Inducible Selectable Marker Genes to Improve Aspergillus fumigatus Genetic Manipulation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Media and Growth Conditions
| Strains | Genotype | Reference |
|---|---|---|
| A1160P+(wt) | ∆ku80, pyrG+ | Fraczek et al. [23] |
| A52 | FGSC A52 (bi1 thi4) | ATCC® 24761™ |
| ΔfcyB-hph | ΔfcyB::hph | Gsaller et al. [25] |
| ΔfcyB-ptrA | ΔfcyB::ptrA | This study |
| ΔfcyB-hphxyl | ΔfcyB::PxylP-hph | This study |
| ΔfcyB-ptrAxyl | ΔfcyB::PxylP-ptrA | This study |
| ΔpksP-hphxyl | ΔpksP::PxylP-hph | This study |
| ΔpksP-ptrAxyl | ΔpksP::PxylP-ptrA | This study |
2.2. Generation of Transformation Constructs
2.3. Fungal Transformation
2.4. Nucleic Acid Manipulation, Southern and Northern Blot Analysis
2.5. Spot Assays on Plate
2.6. Microtiter-Plate Assays
3. Results and Discussion
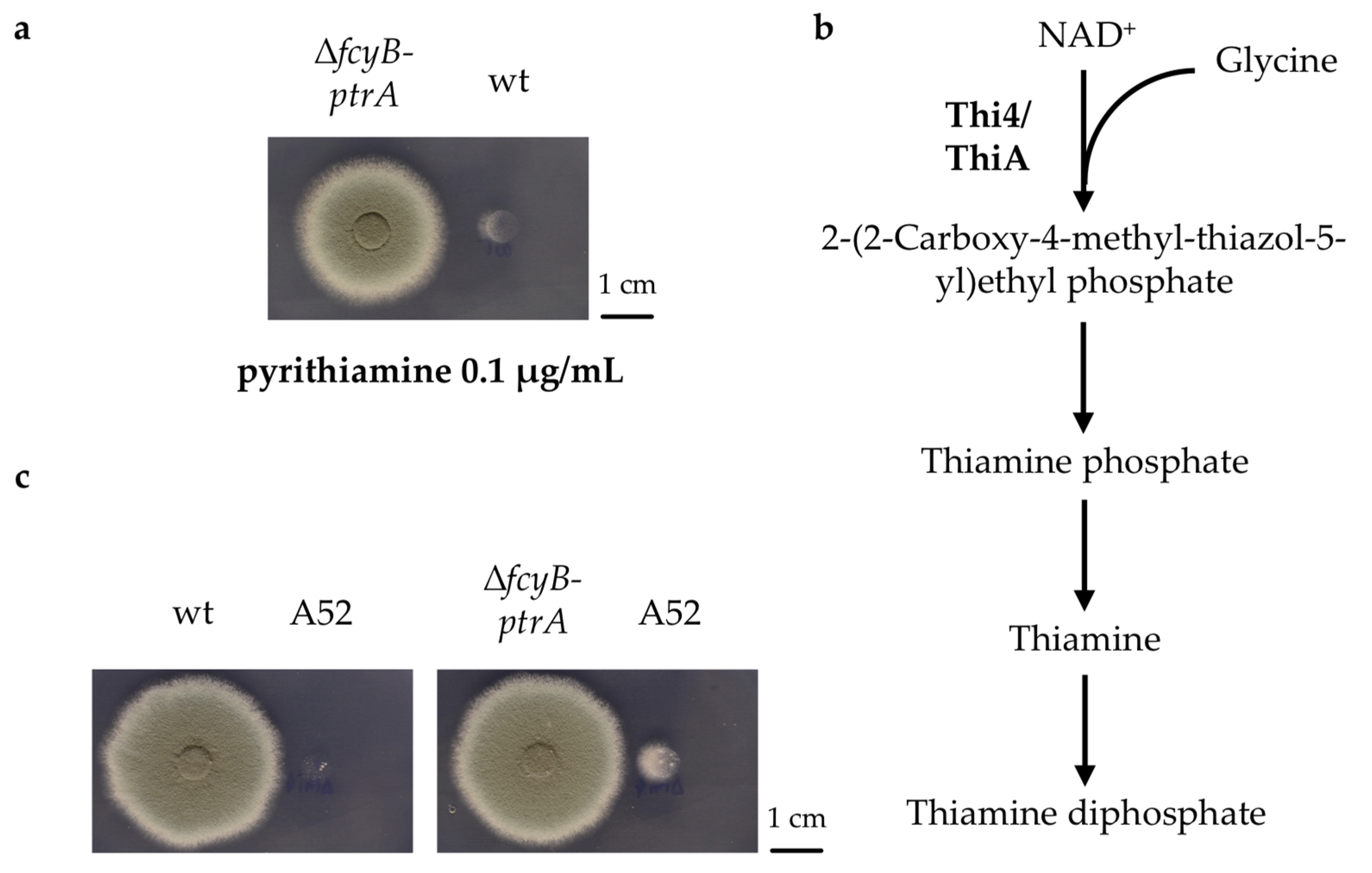
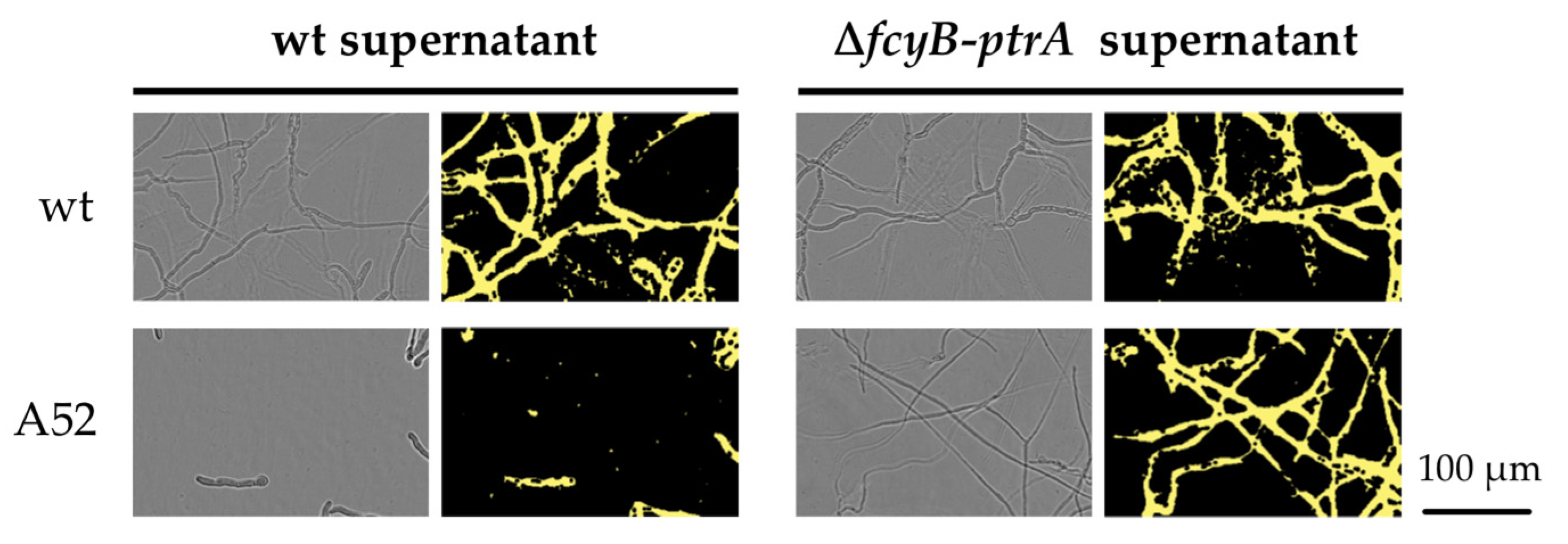
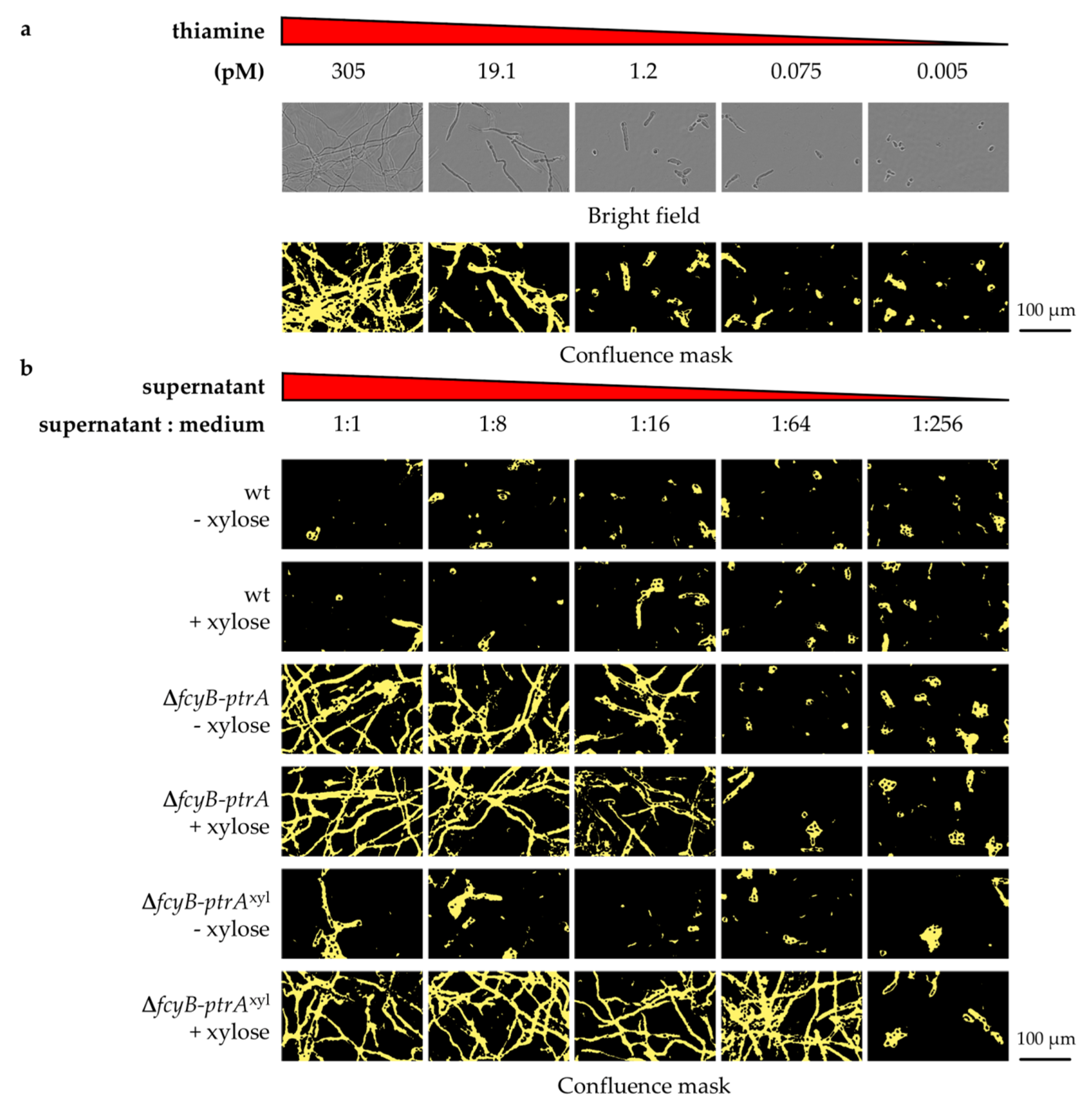
3.1. The Pyrithiamine Resistance Cassette Induces the Production of One or More Metabolites Able to Complement Thiamine Auxotrophy
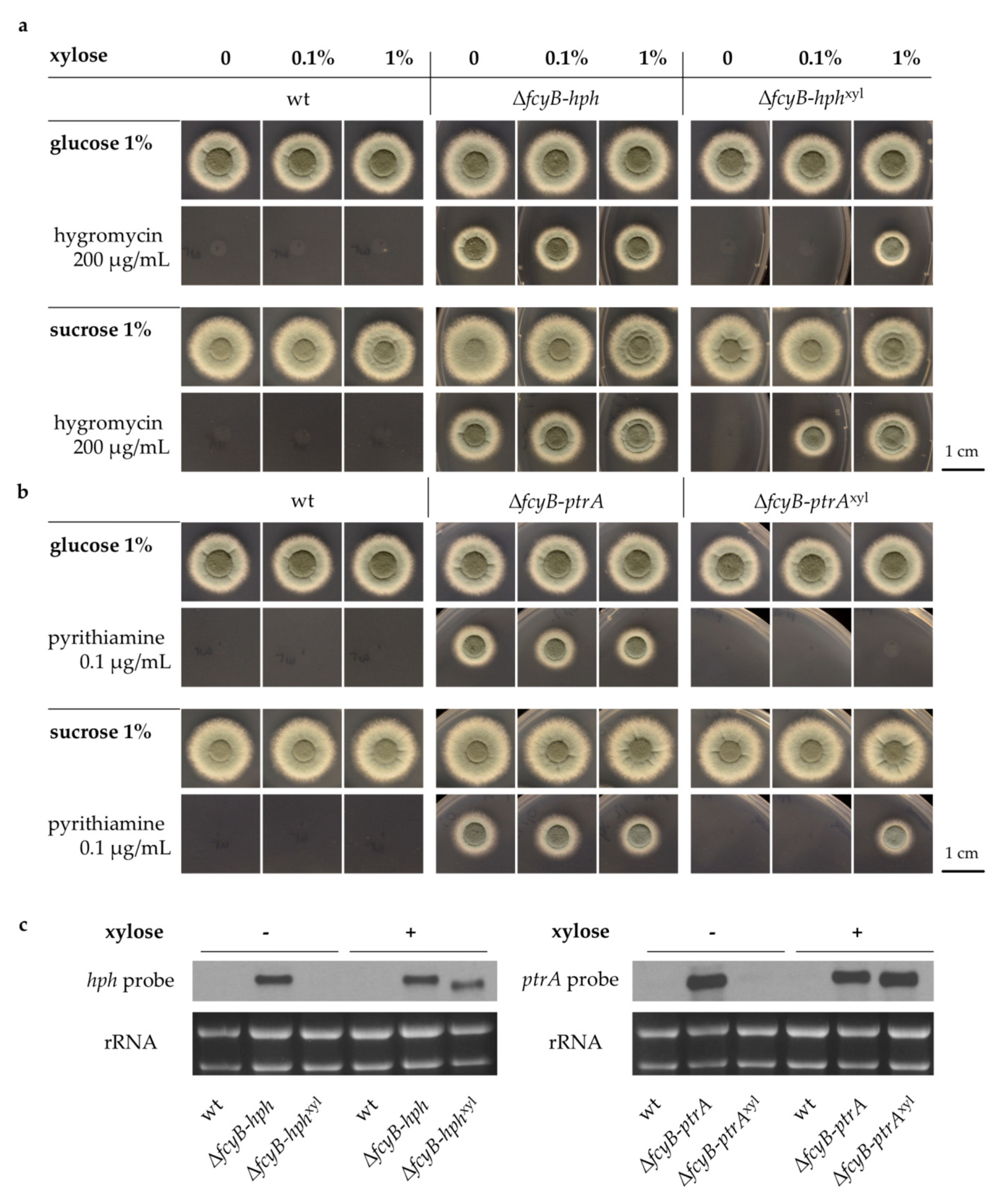
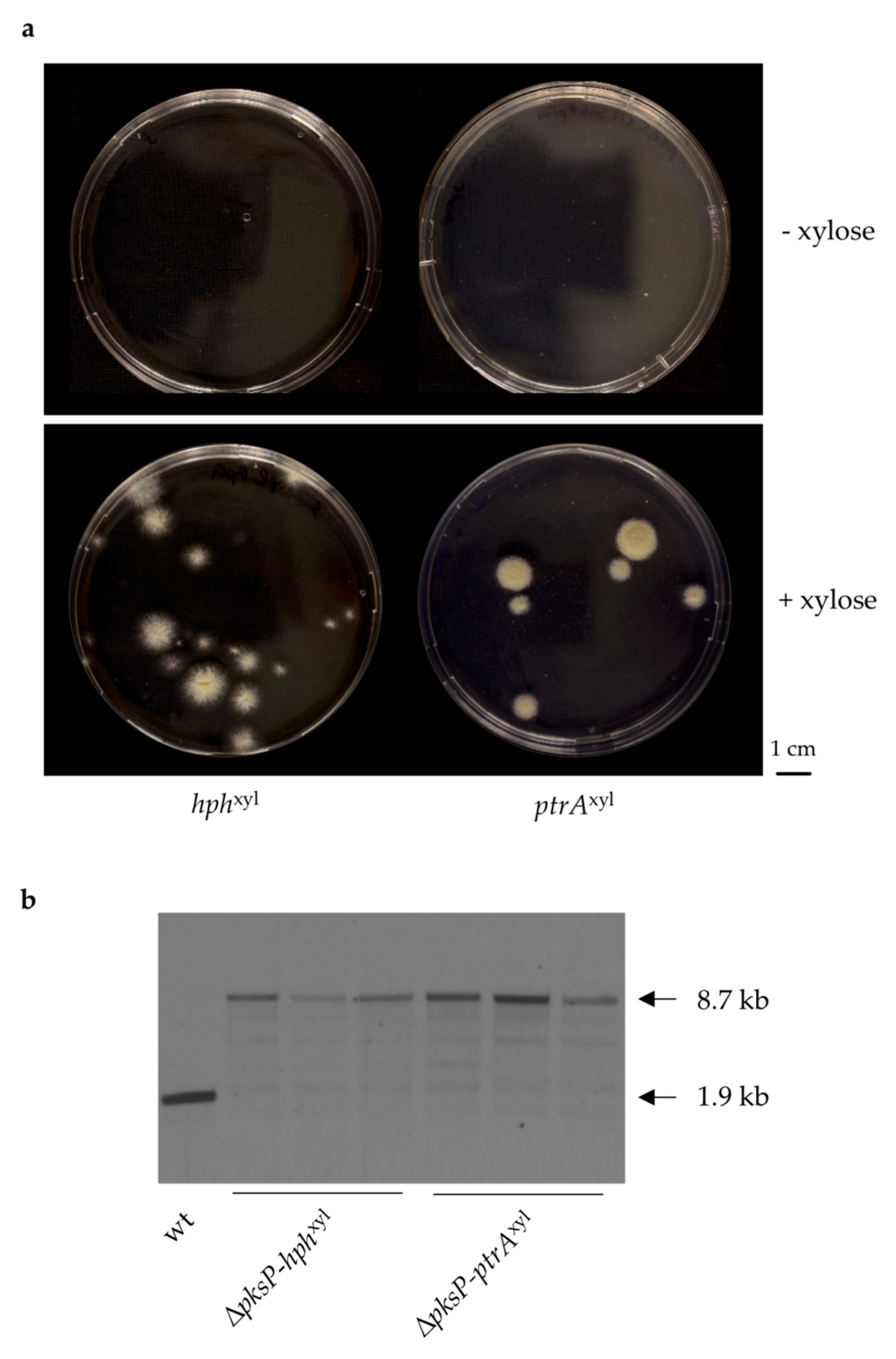
3.2. Generation of A. fumigatus Strains with Inducible Resistance Marker Cassettes
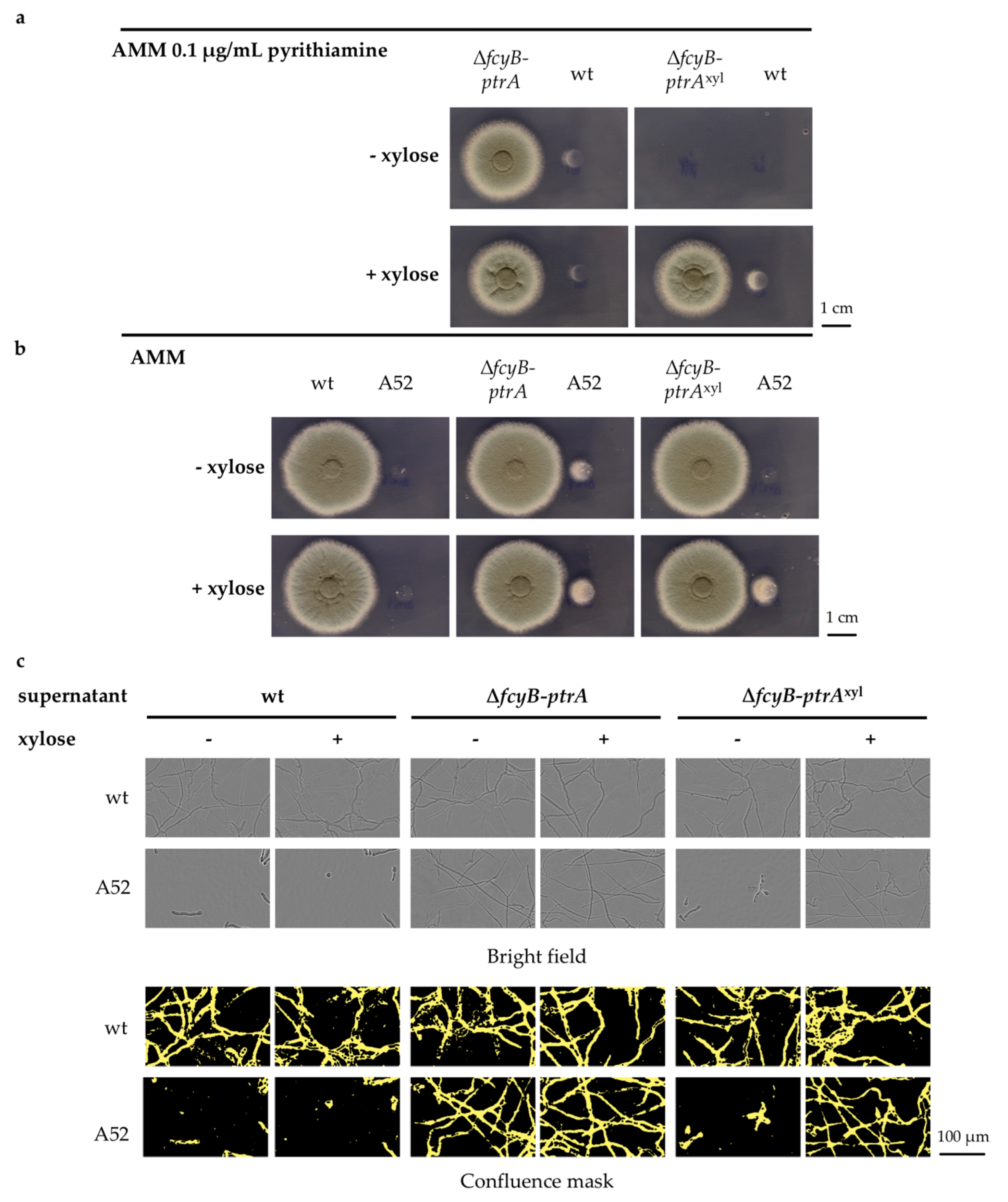
3.3. Validation of the Inducible Marker Cassettes as Selectable Marker in A. fumigatus Transformation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bongomin, F.; Gago, S.; Oladele, R.O.; Denning, D.W. Global and Multi-National Prevalence of Fungal Diseases—Estimate Precision. J. Fungi 2017, 3, 57. [Google Scholar] [CrossRef] [PubMed]
- Fickers, P.; Le Dall, M.; Gaillardin, C.; Thonart, P.; Nicaud, J. New disruption cassettes for rapid gene disruption and marker rescue in the yeast Yarrowia lipolytica. J. Microbiol. Methods 2003, 55, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Gaken, J.; Farzaneh, F.; Stocking, C. Construction of a versatile set of retroviral vectors conferring hygromycin resistance. Biotechniques 1992, 13, 32–34. [Google Scholar]
- Goldstein, A.L.; McCusker, J.H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 1999, 15, 1541–1553. [Google Scholar] [CrossRef]
- Hua, J.; Meyer, J.D.; Lodge, J.K. Development of Positive Selectable Markers for the Fungal Pathogen Cryptococcus neoformans. Clin. Diagn. Lab. Immunol. 2000, 7, 125–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lama, J.; Carrasco, L. Expression of poliovirus nonstructural proteins in Escherichia coli cells. Modification of membrane permeability induced by 2B and 3A. J. Biol. Chem. 1992, 267, 15932–15937. [Google Scholar] [CrossRef]
- Poyedinok, N.L.; Blume, Y.B. Advances, Problems, and Prospects of Genetic Transformation of Fungi. Cytol. Genet. 2018, 52, 139–154. [Google Scholar] [CrossRef]
- Smulian, A.G.; Gibbons, R.S.; Demland, J.A. Expression of hygromycin phosphotransferase alters virulence of Histoplasma capsulatum. Eukaryot. Cell 2007, 6, 2066–2071. [Google Scholar] [CrossRef] [Green Version]
- Soares Rde, B.; Velho, T.A.; De Moraes, L.M. Hygromycin B-resistance phenotype acquired in Paracoccidioides brasiliensis via plasmid DNA integration. Med. Mycol. 2005, 43, 719–723. [Google Scholar] [CrossRef] [Green Version]
- Punt, P.J.; Oliver, R.P.; Dingemanse, M.A.; Pouwels, P.H.; Hondel, C.A.V.D. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene 1987, 56, 117–124. [Google Scholar] [CrossRef]
- Kubodera, T.; Yamashita, N.; Nishimura, A. Pyrithiamine Resistance Gene (ptrA) of Aspergillus oryzae: Cloning, Characterization and Application as a Dominant Selectable Marker for Transformation. Biosci. Biotechnol. Biochem. 2000, 64, 1416–1421. [Google Scholar] [CrossRef] [PubMed]
- Kubodera, T.; Watanabe, M.; Yoshiuchi, K.; Yamashita, N.; Nishimura, A.; Nakai, S.; Gomi, K.; Hanamoto, H. Thiamine-regulated gene expression of Aspergillus oryzae thiA requires splicing of the intron containing a riboswitch-like domain in the 5′-UTR. FEBS Lett. 2003, 555, 516–520. [Google Scholar] [CrossRef] [Green Version]
- Todokoro, T.; Bando, H.; Kotaka, A.; Tsutsumi, H.; Hata, Y.; Ishida, H. Identification of a novel pyrithiamine resistance marker gene thiI for genome co-editing in Aspergillus oryzae. J. Biosci. Bioeng. 2020, 130, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Iwakuma, H.; Koyama, Y.; Miyachi, A.; Nasukawa, M.; Matsumoto, H.; Yano, S.; Ogihara, J.; Kasumi, T. Generation of a glucose de-repressed mutant of Trichoderma reesei using disparity mutagenesis. Biosci. Biotechnol. Biochem. 2016, 80, 486–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubodera, T.; Yamashita, N.; Nishimura, A. Transformation of Aspergillus sp.; Trichoderma reesei using the pyrithiamine resistance gene (ptrA) of Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2002, 66, 404–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Son, Y.E.; Park, H.S. Genetic Manipulation and Transformation Methods for Aspergillus spp. Mycobiology 2020. [Google Scholar] [CrossRef]
- Shoji, J.Y.; Maruyama, J.; Arioka, M. Development of Aspergillus oryzae thiA promoter as a tool for molecular bio-logical studies. FEMS Microbiol. Lett. 2005, 244, 41–46. [Google Scholar] [CrossRef] [Green Version]
- He, B.; Tu, Y.; Jiang, C.; Zhang, Z.; Li, Y.; Zeng, B. Functional Genomics of Aspergillus oryzae: Strategies and Progress. Microorganisms 2019, 7, 103. [Google Scholar] [CrossRef] [Green Version]
- Mignon, C.; Sodoyer, R.; Werle, B. Antibiotic-Free Selection in Biotherapeutics: Now and Forever. Pathogens 2015, 4, 157–181. [Google Scholar] [CrossRef]
- Zuo, J.; Niu, Q.W.; Ikeda, Y. Marker-free transformation: Increasing transformation frequency by the use of regeneration-promoting genes. Curr. Opin. Biotechnol. 2002, 13, 173–180. [Google Scholar] [CrossRef]
- Zadra, I.; Abt, B.; Parson, W.; Haas, H. xylP Promoter-Based Expression System and Its Use for Antisense Downregulation of the Penicillium chrysogenum Nitrogen Regulator NRE. Appl. Environ. Microbiol. 2000, 66, 4810–4816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bauer, I.; Misslinger, M.; Shadkchan, Y. The Lysine Deacetylase RpdA Is Essential for Virulence in Aspergillus fumigatus. Front. Microbiol. 2019, 10, 2773. [Google Scholar] [CrossRef] [Green Version]
- Fraczek, M.G.; Bromley, M.; Buied, A. The cdr1B efflux transporter is associated with non-cyp51a-mediated itraconazole resistance in Aspergillus fumigatus. J. Antimicrob. Chemother. 2013, 68, 1486–1496. [Google Scholar] [CrossRef]
- Pontecorvo, G.; Roper, J.; Chemmons, L.; Macdonald, K.; Bufton, A. The Genetics of Aspergillus nidulans. Adv. Genet. 1953, 5, 141–238. [Google Scholar]
- Gsaller, F.; Furukawa, T.; Carr, P.D. Mechanistic Basis of pH-Dependent 5-Flucytosine Resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef] [Green Version]
- Birštonas, L.; DalleMulle, A.; López-Berges, M.S.; Jacobsen, I.D.; Offterdinger, M.; Abt, B.; Straßburger, M.; Bauer, I.; Schmidt, O.; Sarg, B.; et al. Multiplex Genetic Engineering Exploiting Pyrimidine Salvage Pathway-Based Endogenous Counterselectable Markers. mBio 2020, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hortschansky, P.; Eisendle, M.; Al-Abdallah, Q.; Schmidt, A.D.; Bergmann, S.; Thön, M.; Kniemeyer, O.; Abt, B.; Seeber, B.; Werner, E.R.; et al. Interaction of HapX with the CCAAT-binding complex—A novel mechanism of gene regulation by iron. EMBO J. 2007, 26, 3157–3168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dietl, A.-M.; Meir, Z.; Shadkchan, Y.; Osherov, N.; Haas, H. Riboflavin and pantothenic acid biosynthesis are crucial for iron homeostasis and virulence in the pathogenic mold Aspergillus fumigatus. Virulence 2018, 9, 1036–1049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baldin, C.; Valiante, V.; Krüger, T.; Schafferer, L.; Haas, H.; Kniemeyer, O.; Brakhage, A.A. Comparative proteomics of a tor inducible Aspergillus fumigatus mutant reveals involvement of the Tor kinase in iron regulation. Proteomics 2015, 15, 2230–2243. [Google Scholar] [CrossRef]
- Langfelder, K.; Jahn, B.; Gehringer, H.; Schmidt, A.; Wanner, G.; Brakhage, A.A. Identification of a polyketide synthase gene (pksP) of Aspergillus fumigatus involved in conidial pigment biosynthesis and virulence. Med. Microbiol. Immunol. 1998, 187, 79–89. [Google Scholar] [CrossRef]
- Tsai, H.F.; Wheeler, M.H.; Chang, Y.C. A developmentally regulated gene cluster involved in conidial pigment bio-synthesis in Aspergillus fumigatus. J. Bacteriol. 1999, 181, 6469–6477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Rhijn, N.; Furukawa, T.; Zhao, C.; McCann, B.L.; Bignell, E.; Bromley, M.J. Development of a marker-free mutagenesis system using CRISPR-Cas9 in the pathogenic mould Aspergillus fumigatus. Fungal Genet. Biol. 2020, 145, 103479. [Google Scholar] [CrossRef]
- Morio, F.; Lombardi, L.; Butler, G. The CRISPR toolbox in medical mycology: State of the art and perspectives. PLoS Pathog. 2020, 16, e1008201. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, T.; Dumig, M.; Jaber, B.M. Validation of a self-excising marker in the human pathogen Aspergillus fumigatus by employing the beta-rec/six site-specific recombination system. Appl. Environ. Microbiol. 2010, 76, 6313–6317. [Google Scholar] [CrossRef] [Green Version]
- Balabanova, L.A.; Shkryl, Y.N.; Slepchenko, L.V.; Yugay, Y.A.; Gorpenchenko, T.Y.; Kirichuk, N.N.; Khudyakova, Y.V.; Bakunina, I.Y.; Podvolotskaya, A.; Bulgakov, V. Development of host strains and vector system for an efficient genetic transformation of filamentous fungi. Plasmid 2019, 101, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yau, Y.Y.; Stewart, C.N., Jr. Less is more: Strategies to remove marker genes from transgenic plants. BMC Biotechnol. 2013, 13, 36. [Google Scholar] [CrossRef] [Green Version]
- Bertuzzi, M.; Van Rhijn, N.; Krappmann, S.; Bowyer, P.; Bromley, M.J.; Bignell, E.M. On the lineage of Aspergillus fumigatus isolates in common laboratory use. Med. Mycol. 2021, 59, 7–13. [Google Scholar] [CrossRef]
- Cheng, S.; Nguyen, M.H.; Zhang, Z.; Jia, H.; Handfield, M.; Clancy, C.J. Evaluation of the Roles of Four Candida albicans Genes in Virulence by Using Gene Disruption Strains That Express URA3 from the Native Locus. Infect. Immun. 2003, 71, 6101–6103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baldin, C.; Kühbacher, A.; Merschak, P.; Sastré-Velásquez, L.E.; Abt, B.; Dietl, A.-M.; Haas, H.; Gsaller, F. Inducible Selectable Marker Genes to Improve Aspergillus fumigatus Genetic Manipulation. J. Fungi 2021, 7, 506. https://doi.org/10.3390/jof7070506
Baldin C, Kühbacher A, Merschak P, Sastré-Velásquez LE, Abt B, Dietl A-M, Haas H, Gsaller F. Inducible Selectable Marker Genes to Improve Aspergillus fumigatus Genetic Manipulation. Journal of Fungi. 2021; 7(7):506. https://doi.org/10.3390/jof7070506
Chicago/Turabian StyleBaldin, Clara, Alexander Kühbacher, Petra Merschak, Luis Enrique Sastré-Velásquez, Beate Abt, Anna-Maria Dietl, Hubertus Haas, and Fabio Gsaller. 2021. "Inducible Selectable Marker Genes to Improve Aspergillus fumigatus Genetic Manipulation" Journal of Fungi 7, no. 7: 506. https://doi.org/10.3390/jof7070506
APA StyleBaldin, C., Kühbacher, A., Merschak, P., Sastré-Velásquez, L. E., Abt, B., Dietl, A.-M., Haas, H., & Gsaller, F. (2021). Inducible Selectable Marker Genes to Improve Aspergillus fumigatus Genetic Manipulation. Journal of Fungi, 7(7), 506. https://doi.org/10.3390/jof7070506






