Role of RIM101 for Sporulation at Alkaline pH in Ashbya gossypii
Abstract
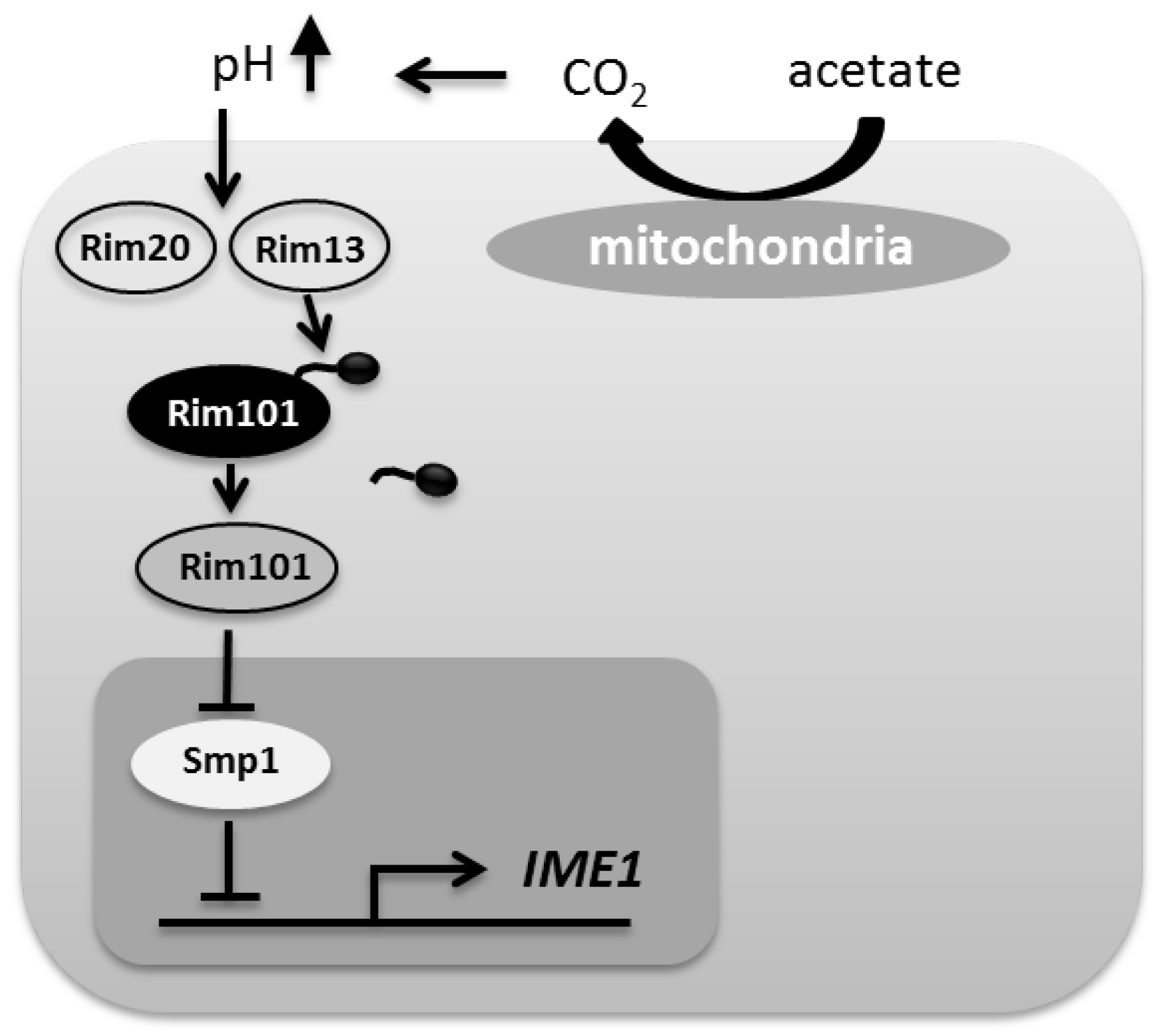
:1. Introduction
2. Materials and Methods
2.1. Strains and Media
2.2. Transformation of A. gossypii
2.3. Sporulation Assay
2.4. Microscopy
3. Results
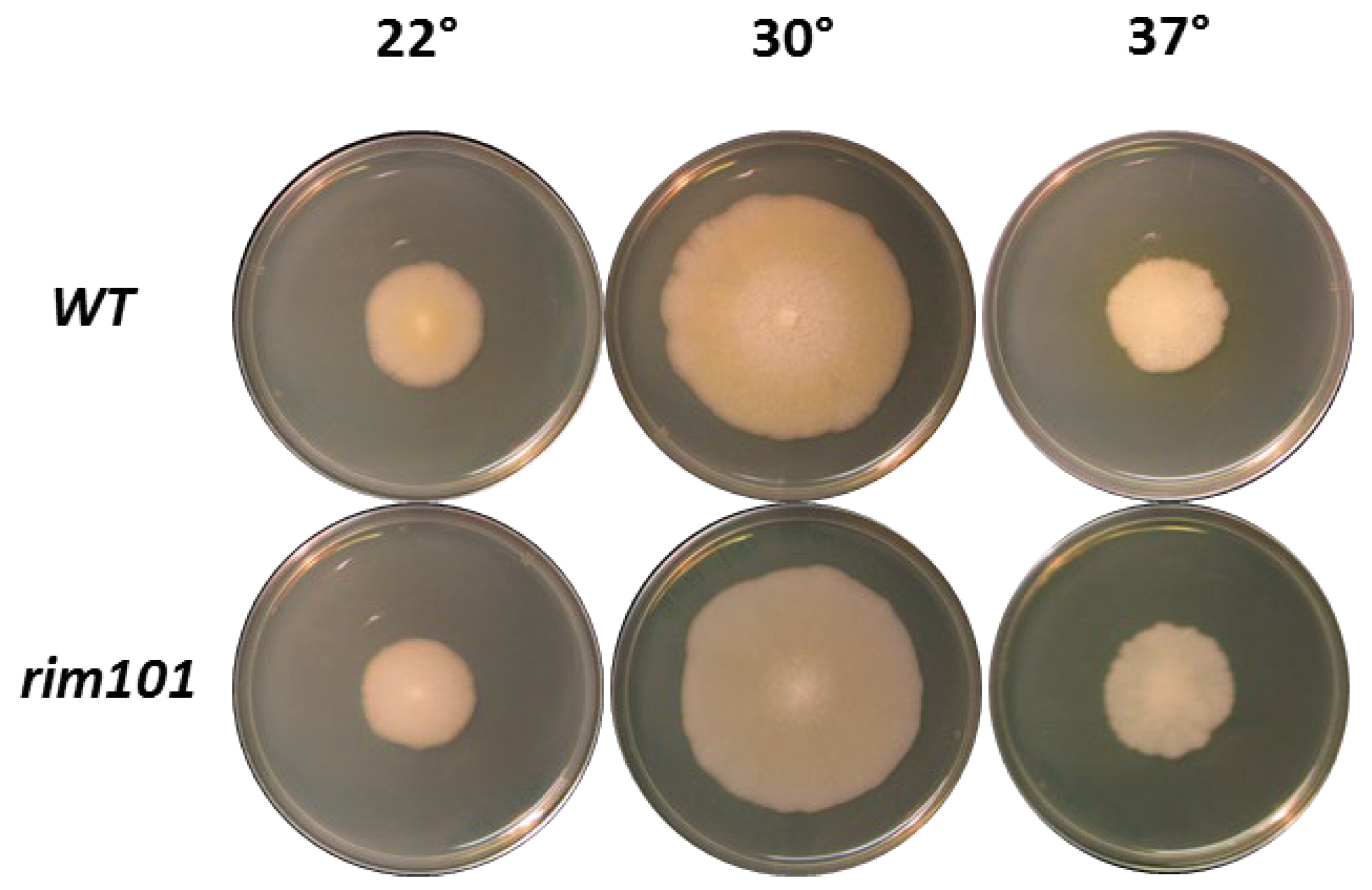
3.1. Rim101 Has No Role in Vegetative Growth in A. gossypii
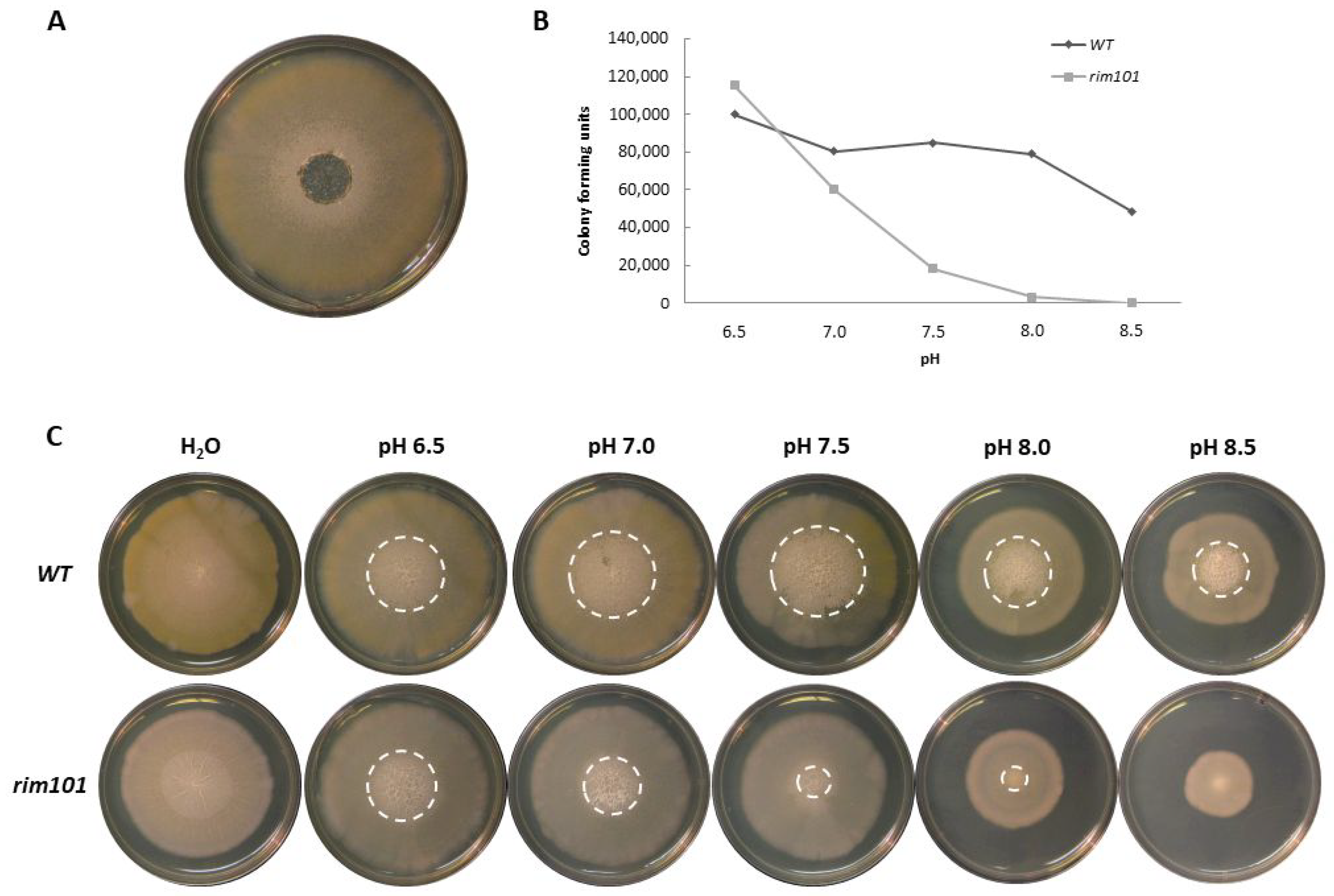
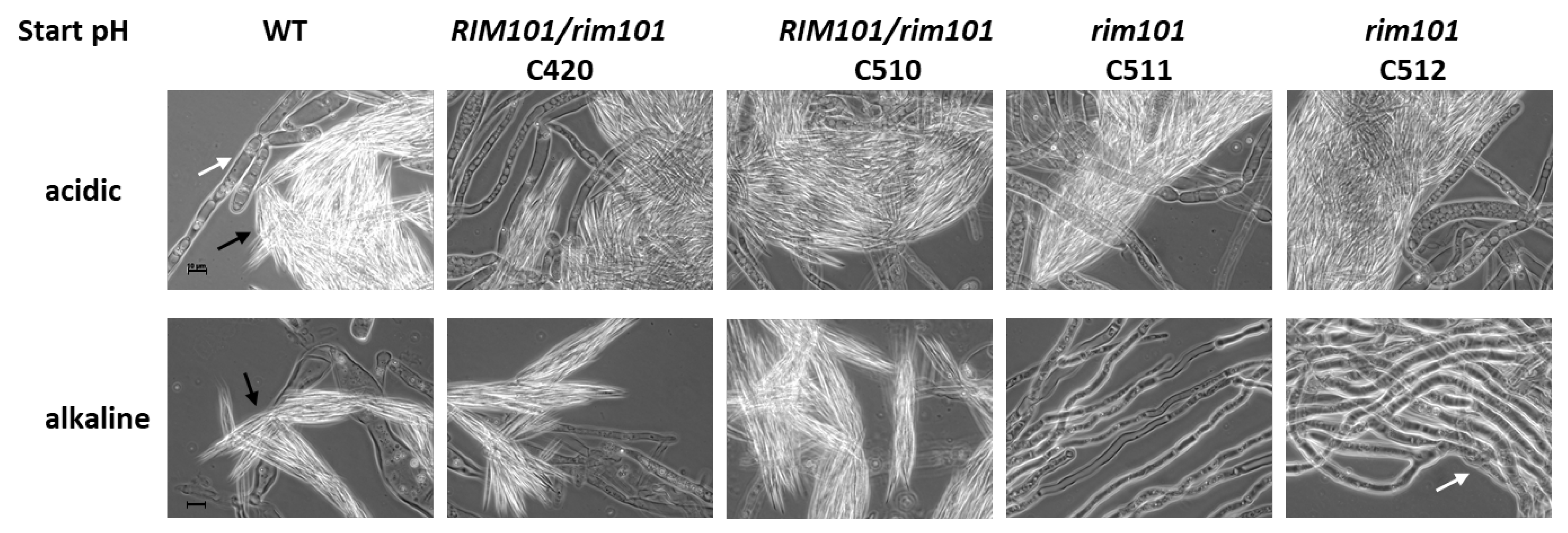
3.2. Sporulation at High pH in A. gossypii Requires Rim101
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Penalva, M.A.; Arst, H.N., Jr. Recent advances in the characterization of ambient pH regulation of gene expression in filamentous fungi and yeasts. Annu. Rev. Microbiol. 2004, 58, 425–451. [Google Scholar] [CrossRef]
- Penalva, M.A.; Tilburn, J.; Bignell, E.; Arst, H.N., Jr. Ambient pH gene regulation in fungi: Making connections. Trends Microbiol. 2008, 16, 291–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bignell, E.; Negrete-Urtasun, S.; Calcagno, A.M.; Haynes, K.; Arst, H.N., Jr.; Rogers, T. The Aspergillus ph-responsive transcription factor PacC regulates virulence. Mol. Microbiol. 2005, 55, 1072–1084. [Google Scholar] [CrossRef]
- Tilburn, J.; Sarkar, S.; Widdick, D.A.; Espeso, E.A.; Orejas, M.; Mungroo, J.; Penalva, M.A.; Arst, H.N., Jr. The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 1995, 14, 779–790. [Google Scholar] [CrossRef] [PubMed]
- Davis, D.; Edwards, J.E., Jr.; Mitchell, A.P.; Ibrahim, A.S. Candida albicans Rim101 pH response pathway is required for host-pathogen interactions. Infect. Immun. 2000, 68, 5953–5959. [Google Scholar] [CrossRef] [Green Version]
- Ramon, A.M.; Porta, A.; Fonzi, W.A. Effect of environmental pH on morphological development of Candida albicans is mediated via the PacC-related transcription factor encoded by PRR2. J. Bacteriol. 1999, 181, 7524–7530. [Google Scholar] [CrossRef] [Green Version]
- Caracuel, Z.; Roncero, M.I.; Espeso, E.A.; Gonzalez-Verdejo, C.I.; Garcia-Maceira, F.I.; Di Pietro, A. The pH signalling transcription factor PacC controls virulence in the plant pathogen Fusarium oxysporum. Mol. Microbiol. 2003, 48, 765–779. [Google Scholar] [CrossRef]
- Landraud, P.; Chuzeville, S.; Billon-Grande, G.; Poussereau, N.; Bruel, C. Adaptation to pH and role of PacC in the rice blast fungus Magnaporthe oryzae. PLoS ONE 2013, 8, e69236. [Google Scholar] [CrossRef] [Green Version]
- Lamb, T.M.; Xu, W.; Diamond, A.; Mitchell, A.P. Alkaline response genes of Saccharomyces cerevisiae and their relationship to the RIM101 pathway. J. Biol. Chem. 2001, 276, 1850–1856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamb, T.M.; Mitchell, A.P. The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol. Cell Biol. 2003, 23, 677–686. [Google Scholar] [CrossRef] [Green Version]
- Zaman, S.; Lippman, S.I.; Zhao, X.; Broach, J.R. How Saccharomyces responds to nutrients. Annu. Rev. Genet. 2008, 42, 27–81. [Google Scholar] [CrossRef]
- Xu, W.; Mitchell, A.P. Yeast PalA/AIP1/Alix homolog Rim20p associates with a pest-like region and is required for its proteolytic cleavage. J. Bacteriol. 2001, 183, 6917–6923. [Google Scholar] [CrossRef] [Green Version]
- Diez, E.; Alvaro, J.; Espeso, E.A.; Rainbow, L.; Suarez, T.; Tilburn, J.; Arst, H.N., Jr.; Penalva, M.A. Activation of the Aspergillus PacC zinc finger transcription factor requires two proteolytic steps. EMBO J. 2002, 21, 1350–1359. [Google Scholar] [CrossRef] [Green Version]
- Broach, J.R. Nutritional control of growth and development in yeast. Genetics 2012, 192, 73–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dietrich, F.S.; Voegeli, S.; Brachat, S.; Lerch, A.; Gates, K.; Steiner, S.; Mohr, C.; Pohlmann, R.; Luedi, P.; Choi, S.; et al. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science 2004, 304, 304–307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wasserstrom, L.; Lengeler, K.B.; Walther, A.; Wendland, J. Molecular determinants of sporulation in Ashbya gossypii. Genetics 2013, 195, 87–99. [Google Scholar] [CrossRef] [Green Version]
- Wasserstrom, L.; Lengeler, K.; Walther, A.; Wendland, J. Developmental growth control exerted via the protein a kinase Tpk2 in Ashbya gossypii. Eukaryot. Cell 2015, 14, 593–601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wendland, J. Sporulation in Ashbya gossypii. J. Fungi 2020, 6, 157. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wang, Q.; Liu, C.; Shang, Y.; Wen, F.; Wang, F.; Liu, W.; Xiao, W.; Li, W. A role for the respiratory chain in regulating meiosis initiation in Saccharomyces cerevisiae. Genetics 2018, 208, 1181–1194. [Google Scholar] [CrossRef] [Green Version]
- Wendland, J.; Ayad-Durieux, Y.; Knechtle, P.; Rebischung, C.; Philippsen, P. PCR-based gene targeting in the filamentous fungus Ashbya gossypii. Gene 2000, 242, 381–391. [Google Scholar] [CrossRef]
- Walther, A.; Wendland, J. PCR-based gene targeting in Candida albicans. Nat. Protoc. 2008, 3, 1414–1421. [Google Scholar] [CrossRef]
- Gattiker, A.; Rischatsch, R.; Demougin, P.; Voegeli, S.; Dietrich, F.S.; Philippsen, P.; Primig, M. Ashbya genome database 3.0: A cross-species genome and transcriptome browser for yeast biologists. BMC Genom. 2007, 8, 9. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Mitchell, A.P. Proteolytic activation of Rim1p, a positive regulator of yeast sporulation and invasive growth. Genetics 1997, 145, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Su, S.S.; Mitchell, A.P. Molecular characterization of the yeast meiotic regulatory gene RIM1. Nucleic Acids Res. 1993, 21, 3789–3797. [Google Scholar] [CrossRef] [Green Version]
- Marques, M.C.; Zamarbide-Fores, S.; Pedelini, L.; Llopis-Torregrosa, V.; Yenush, L. A functional Rim101 complex is required for proper accumulation of the Ena1 na+-atpase protein in response to salt stress in Saccharomyces cerevisiae. FEMS Yeast Res. 2015, 15, fov017. [Google Scholar] [CrossRef] [Green Version]
- Lengeler, K.B.; Wasserstrom, L.; Walther, A.; Wendland, J. Analysis of the cell wall integrity pathway of Ashbya gossypii. Microbiol Res. 2013, 168, 607–614. [Google Scholar] [CrossRef]
- Castrejon, F.; Gomez, A.; Sanz, M.; Duran, A.; Roncero, C. The Rim101 pathway contributes to yeast cell wall assembly and its function becomes essential in the absence of mitogen-activated protein kinase Slt2p. Eukaryot. Cell 2006, 5, 507–517. [Google Scholar] [CrossRef] [Green Version]
- Piccirillo, S.; White, M.G.; Murphy, J.C.; Law, D.J.; Honigberg, S.M. The Rim101p/PacC pathway and alkaline pH regulate pattern formation in yeast colonies. Genetics 2010, 184, 707–716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wasserstrom, L.; Dunkler, A.; Walther, A.; Wendland, J. The APSES protein Sok2 is a positive regulator of sporulation in Ashbya gossypii. Mol. Microbiol. 2017, 106, 949–960. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| No | Strain | Genotype | Source |
|---|---|---|---|
| 71 | ATCC10895 | Agleu2 | Lab collection |
| C420 | AWL13 | leu2, rim101:: GEN3/RIM101 heterokaryon | This study |
| C510 | AWL62 | leu2, rim101:: GEN3/RIM101 heterokaryon | This study |
| C511 | AWL63 | leu2, rim101:: GEN3 homokaryon derived from AWL62 | This study |
| C512 | AWL64 | leu2, rim101:: GEN3 homokaryon derived from C420 | This study |
| Oligonucleotide | Sequence 5′-to-3′ * |
|---|---|
| #1214-G2 | GGGTAATTTGTCGCGGTCTGGG |
| #1215-G3 | GCCCATCAGATTGATGTCCTCC |
| #4603-G1-RIM101 | GCTGCTATCGGACGCAGC |
| #4604-G4-RIM101 | GTGGAGTGCGACTTCTCC |
| #4605-S1-RIM101 | ACGATTATACGGCCAGCCAAATAGCAAGCGCGTTACTGCATGAACgaagcttcgtacgctgcaggtc |
| #4606-S2-RIM101 | ATTATGTTCACAGTCTGGAGTTCTGCACGGAGGGCATCAGTGTCGctgatatcatcgatgaattcgag |
| #4607-I1-RIM101 | CACTTATGCCAGGATCACG |
| #4608-I2-RIM101 | GCTCCCGGAGAGGTCACC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wasserstrom, L.; Wendland, J. Role of RIM101 for Sporulation at Alkaline pH in Ashbya gossypii. J. Fungi 2021, 7, 527. https://doi.org/10.3390/jof7070527
Wasserstrom L, Wendland J. Role of RIM101 for Sporulation at Alkaline pH in Ashbya gossypii. Journal of Fungi. 2021; 7(7):527. https://doi.org/10.3390/jof7070527
Chicago/Turabian StyleWasserstrom, Lisa, and Jürgen Wendland. 2021. "Role of RIM101 for Sporulation at Alkaline pH in Ashbya gossypii" Journal of Fungi 7, no. 7: 527. https://doi.org/10.3390/jof7070527






