation network for control of multicellular development. Mol. Microbiol. 2013, 90, 1125–1145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szewczyk, E.; Chiang, Y.-M.; Oakley, C.E.; Davidson, A.D.; Wang, C.C.; Oakley, B.R. Identification and characterization of the asperthecin gene cluster of Aspergillus nidulans. Appl. Environ. Microbiol. 2008, 74, 7607–7612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdallah, Q.A.; Fortwendel, J.R. Exploration of Aspergillus fumigatus Ras pathways for novel antifungal drug targets. Front. Microbiol. 2015, 6, 128–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, C. Protein glycosylation in Aspergillus fumigatus is essential for cell wall synthesis and serves as a promising model of multicellular eukaryotic development. Int. J. Microbiol. 2012, 12, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Nie, X.; Li, B.; Wang, S. Epigenetic and posttranslational modifications in regulating the biology of Aspergillus species. Adv. Appl. Microbiol. 2018, 105, 191–226. [Google Scholar] [PubMed]
- Eliahu, N.; Igbaria, A.; Rose, M.S.; Horwitz, B.A.; Lev, S. Melanin biosynthesis in the maize pathogen Cochliobolus heterostrophus depends on two mitogen-activated protein kinases, Chk1 and Mps1, and the transcription factor Cmr1. Eukaryot. Cell 2007, 6, 421–429. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Qin, L.; Wang, Y.; Xie, Q.; Li, N.; Wang, S.; Yuan, J. AflSte20 Regulates Morphogenesis, Stress Response, and Aflatoxin Biosynthesis of Aspergillus flavus. Toxins 2019, 11, 730. [Google Scholar] [CrossRef] [Green Version]
- Yuan, J.; Chen, Z.; Guo, Z.; Li, D.; Zhang, F.; Shen, J.; Zhang, Y.; Wang, S.; Zhuang, Z. PbsB regulates morphogenesis, Aflatoxin B1 biosynthesis and pathogenicity of Aspergillus flavus. Front. Cell. Infect. Microbiol. 2018, 8, 162–172. [Google Scholar] [CrossRef] [Green Version]
- Qin, L.; Li, D.; Zhao, J.; Yang, G.; Wang, Y.; Yang, K.; Tumukunde, E.; Wang, S.; Yuan, J. The membrane mucin Msb2 regulates aflatoxin biosynthesis and pathogenicity in fungus Aspergillus flavus. Microb. Biotechnol. 2021, 14, 628–642. [Google Scholar] [CrossRef]
- Zhang, F.; Geng, L.; Deng, J.; Huang, L.; Zhong, H.; Xin, S.; Fasoyin, O.E.; Wang, S. The MAP kinase AflSlt2 modulates aflatoxin biosynthesis and peanut infection in the fungus Aspergillus flavus. Int. J. Food Microbiol. 2020, 322, 108576–108587. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Huang, L.; Deng, J.; Tan, C.; Geng, L.; Liao, Y.; Yuan, J.; Wang, S. A Cell Wall Integrity–Related MAP Kinase Kinase Kinase AflBck1 Is Required for Growth and Virulence in Fungus Aspergillus flavus. Mol. Plant. Microbe Interact. 2020, 33, 680–692. [Google Scholar] [CrossRef]
- Qi, M.; Elion, E.A. MAP kinase pathways. J. Cell Sci. 2005, 118, 3569–3572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, K.; Qin, Q.; Liu, Y.; Zhang, L.; Liang, L.; Lan, H.; Chen, C.; You, Y.; Zhang, F.; Wang, S. Adenylate cyclase AcyA regulates development, aflatoxin biosynthesis and fungal virulence in Aspergillus flavus. Front. Cell. Infect. Microbiol. 2016, 6, 190–204. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Yang, K.; Qin, Q.; Lin, G.; Hu, T.; Xu, Z.; Wang, S. G protein α subunit GpaB is required for asexual development, aflatoxin biosynthesis and pathogenicity by regulating cAMP signaling in Aspergillus flavus. Toxins 2018, 10, 117. [Google Scholar] [CrossRef] [Green Version]
- Yang, K.; Liu, Y.; Wang, S.; Wu, L.; Xie, R.; Lan, H.; Fasoyin, O.E.; Wang, Y.; Wang, S. Cyclase-associated protein cap with multiple domains contributes to mycotoxin biosynthesis and fungal virulence in Aspergillus flavus. J. Agric. Food Chem. 2019, 67, 4200–4213. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).