DNA Methyltransferases Regulate Pathogenicity of Botrytis cinerea to Horticultural Crops
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Phylogenetic Analysis
2.3. 5-Azacytidine Treatment
2.4. Construction of Knockout Mutants
2.5. Expression Analysis
2.6. Virulence Assay
2.7. ROS Detection
2.8. Quantification of Global DNA Methylation
2.9. Whole-Genome Bisulfite Sequencing
2.10. Statistical Analysis of Data
3. Results
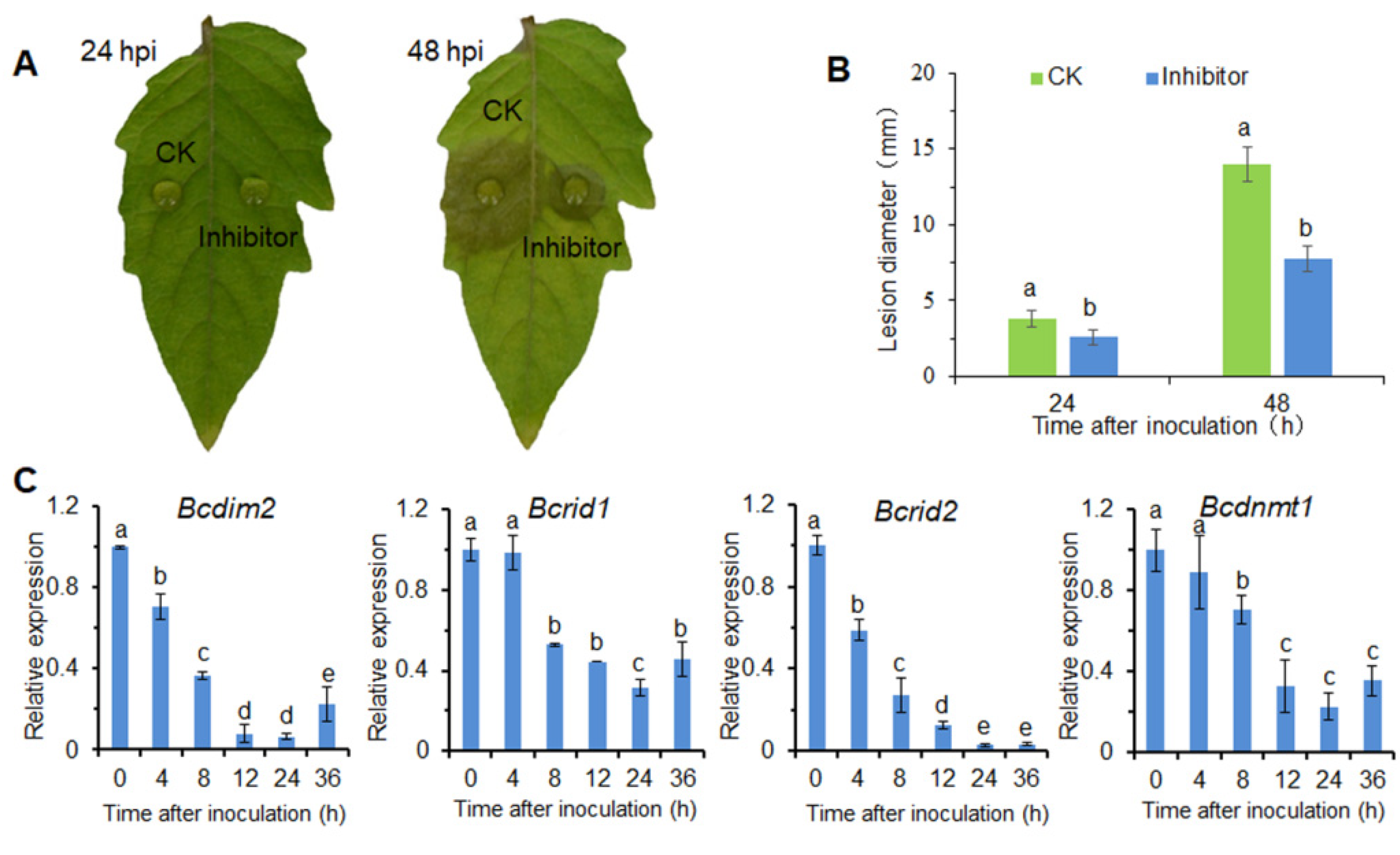
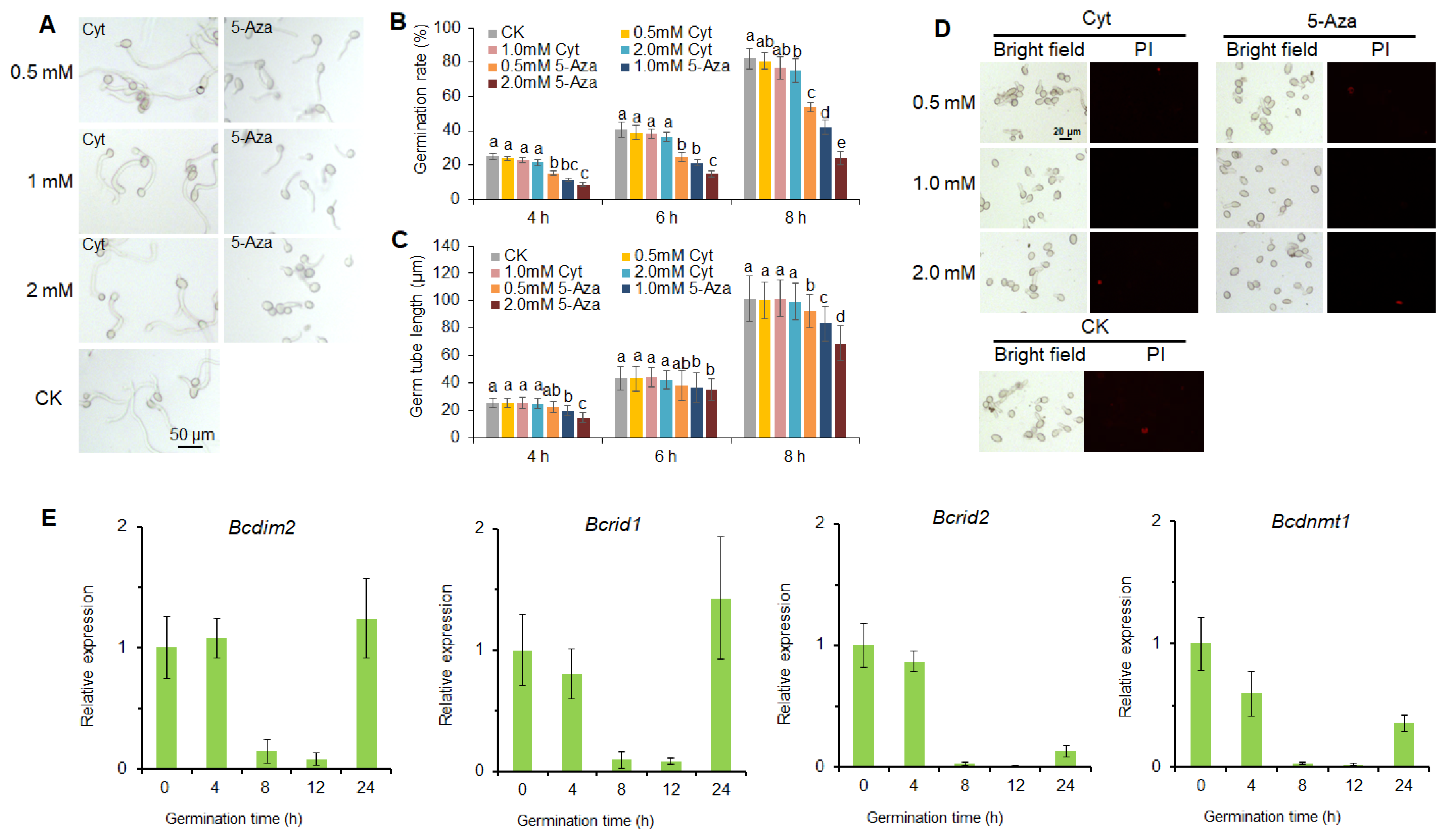
3.1. DNA Methylation Is Involved in the Infection Process of B. cinerea
3.2. DNA Methylation Is Involved in the Development of B. cinerea
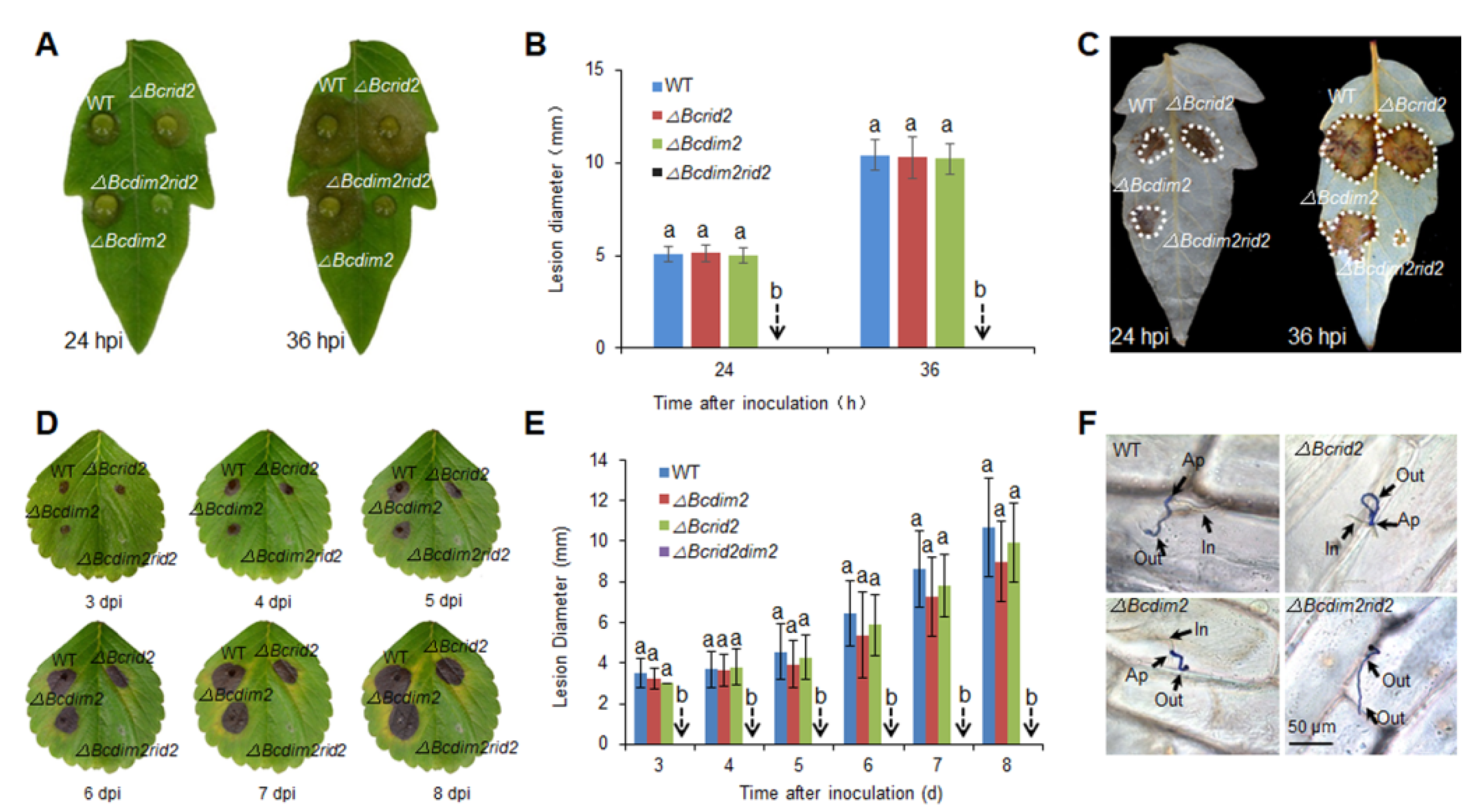
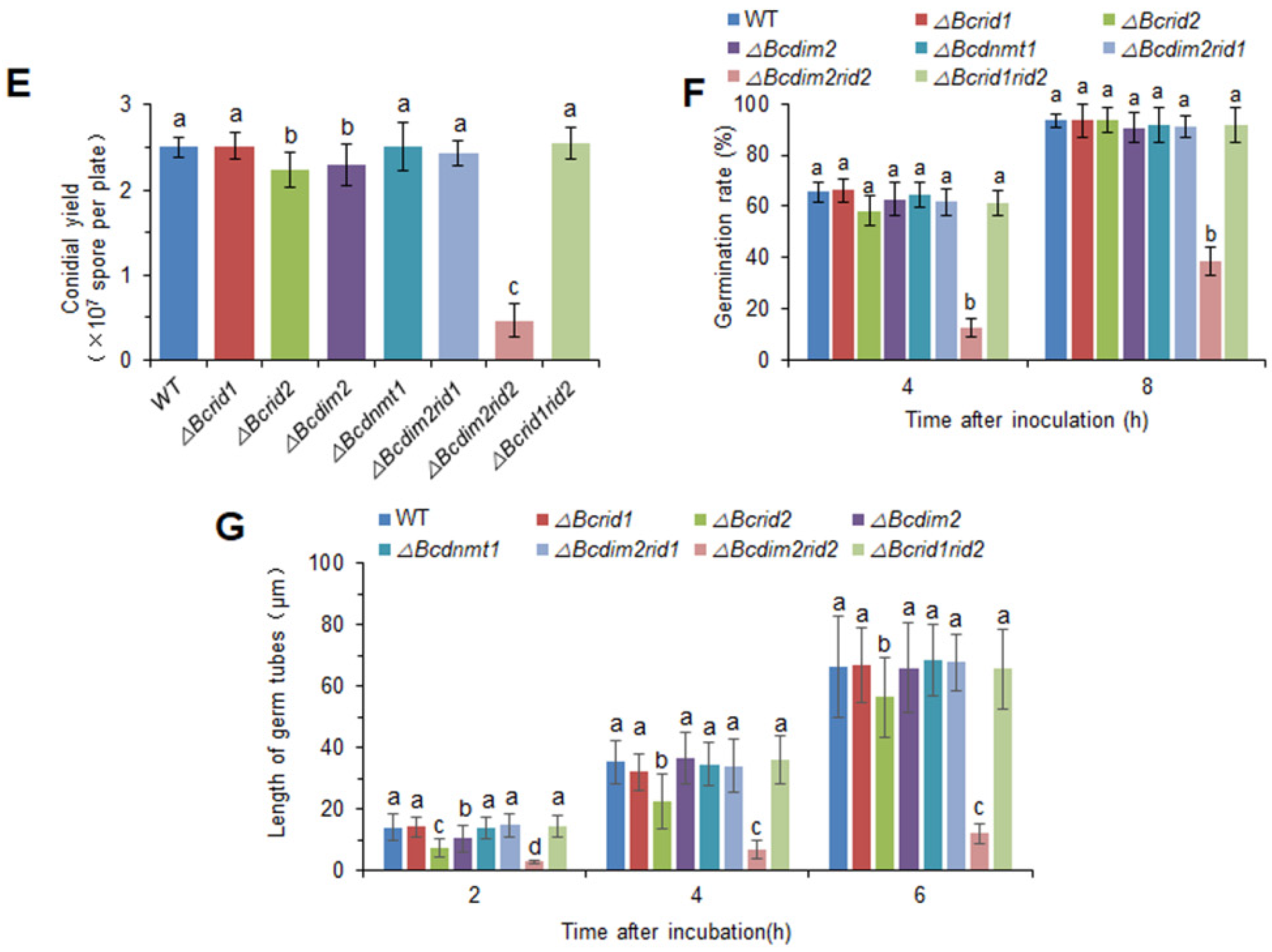
3.3. Virulence Assay of DNA MTase Mutants on Fruit Hosts
3.4. Verification of the Synergistic Effect of Bcdim2 and Bcrid2 on Leaf Hosts
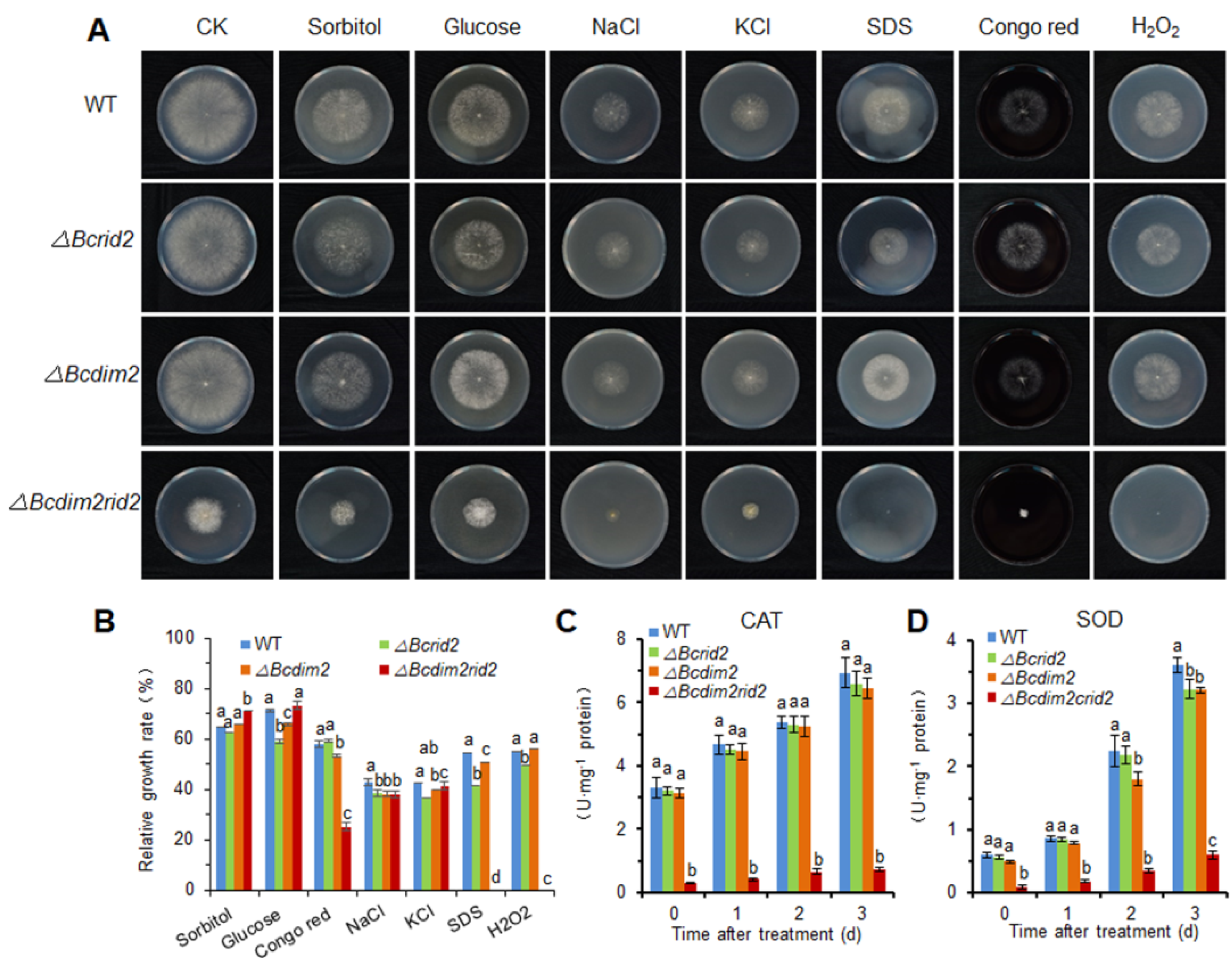
3.5. DNA MTases Ais Attenuatedre Involved in the Development of B. cinerea
3.6. The Oxidative Tolerance of ΔBcdim2rid2 Is Attenuated
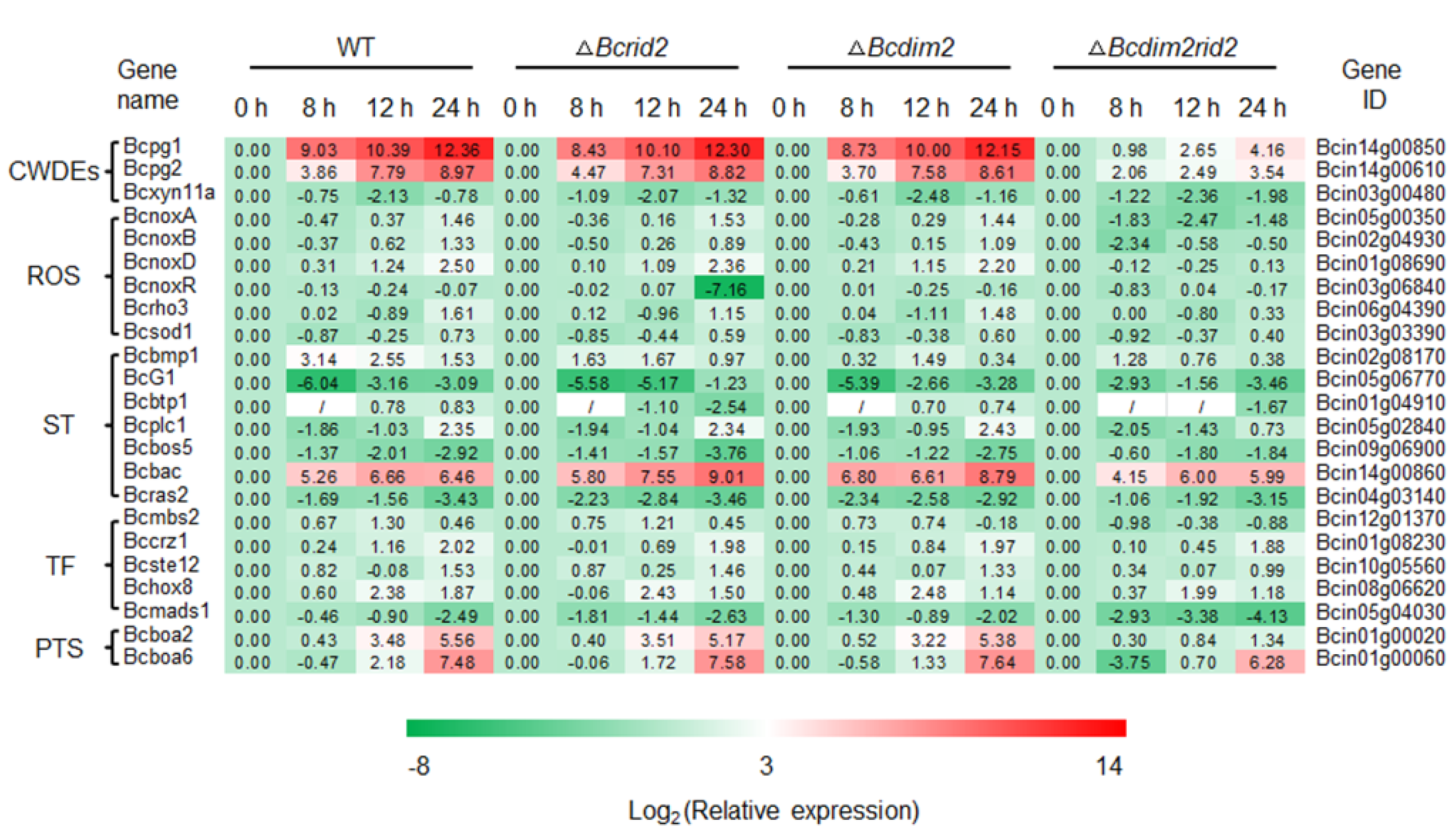
3.7. Expression of Pathogenic Genes in ΔBcdim2rid2 Is Inhibited
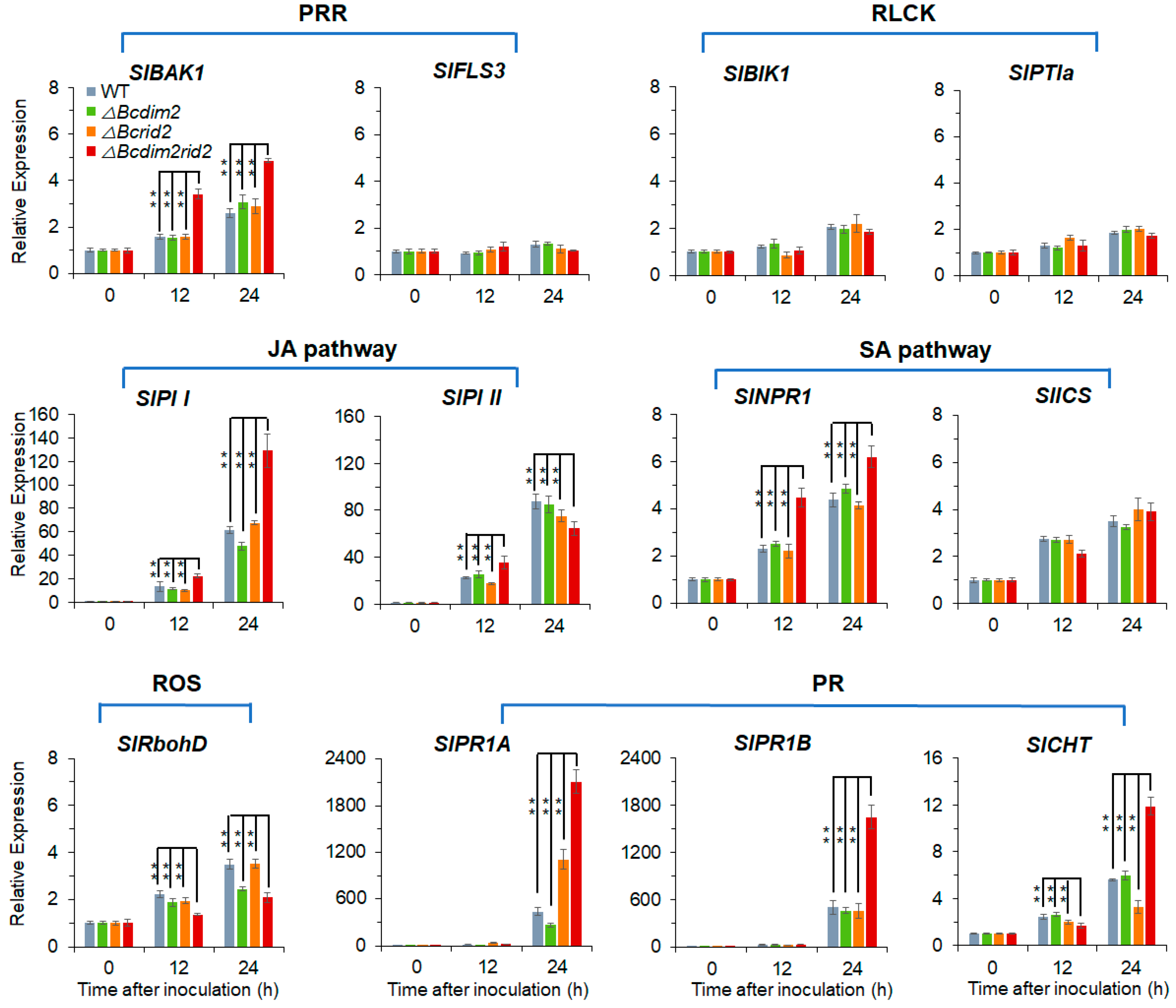
3.8. ΔBcdim2rid2 Induces the Resistant of Host
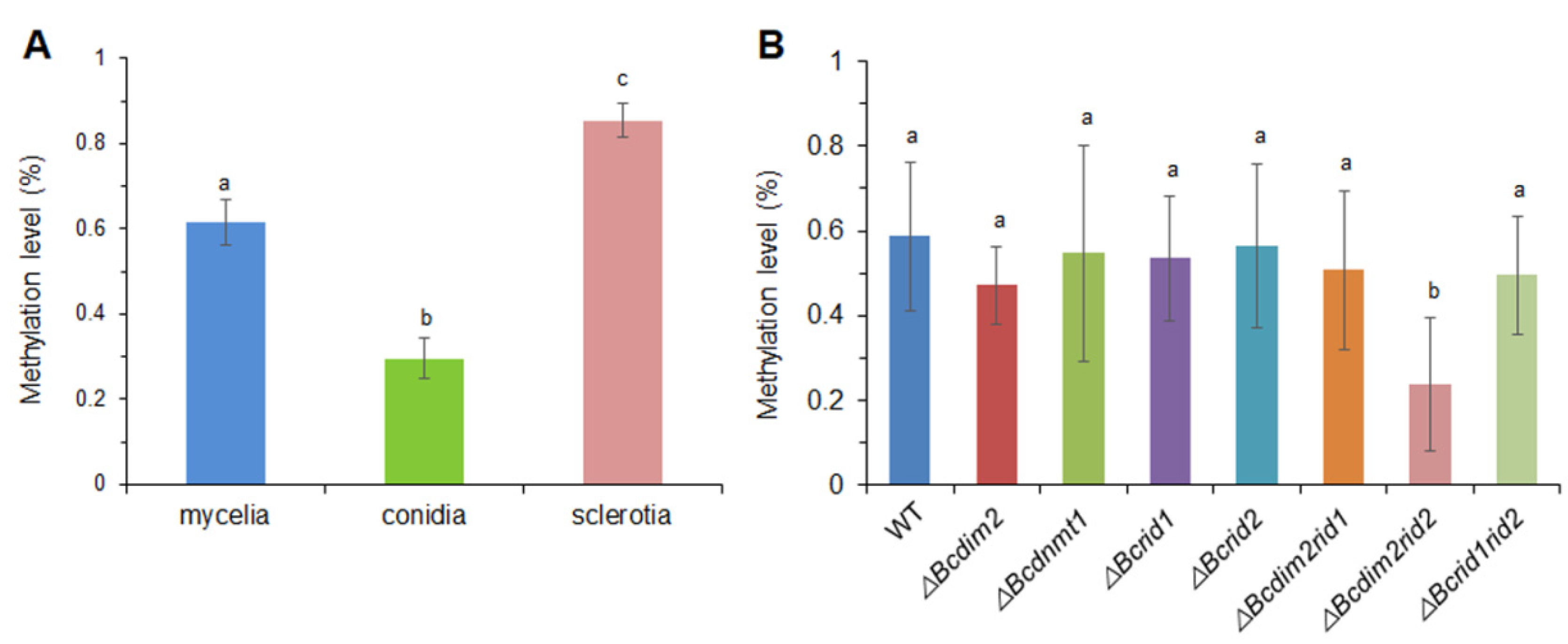
3.9. DNA MTases Affect Genomic DNA Methylation of B. cinerea
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weiberg, A.; Wang, M.; Lin, F.M.; Zhao, H.W.; Zhang, Z.H.; Kaloshian, I.; Huang, H.D.; Jin, H.L. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 2013, 342, 118–123. [Google Scholar] [CrossRef] [Green Version]
- Williamson, B.; Tudzynski, B.; Tudzynski, P.; van Kan, J.A.L. Botrytis cinerea: The cause of grey mould disease. Mol. Plant Pathol. 2007, 8, 561–580. [Google Scholar] [CrossRef]
- Dean, R.; van Kan, J.A.L.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Pietro, A.; Spanu, P.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segmüller, N.; Ellendorf, U.; Tudzynski, B.; Tudzynski, P. BcSAK1, a stress-activated MAP kinase is involved in vegetative differentiation and pathogenicity in Botrytis cinerea. Eukaryot. Cell 2007, 6, 211–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minz-Dub, A.; Kokkelink, L.; Tudzynski, B.; Tudzynski, P.; Sharon, A. Involvement of Botrytis cinerea small GTPases BcRAS1 and BcRAC in differentiation, virulence, and the cell cycle. Eukaryot. Cell 2013, 12, 1609–1618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segmüller, N.; Kokkelink, L.; Giesbert, S.; Odinius, D.; van Kan, J.A.N.; Tudzynski, P. NADPH oxidases are involved in differentiation and pathogenicity in Botrytis cinerea. Mol. Plant Microbe Interact. 2008, 21, 808–819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, B.; Li, B.Q.; Qin, G.Z.; Tian, S.P. Functions of small GTPase Rho3 in regulating growth, conidiation and virulence of Botrytis cinerea. Fungal Genet. Biol. 2015, 75, 46–55. [Google Scholar] [CrossRef]
- An, B.; Li, B.Q.; Li, H.; Zhang, Z.Q.; Qin, G.Z.; Tian, S.P. Aquaporin8 regulates cellular development and ROS production, a critical component of virulence in Botrytis cinerea. New Phytol. 2016, 209, 1668–1680. [Google Scholar] [CrossRef]
- Li, H.; Zhang, Z.Q.; He, C.; Qin, G.Z.; Tian, S.P. Comparative proteomics reveals the potential targets of BcNoxR, a putative regulatory subunit of NADPH oxidase of Bortytis cinerea. Mol. Plant Microbe Interact. 2016, 29, 990–1003. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Tian, S.P.; Qin, G.Z. NADPH oxidase is crucial for the cellular redox homeostasis in fungal pathogen Botrytis cinerea. Mol. Plant Microbe Interact. 2019, 32, 1508–1516. [Google Scholar] [CrossRef]
- Zhang, Z.Q.; Qin, G.Z.; Li, B.Q.; Tian, S.P. Knocking out Bcsas1 in Botrytis cinerea impacts growth, development, and secretion of extracellular proteins, which decreases virulence. Mol. Plant Microbe Interact. 2014, 27, 590–600. [Google Scholar] [CrossRef]
- Li, H.; Zhang, Z.Q.; Qin, G.Z.; He, C.; Li, B.Q.; Tian, S.P. Actin is required for cellular development and virulence of Botrytis cinerea via the mediation of secretory of proteins. mSystems 2020, 5, e00732-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvarez, M.E.; Nota, F.; Cambiagno, D.A. Epigenetic control of plant immunity. Mol. Plant Pathol. 2010, 11, 563–576. [Google Scholar] [CrossRef]
- Boyko, A.; Kovalchuk, I. Genetic and epigenetic effects of plant-pathogen interactions: An evolutionary perspective. Mol. Plant 2011, 4, 1014–1023. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.Y.; Chen, X.S.; Wang, Z.X.; Sun, Q.; Hong, A.; Zhang, A.Q.; Zhong, X.H.; Hua, J. HOS15 and HDA9 negatively regulate immunity through histone deacetylation of intracellular immune receptor NLR genes in Arabidopsis. New Phytol. 2020, 226, 507–522. [Google Scholar] [CrossRef] [PubMed]
- Aramayo, R.; Selker, E.U. Neurospora crassa, a model system for epigenetics research. Cold Spring Harb. Perspect. Biol. 2013, 5, a017921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grunstein, M.; Gasser, S.M. Epigenetics in Saccharomyces cerevisiae. Cold Spring Harb. Perspect. Biol. 2013, 5, a017491. [Google Scholar] [CrossRef] [Green Version]
- Allshire, R.C.; Ekwall, K. Epigenetic regulation of chromatin states in Schizosaccharomyces pombe. Cold Spring Harb. Perspect. Biol. 2015, 7, a018770. [Google Scholar] [CrossRef] [Green Version]
- He, C.; Zhang, Z.Q.; Li, B.Q.; Tian, S.P. The pattern and function of DNA methylation in fungal plant pathogen. Microorganisms 2020, 8, 227. [Google Scholar] [CrossRef] [Green Version]
- Hüberli, D.; Garbelotto, M. Phytophthora ramorum is a generalist plant pathogen with differences in virulence between isolates from infectious and dead-end hosts. Forest Pathol. 2012, 42, 8–13. [Google Scholar] [CrossRef] [Green Version]
- Dou, D.; Kale, S.D.; Liu, T.; Tang, Q.; Wang, X.; Arredondo, F.D.; Basnayake, S.; Whisson, S.; Drenth, A.; Maclean, D.; et al. Different domains of Phytophthora sojae effector Avr4/6 are recognized by soybean resistance genes Rps4 and Rps6. Mol. Plant Microbe Interact. 2010, 23, 425–435. [Google Scholar] [CrossRef] [Green Version]
- Dong, S.; Yu, D.; Cui, L.; Qutob, D.; Tedman-Jones, J.; Kale, S.D.; Tyler, B.M.; Wang, Y.C.; Gijzen, M. Sequence variants of the Phytophthora sojae RXLR effector Avr3a/5 are differentially recognized by Rps3a and Rps5 in soybean. PLoS ONE 2011, 6, e20172. [Google Scholar] [CrossRef]
- Gilroy, E.M.; Breen, S.; Whisson, S.C.; Squires, J.; Hein, I.; Kaczmarek, M.; Dionne, T.; Boevink, P.C.; Lokossou, A.; Cano, L.M.; et al. Presence/absence, differential expression and sequence polymorphisms between PiAVR2 and PiAVR2-like in Phytophthora infestans determine virulence on R2 plants. New Phytol. 2011, 191, 763–776. [Google Scholar] [CrossRef]
- Qutob, D.; Chapman, B.P.; Gijzen, M. Transgenerational gene silencing causes gain of virulence in a plant pathogen. Nat. Commun. 2013, 4, 1349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, S.; Laurie, J.D.; Linning, R.; Cervantes-Chavez, J.A.; Gaudet, D.; Bakkeren, G. An immunity-triggering effector from the Barley smut fungus Ustilago hordei resides in an Ustilaginaceae-specific cluster bearing signs of transposable element-assisted evolution. PLoS Pathog. 2014, 10, e1004223. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.Y.; Chen, H.; Li, J.J.; Shu, H.D.; Zhang, X.X.; Wang, Y.C.; Tyler, B.M.; Dong, S.M. Effector gene silencing mediated by histone methylation underpins host adaptation in an oomycete plant pathogen. Nucleic Acids Res. 2020, 48, 1790–1799. [Google Scholar] [CrossRef] [PubMed]
- Martienssen, R.A.; Colot, V. DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science 2001, 293, 1070–1074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reik, W.; Dean, W.; Walter, J. Epigenetic reprogramming in mammalian development. Science 2001, 293, 1089–1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heard, E.; Disteche, C.M. Dosage compensation in mammals: Fine-tuning the expression of the X chromosome. Gene Dev. 2006, 20, 1848–1867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weber, M.; Schubeler, D. Genomic patterns of DNA methylation: Targets and function of an epigenetic mark. Curr. Opin. Cell Biol. 2007, 19, 273–280. [Google Scholar] [CrossRef]
- Reyna-López, G.E.; Simpson, J.; Ruiz-Herrera, J. Differences in DNA methylation patterns are detectable during the dimorphic transition of fungi by amplification of restriction polymorphisms. Mol. Gen. Genet. 1997, 253, 703–710. [Google Scholar] [CrossRef]
- So, K.K.; Ko, Y.H.; Chun, J.; Bal, J.; Jeno, J.; Kim, J.M.; Choi, J.; Lee, Y.H.; Jin, H.H.; Kim, D.H. Global DNA methylation in the chestnut blight fungus Cryphonectria parasitica and genome-wide changes in DNA methylation accompanied with sectorization. Front. Plant. Sci. 2018, 9, 103. [Google Scholar] [CrossRef] [Green Version]
- Jeon, J.; Choi, J.; Lee, G.W.; Park, S.Y.; Huh, A.; Dean, R.A.; Lee, Y.H. Genome- wide profiling of DNA methylation provides insights into epigenetic regulation of fungal development in a plant pathogenic fungus, Magnaporthe oryzae. Sci. Rep. 2015, 5, e8567. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, M.M.; Bird, A. DNA methylation landscapes: Provocative insights from epigenomics. Nat. Rev. Genet. 2008, 9, 465–476. [Google Scholar] [CrossRef]
- Law, J.A.; Jacobsen, S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010, 11, 204–220. [Google Scholar] [CrossRef]
- Feng, S.; Jacobsen, S.E. Epigenetic modifications in plants: An evolutionary perspective. Curr. Opin. Cell Biol. 2011, 14, 179–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishra, P.K.; Baum, M.; Carbon, J. DNA methylation regulates phenotype- dependent transcriptional activity in Candida albicans. Proc. Natl. Acad. Sci. USA 2011, 108, 11965–11970. [Google Scholar] [CrossRef] [Green Version]
- Bestor, T.H. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000, 9, 2395–2402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.; Ohr, H.; Lee, J.W.; Hyun, Y.; Fischer, R.L.; Choi, Y. Temporal and spatial downregulation of Arabidopsis MET1 activity results in global DNA hypomethylation and developmental defects. Mol. Cell. 2008, 26, 611–615. [Google Scholar]
- Kouzminova, E.; Selker, E.U. dim-2 encodes a DNA methyltransferase responsible for all known cytosine methylation in Neurospora. EMBO J. 2001, 20, 4309–4323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freitag, M.; Williams, R.L.; Kothe, G.O.; Selker, E.U. A cytosine methyl-transferase homologue is essential for repeat-induced point mutation in Neurospora crassa. Proc. Natl. Acad. Sci. USA 2002, 99, 8802–8807. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.L.; Wang, T.T.; Qiao, L.T.; Zhu, J.Y.; Fan, J.R.; Zhang, T.T.; Wang, Z.X.; Li, W.Z.; Chen, A.H.; Huang, B. DNA methyltransferases contribute to the fungal development, stress tolerance and virulence of the entomopathogenic fungus Metarhizium robertsii. Appl. Microbiol. Biotechnol. 2017, 101, 4215–4266. [Google Scholar] [CrossRef]
- Wang, X.L.; Song, S.H.; Wu, Y.S.; Li, Y.L.; Chen, T.T.; Huang, Z.Y.; Liu, S.; Dunwell, T.L.; Pfeifer, G.P.; Dunwell, J.M.; et al. Genome-wide mapping of 5-hydroxymethylcytosine in three rice cultivars reveals its preferential localization in transcriptionally silent transposable element genes. J. Exp. Bot. 2015, 66, 6651–6663. [Google Scholar] [CrossRef] [Green Version]
- Yang, K.L.; Liang, L.L.; Ran, F.L.; Liu, Y.H.; Li, Z.G.; Lan, H.H.; Gao, P.L.; Zhuang, Z.H.; Zhang, F.; Nie, X.Y.; et al. The DmtA methyltransferase contributes to Aspergillus flavus conidiation, sclerotial production, aflatoxin biosynthesis and virulence. Sci. Rep. 2016, 6, 23259. [Google Scholar] [CrossRef] [Green Version]
- Ko, Y.H.; So, K.K.; Chun, J.; Kim, D.H. Distinct roles of two DNA methyltransferases from Cryphonectria parasitica in fungal virulence, responses to hypovirus infection, and viral clearance. mBio 2021, 12, e02890-20. [Google Scholar] [CrossRef]
- Lee, D.W.; Freitag, M.; Selker, E.U.; Aramayo, R. A cytosine methyltransferase homologue is essential for sexual development in Aspergillus nidulans. PLoS ONE 2008, 3, e2531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamada, W.; Reignault, P.; Bompeix, G.; Boccara, M. Transformation of Botrytis cinerea with the hygromycin B resistance gene, hph. Curr. Genet. 1994, 26, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Liva, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Daudi, A.; Cheng, Z.; O’Brien, J.A.; Mammarella, N.; Khan, S.; Ausube, F.M.; Bolwell, G.P. The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern-triggered immunity. Plant Cell 2012, 24, 275–287. [Google Scholar] [CrossRef] [Green Version]
- Bewick, A.J.; Hofmeister, B.T.; Powers, R.A.; Mondo, S.J.; Grigoriev, I.V.; James, T.Y.; Stajich, J.E.; Schmitz, R.J. Diversity of cytosine methylation across the fungal tree of life. Nat. Ecol. Evol. 2019, 3, 479–490. [Google Scholar] [CrossRef]
- Michielse, C.B.; Becker, M.; Heller, J.; Moraga, J.; Collado, I.G.; Tudzynski, P. The Botrytis cinerea Reg1 protein, a putative transcriptional regulator, is required for pathogenicity, conidiogenesis, and the production of secondary metabolites. Mol. Plant Microbe Interact. 2011, 24, 1074–1085. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.Q.; Li, H.; Qin, G.Z.; He, C.; Li, B.Q.; Tian, S.P. The MADS-box transcription factor Bcmads1 is required for growth, sclerotia production and pathogenicity of Botrytis cinerea. Sci. Rep. 2016, 6, 33901. [Google Scholar] [CrossRef]
- Li, B.Q.; Wang, W.H.; Zong, Y.Y.; Qin, G.Z.; Tian, S.P. Exploring pathogenic mechanisms of Botrytis cinerea secretome under different ambient pH based on comparative proteomic analysis. J. Proteome Res. 2012, 11, 4249–4260. [Google Scholar] [CrossRef]
- Tamaru, H.; Zhang, X.; McMillen, D.; Singh, P.B.; Nakayama, J.; Grewal, S.I.; Cheng, X.; Selker, E.U. Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat. Genet. 2003, 34, 75–79. [Google Scholar] [CrossRef]
- Schübeler, D. Epigenetic islands in a genetic ocean. Science 2012, 338, 756–757. [Google Scholar] [CrossRef] [PubMed]
- Goll, M.G.; Bestor, T.H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005, 74, 481–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999, 99, 247–257. [Google Scholar] [CrossRef] [Green Version]
- Finnegan, E.J.; Peacock, W.J.; Dennis, E.S. Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc. Natl. Acad. Sci. USA 1996, 93, 8449–8454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Jacobsen, S.E. Genetic analyses of DNA methyltransferases in Arabidopsis thaliana. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 439–447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reik, W.; Walter, J. Genomic imprinting: Parental influence on the genome. Nat. Rev. Genet. 2001, 2, 21–32. [Google Scholar] [CrossRef]
- Miura, A.; Yonebayashi, S.; Watanabe, K.; Toyama, T.; Shimada, H.; Kakutani, T. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 2001, 411, 212–214. [Google Scholar] [CrossRef]
- Wolffe, A.P.; Matzke, M.A. Epigenetics: Regulation through repression. Science 1999, 286, 481–486. [Google Scholar] [CrossRef]
- Li, E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 2002, 3, 662–673. [Google Scholar] [CrossRef] [PubMed]
- Lei, H.; Oh, S.P.; Okano, M.; Jüttermann, R.; Goss, K.A.; Jaenisch, R.; Li, E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 1996, 122, 3195–3205. [Google Scholar] [CrossRef]
- Govrin, E.M.; Levine, A. The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr. Biol. 2000, 10, 751–757. [Google Scholar] [CrossRef] [Green Version]
- ten Have, A.; Mulder, W.; Visser, J.; van Kan, J.A.L. The endopoly- galacturonase gene Bcpg1 is required for full virulence of Botrytis cinerea. Mol. Plant Microbe Interact. 1998, 11, 1009–1016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Oirdi, M.; Bouarab, K. Plant signaling components EDS1 and SGT1 enhance disease caused by the necrotrophic pathogen Botrytis cinerea. New Phytol. 2007, 175, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Buxdorf, K.; Rahat, I.; Gafni, A.; Levy, M. The epiphytic fungus Pseudozyma aphidis induces jasmonic acid- and salicylic acid/nonexpressor of PR1-independent local and systemic resistance. Plant Physiol. 2013, 161, 2014–2022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clutterbuck, A.J. Genomic evidence of repeat-induced point mutation (RIP) in filamentous ascomycetes. Fungal Genet. Biol. 2011, 48, 306–326. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; He, C.; Chen, Y.; Li, B.; Tian, S. DNA Methyltransferases Regulate Pathogenicity of Botrytis cinerea to Horticultural Crops. J. Fungi 2021, 7, 659. https://doi.org/10.3390/jof7080659
Zhang Z, He C, Chen Y, Li B, Tian S. DNA Methyltransferases Regulate Pathogenicity of Botrytis cinerea to Horticultural Crops. Journal of Fungi. 2021; 7(8):659. https://doi.org/10.3390/jof7080659
Chicago/Turabian StyleZhang, Zhanquan, Chang He, Yong Chen, Boqiang Li, and Shiping Tian. 2021. "DNA Methyltransferases Regulate Pathogenicity of Botrytis cinerea to Horticultural Crops" Journal of Fungi 7, no. 8: 659. https://doi.org/10.3390/jof7080659
APA StyleZhang, Z., He, C., Chen, Y., Li, B., & Tian, S. (2021). DNA Methyltransferases Regulate Pathogenicity of Botrytis cinerea to Horticultural Crops. Journal of Fungi, 7(8), 659. https://doi.org/10.3390/jof7080659







