Abstract
Mitogen activated protein kinase (MAPK) signaling pathways execute essential functions in eukaryotic organisms by transducing extracellular stimuli into adaptive cellular responses. In the fission yeast model Schizosaccharomyces pombe the cell integrity pathway (CIP) and its core effector, MAPK Pmk1, play a key role during regulation of cell integrity, cytokinesis, and ionic homeostasis. Schizosaccharomyces japonicus, another fission yeast species, shows remarkable differences with respect to S. pombe, including a robust yeast to hyphae dimorphism in response to environmental changes. We show that the CIP MAPK module architecture and its upstream regulators, PKC orthologs Pck1 and Pck2, are conserved in both fission yeast species. However, some of S. pombe’s CIP-related functions, such as cytokinetic control and response to glucose availability, are regulated differently in S. japonicus. Moreover, Pck1 and Pck2 antagonistically regulate S. japonicus hyphal differentiation through fine-tuning of Pmk1 activity. Chimeric MAPK-swapping experiments revealed that S. japonicus Pmk1 is fully functional in S. pombe, whereas S. pombe Pmk1 shows a limited ability to execute CIP functions and promote S. japonicus mycelial development. Our findings also suggest that a modified N-lobe domain secondary structure within S. japonicus Pmk1 has a major influence on the CIP signaling features of this evolutionarily diverged fission yeast.
Keywords:
fission yeast; MAPK; cell integrity pathway; S. japonicus; S. pombe; protein kinase C; Pmk1; dimorphism; hyphae 1. Introduction
The evolutionarily conserved MAPK signaling pathways orchestrate multiple and fundamental cellular functions in eukaryotic organisms that assure cell proliferation and survival during unperturbed conditions and in response to environmental stimuli. The intensely studied fission yeast model Schizosaccharomyces pombe has three MAPK cascades, known as the stress-activated protein kinase (SAPK) pathway, the cell integrity pathway (CIP), and the pheromone sexual differentiation pathway [1]. Specifically, the CIP regulates a wide range of processes in S. pombe, including cell wall composition and integrity, vacuole fusion, cytokinesis, and ionic homeostasis [1,2,3,4]. The core effector of the CIP is MAPK Pmk1, an ERK1/2 ortholog that is the downstream target of a MAPK module composed by MAPKKK Mkh1 and MAPKK Pek1 [1,5]. S. pombe Pmk1 becomes activated in a cell-cycle-dependent manner during cytokinesis [6], during nutritional changes such as glucose fasting [3], or in response to environmental insults including heat, cell wall damage, osmotic-saline, and oxidative stresses [5,6]. The architecture of the CIP upstream of the Mkh1–Pek1–Pmk1 MAPK module includes two redundant protein kinase C (PKC) orthologs named Pck1 and Pck2 [6,7,8,9,10,11]. Pck1 and Pck2 share overlapping roles in cell viability and partially complement each other, and their simultaneous deletion elicits a synthetic lethal phenotype [1]. Pck2 operates downstream of the Rho GTPases Rho1 and Rho2, and is the major positive regulator of CIP activity during unperturbed growth and stress, whereas a Rho1–Pck1 branch contributes scarcely to Pmk1 activation during vegetative growth and in response to cell wall stress [6,7,8,9,10,11,12]. A complex cross-regulatory relationship between the CIP and TOR signaling that involves the AKT (Gad8)-dependent translation of both Pck1 and Pck2, allows activation of the CIP during growth and stress [13]. However, Rho1 and Rho2 can also synergistically regulate the biosynthesis of the two major S. pombe cell wall polymers α- and β-D-glucan through Pck1 and Pck2 and independently of CIP signaling [1]. The CIP and SAPK pathways functionally interact In Vivo, and share common targets including the transcription factor Atf1 and the RNA binding protein Rnc1 [14].
S. japonicus, another fission yeast species, is becoming an emerging model organism to explore developmental and physiological evolutionary changes within the genus Schizosaccharomyces thanks to its distinctive features with respect to S. pombe. Indeed, S. japonicus cells display unusual membrane lipid composition [15], and divide through semi-open mitosis, as opposed to the typical closed mitosis of S. pombe cells [16]. In addition, S. japonicus utilizes glucose exclusively via fermentation and is essentially respiration defective, whereas S. pombe is a respiro-fermentative organism [17]. Most importantly, and unlike the remaining species of the genus, S. japonicus yeast cells have the ability to undergo a vigorous and reversible dimorphic switch to hyphae in response to environmental stimuli. Known inducers of hyphal differentiation in this organism include changes in the composition of the growth medium, and DNA damage triggered with camptothecin (CPT), a specific topoisomerase I inhibitor [18,19,20,21].
The S. japonicus proteome carries gene orthologs encoding the main components of the three MAP kinase pathways present in S. pombe [22]. We have recently described that the stress-responsive functions of the SAPK pathway in S. pombe are largely conserved in S. japonicus, and include regulation of G2/M progression during the cell cycle, cellular adaptation to multiple stress conditions, and the positive control of chronological lifespan and sexual differentiation [23]. Notably, the SAPK pathway acts in concert with a quorum sensing mechanism mediated by aromatic alcohols that represses S. japonicus yeast to hypha differentiation in response to increased population density [23]. In contrary, S. japonicus mutants lacking the putative CIP MAP kinase module components Mkh1, Pek1, and MAPK Pmk1 display defective mycelia formation in solid medium [22], suggesting that this signaling pathway may play instead a positive role to modulate yeast to hypha dimorphic transition. However, the precise biological functions of the S. japonicus CIP during growth, stress, and mycelial differentiation, together with the possible role of PKC orthologs Pck1 and Pck2 as upstream regulators of this signaling cascade, are currently unknown. In this work we show that, despite the existence of some degree of functional conservation, certain biological features of CIP signaling have evolved separately in S. japonicus and S. pombe, and that Pck1 and Pck2 modulate Pmk1 MAPK activity during S. japonicus dimorphic switch in an opposite fashion. Finally, structural changes at the N-lobe of S. japonicus Pmk1 likely played a significant role in modulating CIP signaling to cope with the specific developmental requirements of this fission yeast species.
2. Materials and Methods
2.1. Strains, Growth Conditions, and Reagents
References and genotypes of S. pombe and S. japonicus strains employed in this work are listed in Table S1. They were grown with shaking at 30 °C in yeast extract plus supplements (YES) or Edinburgh minimal medium (EMM2) with 2% glucose, and supplemented with adenine, leucine, histidine, or uracil (100 mg/L, Sigma-Aldrich, St. Louis, MO, USA) [24]. Mutant strains with single/multiple gene deletions, or expressing genomic fusions, were obtained by transformation, or after tetrad or random spore dissection and analysis of appropriate crosses in sporulation medium (SPA) [25]. In stress treatments, log-phase cultures (OD600 = 0.5; ~106 cells/mL) were supplemented with either KCl (osmotic-saline stress; Sigma-Aldrich), caspofungin (cell wall stress; Sigma-Aldrich), or hydrogen peroxide (oxidative stress; Sigma-Aldrich). In glucose starvation experiments, cells grown in YES medium with 7% glucose were recovered by filtration, and resuspended in the same medium lacking glucose and osmotically equilibrated with 3% glycerol. In yeast to hyphae induction experiments with S. japonicus, log-phase cultures grown in YES medium with 6% glucose were treated with camptothecin (CPT; Sigma-Aldrich) to a final concentration of 0.2 μM. YEMA [18] and RGE [26] media supplemented with 2% agar were used to quantify S. japonicus mycelial growth. S. japonicus transformation by electroporation was performed exactly as described [27].
2.2. Gene Disruption and Gene Fusion
Sequences of S. japonicus genes were obtained from the annotated database at EnsemblFungi (http://fungi.ensembl.org/Schizosaccharomyces_japonicus/Info/Index?db=core (accessed on 14 June 2021)). The S. japonicus pck1+, pck2+, and pmk1+ null mutants were obtained by entire deletion of the corresponding coding sequences by PCR-mediated strategy using plasmids pFK14 (S. japonicus ura4+ gene cloned into pGEMT-easy vector; [28]) or pFA6a-natMX6 [29] as templates, and their replacement with either ura4+ or natMX6 cassettes flanked by long 5′ and 3′UTRs of respective genes following a PCR approach [30]. Oligonucleotides employed to obtain each one of the transformation cassettes are shown in Table S2.
To construct the integrative plasmid pFA6a-Pmk1Sj:GFP-natMX6, the pmk1+Sj ORF plus 5′ regulatory sequences were amplified by PCR using S. japonicus genomic DNA as a template, the 5’-oligonucleotide Pmk1Sj:GFP-SalI-5′, which hybridizes −309 to −288 bp upstream of the pmk1+Sj start codon and contains a SalI site, and the 3′-oligonucleotide Pmk1Sj:GFP-PacI-3’, which hybridizes at the 3′ end of pmk1+Sj ORF before the stop codon, and contains a PacI site. The obtained DNA fragment was cloned into plasmid pFA6a-GFP:natMX6 [29] digested with SalI and PacI. Plasmid pFA6a-Pmk1Sj:GFP-natMX6 was then digested at the unique SwaI site within pmk1+Sj ORF, and transformed into strain NIG2028. NatR transformants were obtained, and the correct integration and expression of the fusion was verified by DNA sequencing and Western blot analysis.
2.3. Construction of S. pombe and S. japonicus Strains Expressing Wild-Type and Chimeric Pmk1–HA Fusions
To obtain the integrative plasmid pSL-Pmk1Sj:HA, the pmk1+Sj ORF plus 5′ regulatory sequences were amplified by PCR using S. japonicus genomic DNA as a template, the 5′-oligonucleotide PromPmk1Jp-NotI-5′, which hybridizes at positions −498 to −475 bp upstream of the pmk1+ start codon and contains a NotI site, and the 3′-oligonucleotide Pmk1Jp:HA-BamHI-3′, which hybridizes at the 3′ end of pmk1+Sj ORF and incorporates an additional 33-nucleotide sequence encoding one HA epitope (sequence -GYPYDVPDYAG), and contains a BamHI site. The obtained PCR fragment was digested with NotI and BamHI and cloned into plasmid pSL, which is a derivative of plasmid pS0550 (kindly provided by S. Oliferenko) that incorporates the nmt1+ 3′ regulatory sequences. To obtain integrative plasmid pSL-Pmk1Sj(ins 82-105)–HA, we followed a modular PCR-based approach employing S. japonicus genomic DNA and different nucleotide pairs. A first DNA fragment was amplified with the oligonucleotide Pmk1Jp-EXON-5’, which hybridizes 620 to 691 bp downstream of pmk1+Sp ORF and incorporates a 71-nucleotide sequence encoding the 24 aa motif present in Pmk1Sp N-lobe, and 3′-oligonucleotide Pmk1Jp:HA-BamHI-3′. A second DNA fragment was amplified with 5′-oligonucleotide PromPmk1Jp-NotI-5′ and 3′-oligonucleotide Pmk1Jp-EXON-3’, which hybridizes 424 to 448 bp downstream of pmk1+Sj ORF plus and incorporates a reverse complement 71-nucleotide sequence from Pmk1Sp ORF described above. Both DNA fragments were gel purified and used as templates in a second-round PCR in the presence of PromPmk1Jp-NotI-5′ and Pmk1Jp:HA-BamHI-3′. The resulting PCR fragment was digested with NotI/BamHI and cloned into plasmid pSL. To obtain integrative plasmid pSL-Pmk1sp:HA6H, the pmk1+Sp ORF plus regulatory sequences were amplified by PCR using genomic DNA from S. pombe strain MM2 as template [5], and 5′-oligonucleotide PromPmk1Sp-XbaI-5′, which hybridizes at positions 330 to 313 upstream of the pmk1+Sp start codon and contains a XbaI site, and the 3′-oligonucleotide Pmk1Sp–HA-3′, which hybridizes at the 3′ end of pmk1+Sp ORF and incorporates a 48-nucleotide sequence encoding one HA epitope plus six consecutive histidine residues, and a SmaI site. The DNA fragment was digested with XbaI and SmaI and cloned into plasmid pSL. For plasmid pSL-Pmk1Sp (Δ82-105):HA, the pmk1+Sp ORF plus regulatory sequences were amplified in by PCR using S. pombe genomic DNA as template, the 5′-oligonucleotide PromPmk1Sp-XbaI-5′, and 3′-oligonucleotide Pmk1Sp-ΔExon-3′, which hybridizes −326 to −346 bp upstream and 774 to 792 bp downstream of pmk1+Sp ORF. A second DNA fragment was amplified with the 5′-oligonucleotide Pmk1Sp-ΔExon-5′, which is complementary and the reverse of Pmk1Sp-ΔExon-3′, and 3′-oligonucleotide Pmk1Sp–HA-3′. Both fragments were gel purified and used in a second-round PCR as templates in the presence of the external oligonucleotides PromPmk1Sp-XbaI-5′ and Pmk1Sp–HA-3′. The resulting PCR fragment was digested with XbaI and SmaI and cloned into plasmid pSL. All of the above plasmids were digested at the unique AfeI site within ura4+Sj, and the linear fragments were transformed into the S. japonicus pmk1+::NatR strain EGJ83. ura4+ transformants were obtained, and the correct integration and expression of the HA fusions was verified by sequencing and Western blot analysis.
To express the chimeric constructs described above in S. pombe, the respective Pmk1–HA fused cassettes plus 5′ regulatory sequences were gel purified from plasmids pSL-Pmk1Sj:HA, pSL-Pmk1Sj(ins 82-105)–HA, pSL-Pmk1Sp:HA, and pSL-Pmk1Sp(Δ82-105):HA after digestion with XbaI and SmaI, and cloned into S. pombe integrative plasmid pJK148. The resulting plasmid constructs were digested at the unique NruI site within Leu1+Sp, and the linear fragments were transformed into the S. pombe pmk1+::NatR strain MM1904. Leu1+ transformants were obtained, and the correct integration and expression of the fusions was verified by sequencing and Western blot analysis.
2.4. Detection and Quantification of Activated and Total Pmk1 Levels
Extracts from S. pombe and S. japonicus cells collected during unperturbed growth, or after stress or CPT treatments were prepared under native conditions employing chilled acid-washed glass beads and lysis buffer (10% glycerol, 50 mM Tris/HCl pH 7.5, 15 mM Imidazole, 150 mM NaCl, 0.1% Nonidet NP-40, plus specific protease and phosphatase inhibitor, Sigma-Aldrich), as described [31]. Dually phosphorylated Pmk1 was detected employing a rabbit polyclonal anti-phospho-p44/42antibody (9191L; Cell Signaling). Total Pmk1 levels were detected with either mouse monoclonal anti-HA (Pmk1–HA fusions; clone 12CA5; Roche Molecular Biochemicals, Basel, Switzerland), or anti-GFP antibody (Pmk1–GFP fusions; Roche Molecular Biochemicals). Rabbit polyclonal anti-Cdk1/Cdc2 antibody (PSTAIR; Millipore) was used for loading control. Immunoreactive bands were revealed with anti-mouse or anti-rabbit peroxidase-conjugated secondary antibodies (both from Sigma-Aldrich), and the ECL system (GE-Healthcare, Uppsala, Sweden). Densitometric quantification of Western blot signals from 16-bit .jpg digital images of blots was performed using ImageJ [32]. Relative units for Pmk1 activation were estimated by determining the signal ratio of the anti-phospho-p44/42 blot (activated Pmk1) with respect to the anti-HA or anti-GFP blot (total Pmk1) at each time point. Unless otherwise stated, results shown correspond to experiments performed as biological duplicates or triplicates. Mean relative units ± SD and/or representative results are shown. p-values were analyzed by unpaired Student’s t-test.
2.5. Co-immunoprecipitation
Whole-cell extracts (5 mg total protein) were prepared in lysis buffer (50 mM Tris/HCl pH 7.5, 0.5% sodium deoxicholate, 150 mM NaCl, 1% NP-40 and protease inhibitor (Sigma-Aldrich), and incubated with anti-HA monoclonal antibody (12CA5; Roche Molecular Biochemicals) and protein A sepharose beads for 4 h at 4 °C. The beads were washed two times with lysis buffer, two times with washing buffer 2 (50 mM Tris/HCl pH 7.5, 0.05% sodium deoxicholate, 500 mM NaCl, 0.1% NP-40), one time with washing buffer 3 (50 mM Tris/HCl pH 7.5, 0.05% sodium deoxicholate, 0.1% NP-40), and resuspended in sample buffer. Proteins were separated by SDS-PAGE, transferred to nitrocellulose filters (GE Healthcare), and hybridized with either mouse monoclonal anti-HA antibody (Pmk1–HA fusions; clone 12CA5; Roche Molecular Biochemicals), or anti-GFP antibody (Pek1–GFP fusions; Roche Molecular Biochemicals), followed by immunodetection with anti-mouse peroxidase-conjugated secondary antibody (Sigma-Aldrich), and the ECL system (GE Healthcare).
2.6. Plate Assay of Stress Sensitivity for Growth
S. pombe and S. japonicus wild-type and mutant strains were grown in YES liquid medium to OD600 = 0.5, and appropriate decimal dilutions were spotted per duplicate on YES solid medium or in the same medium supplemented with varying concentrations of caspofungin (Sigma-Aldrich), and magnesium chloride plus the calcineurin inhibitor FK506 (Tracholimus; Sigma-Aldrich). Plates were incubated at 30 °C for 3 days and then photographed. All the assays were repeated at least three times with similar results. Representative experiments are shown in the corresponding figures.
2.7. Quantification of Mycelial Growth during Nutritional Stress
Approximately 2.106 cells from log-phase cultures (OD600= 0.5) of wild-type and mutant strains growing in YES medium were spotted on YEMA or RGE plates, incubated at 30 °C for 7 days, and then photographed and saved as 16-bit .jpg digital images. The area of mycelial expansion was outlined for each strain (n ≥ 6) by freehand, and measured with ImageJ using the analyze tool [32].
2.8. Microscopy Observation
Fluorescence images in Figure 1C were acquired using a Leica Thunder imager by either fluorescence or DIC microscopy as single medial plane images from a set of 20 stacks (0.3 µm each), and processed through ImageJ [32]. Fluorescence images in Figure 1A, Figure 2D,E and Figure 4E were obtained with a Leica DM4000B microscope and processed using IM500 Image Manager software. Calcofluor white was employed for cell wall/septum staining as described [24]. To quantify the increase in cell length during hyphal induction experiments with CPT, samples were taken at the indicated times and fixed immediately with formaldehyde [33]. After staining with calcofluor white the length of mononuclear late G2 cells was measured. Three biological replicates (n ≥ 400) were scored for each strain genotype.
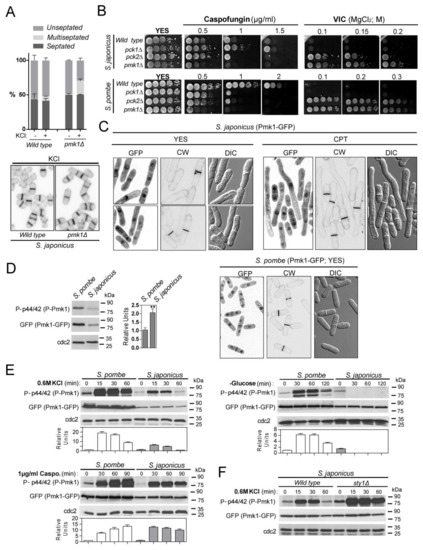
Figure 1.
Functional roles, localization, and activation pattern of the CIP MAPK Pmk1 in S. japonicus. (A) Upper: Exponentially growing S. japonicus wild-type and pmk1Δ strains were incubated for 10 h in YES medium plus 0.6M KCl, and the percentage of unseptated, septated, and multiseptated cells (represented as mean ± SD from biological triplicates; number of total cells ≥ 200) was determined by fluorescence microscopy after calcofluor white staining. Lower: Representative images are shown; (B) Decimal dilutions of the indicated strains growing in YES medium were spotted onto YES plates supplemented with caspofungin (0.5–2 µg/mL) (upper panels), and 0.5 µg/mL FK506 plus 0.1, 0.15, or 0.2 M MgCl2 (VIC assay; lower panels), and incubated for 3 days at 30 °C before being photographed. Representative experiments are shown; (C) S. japonicus and S. pombe wild-type cells expressing genomic Pmk1–GFP fusions were grown in YES medium to mid-log phase or treated for 6 h with 0.2 μM CPT (S. japonicus), and observed both by fluorescence and DIC microscopy after calcofluor white staining. Images were acquired as single medial plane images and are inverted for fluorescence; (D) S. pombe and S. japonicus wild-type strains expressing genomic Pmk1–GFP fusions were grown in YES medium to mid-log phase, and activated/total Pmk1 were detected with anti-phospho-p44/42 and anti-GFP antibodies, respectively. Anti-Cdc2 was used as a loading control. Relative units as mean ± SD (biological triplicates) for Pmk1 phosphorylation (anti-phospho-p44/42 blot) were determined with respect to the internal control (anti-GFP blot). **, p < 0.005; as calculated by unpaired Student’s t-test; (E) S. pombe and S. japonicus wild-type strains expressing genomic Pmk1–GFP fusions were grown in YES medium to mid-log phase, and treated with 0.6 M KCl, 1 µg/mL caspofungin, or recovered by filtration and resuspended in osmotically equilibrated medium lacking glucose for the indicated times. Activated/total Pmk1 was detected with anti-phospho-p44/42 and anti-GFP antibodies, respectively. Anti-Cdc2 was used as a loading control. Relative Pmk1 activity units as mean ± SD (biological duplicates) were determined as described in (C); (F) S. japonicus wild-type and sty1Δ strains expressing a genomic Pmk1–GFP fusion were grown in YES medium to mid-log phase, and treated with 0.6 M KCl for the indicated times. Activated/total Pmk1 were detected with anti-phospho-p44/42 and anti-GFP antibodies, respectively. Relative Pmk1 activity units as mean ± SD (biological duplicates) were determined as above.
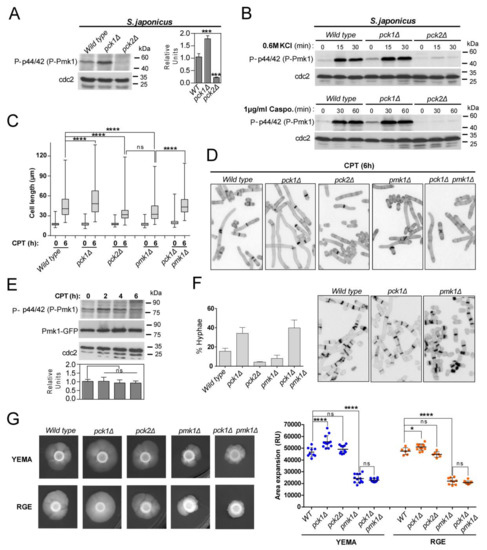
Figure 2.
Antagonistic control of Pmk1 activity by PKC orthologs Pck1 and Pck2 modulates S. japonicus hyphal differentiation. (A) S. japonicus strains of the indicated genotypes were grown in YES medium to mid-log phase, and activated Pmk1 were detected with anti-phospho-p44/42, whereas anti-Cdc2 was used as a loading control. Relative units as mean ± SD (biological triplicates) for Pmk1 phosphorylation (anti-phospho-p44/42 blot) were determined with respect to the internal control (anti-Cdc2 blot). ***, p < 0.001; as calculated by unpaired Student’s t-test; (B) S. japonicus strains of the indicated genotypes were grown in YES medium to mid-log phase, treated with either 0.6 M KCl (upper panels) or 1 µg/mL caspofungin (lower panels) for the indicated times, and activated Pmk1 was detected with anti-phospho-p44/42, whereas anti-Cdc2 was used as a loading control. Results from representative experiments are shown; (C) Exponentially growing S. japonicus cells of the indicated genotypes were inoculated at an initial cell density of 106 cells/mL in YES medium with 6% glucose and incubated for 0 and 6 h with 0.2 μM CPT. Cell length at each time point is represented as box and whisker plots. Data obtained after quantification of one experiment performed per triplicate (n ≥ 400 cells/strain) is shown. ****, p < 0.0001; ns, not significant, as calculated by one-way ANOVA; (D) Representative images of strains analyzed in (C) were obtained by fluorescence microscopy after calcofluor white staining; (E) Exponentially growing S. japonicus wild-type cells (YES medium with 6% glucose) expressing a Pmk1–GFP fusion were treated with 0.2 μM CPT for the indicated times. Activated/total Pmk1 were detected with anti-phospho-p44/42 and anti-GFP antibodies, respectively. Anti-Cdc2 was used as a loading control. Relative units as mean ± SD (biological duplicates) for Pmk1 phosphorylation (anti-phospho-p44/42 blot) were determined with respect to the internal control (anti-GFP blot). ns, not significant, as calculated by unpaired Student’s t-test; (F) Left: S. japonicus strains of the indicated genotypes were grown in YES medium plus 0.2 μM CPT for 12 h, and the percentage of hyphae were quantified. Percentages are expressed as mean ± SD and correspond to biological duplicates (n ≥ 200 cells/sample). Right: Representative images of strains were obtained by fluorescence microscopy after calcofluor white staining; (G) Left: cells from log-phase cultures of the indicated strains growing in YES medium (~2.106 cells in each case) were spotted on YEMA and RGE plates, incubated at 30 °C for 7 days, and then photographed. Right: the total area of mycelial expansion (expressed as relative units, RU) was measured for each strain genotype (n ≥ 6) and is represented as scatter plot. *, p < 0.05; ****, p < 0.0001; ns, not significant, as calculated by one-way ANOVA.
3. Results
3.1. Environmental Control of S. japonicus Cell Integrity MAPK Pathway by the PKC Orthologs Pck1 and Pck2
To explore the evolutionary conservation of the cell integrity pathway within the fission yeast clade, we obtained a S. japonicus strain deleted in MAPK Pmk1 by homologous recombination (see Materials and Methods). S. pombe pmk1Δ cells display a multiseptated phenotype when cultured in medium supplemented with sorbitol, calcofluor, KCl, or caffeine [34,35,36]. While the percentage of septated and multiseptated cells was very similar in S. japonicus wild-type and pmk1Δ cells growing in the presence of sorbitol, calcofluor and caffeine, multiseptation increased significantly in pmk1Δ cells treated with KCl (Figure 1A), suggesting that Pmk1 might play a less meaningful role during control of septation in this organism. As compared to the wild-type strain, S. japonicus pmk1Δ cells were growth sensitive to the β-glucan synthase inhibitor caspofungin but to a lesser extent than the S. pombe pmk1Δ mutant (Figure 1B), indicating that Pmk1 likely regulates cell wall synthesis and/or integrity in both fission yeast species. Calcineurin and Pmk1 play antagonistic roles during control of calcium homeostasis in S. pombe, and lack of Pmk1 activity allows cell growth in the presence of MgCl2 when calcineurin activity is blocked with the specific inhibitor FK506. This phenotype, known as “VIC” (viable in the presence of immunosuppressant and chloride ion) [37], was also displayed by S. japonicus pmk1Δ cells (Figure 1B), revealing that a functional crosstalk between calcineurin and Pmk1 signaling is conserved within the Schizosaccharomyces genus.
Pmk1 MAPKs in S. pombe and S. japonicus share a high amino-acid sequence identity, particularly at the N- and C-lobes (~85% identity within a 336 amino-acid sequence), and decreases gradually towards the C-tail (Figure S2). The glycine-ATP-phosphate-binding loop, the invariant lysine and glutamic acid residues required for full kinase activity, the ERK-type TEY activation loop, and the common docking (CD) domain required for MAPK binding to upstream activators and downstream targets, are conserved in S. japonicus Pmk1 with respect to the corresponding S. pombe counterpart (Figure S2). However, a striking feature of S. japonicus Pmk1 secondary structure is the absence at its N-lobe of a 24 aa motif (sequence –ITCIYDLDIINPYNFNEVYIYEEL-), which encompasses the full subdomain IV and part of subdomain V (hinge region), and includes the putative gatekeeper residue E104 (Figure S2). A global BlastP search of yeast and fungal proteomes indicated that this unusual N-lobe conformation is highly specific of S. japonicus Pmk1, as it was found in only three hypothetical cell integrity MAPKs from ascomycetes belonging to order Chaetothyriales (Gen Pep: DV737_g786 and DV736_g4926), and the species Ophiocordyceps camponoti (Gen Pep: CP532_1836) (Figure S3).
S. pombe Pmk1 displays a nucleo-cytoplasmic localization during unperturbed growth and is targeted to the septum during cytokinesis [5]. We found that, in contrast to S. pombe, in S. japonicus a genomic Pmk1–GFP fusion shows nucleo–cytoplasmic location but it is not targeted to the septum, as confirmed after observation of septum staining with calcofluor white and DIC microscopy of yeast cells growing exponentially in rich medium, or in hyphae produced after treatment with CPT (Figure 1C). Incubation with an anti-phospho-p44/42 ERK antibody, which detects dually phosphorylated Pmk1 in S. pombe [5], revealed a single phosphorylation signal with the predicted molecular weight of Pmk1 (~47 Da) in extracts from S. japonicus wild-type cells, but not in those from the pmk1Δ mutant (Figure S2). We note that the relative basal Pmk1 phosphorylation level was significantly higher (2–2.5 times) in vegetatively growing S. japonicus versus S. pombe cells expressing genomic Pmk1–GFP fusions (Figure 1D). S. pombe Pmk1 becomes activated in response to several stimuli including saline stress, glucose starvation, and cell wall damage induced with caspofungin [3,5,7,12]. Pmk1 also became activated in S. japonicus cells treated with salt (KCl) or caspofungin, but to different magnitudes and dynamics than in S. pombe (Figure 1E). Remarkably, Pmk1 activity decreased quickly during glucose starvation (Figure 1E), suggesting that the CIP is not operative in S. japonicus under this specific stimulus. In S. pombe, transcriptional induction of tyrosine and serine/threonine phosphatases by the SAPK pathway through the Sty1–Atf1 branch promotes the dephosphorylation of activated Pmk1 during vegetative growth and in response to stress, thus revealing a cross-inhibitory mechanism between the two MAPK cascades [4]. Indeed, both basal and the induced phosphorylation of Pmk1 under saline stress increased significantly in a S. japonicus sty1Δ mutant [23], as compared to wild-type cells (Figure 1F), suggesting that the interplay between the SAPK and CIP pathways is conserved in both Schizosaccharomyces species.
The two non-essential PKC orthologs, Pck1 and Pck2 [1,38], are core upstream activators of the CIP during growth and stress in S. pombe [6,7]. S. japonicus proteome also carries two putative orthologs for Pck1 (SJAG_01787) and Pck2 (SJAG_00610), which share ~52 and ~66% amino-acids sequence identity with the respective S. pombe counterparts. Both kinases have the conserved putative HR1 rho-binding repeat domains that in S. pombe mediate their binding to Rho GTPases Rho1 and Rho2 [39], plus the C2 (Ca2+-binding), Ps (pseudosubstrate), and C1 (diacylglycerol-binding) motifs present in S. pombe Pck1 and Pck2 (Figures S4 and S5) [39]. The canonical phosphorylated residues at the kinase domain within the AL (activation loop), TM (Turn motif), and HM (hydrophobic motif), are also conserved (Figures S4 and S5) [39]. S. japonicus single mutants lacking either Pck1 or Pck2 were viable; however, we were unable to obtain a double-mutant strain lacking both kinases, suggesting that they partially complement each other and share essential functional roles for cell viability as in S. pombe [40,41]. As compared to wild-type cells, Pmk1 basal activity was strongly reduced (>80%) in S. japonicus pck2Δ cells during vegetative growth, and increased approximately by two-fold in the pck1Δ mutant (Figure 2A). Accordingly, in pck2Δ cells reduced Pmk1 activity was accompanied by a less severe VIC phenotype than that of pmk1Δ cells (Figure 1B). Thus, S. japonicus Pck2 is a major activator of the Pmk1 MAPK module during unperturbed growth. S. pombe Pmk1 activation during osmotic–saline stress with KCl is fully dependent on Pck2 function, whereas Pck1 does not play a meaningful role during this response [6]. This signaling mechanism is also conserved within the S. japonicus CIP, since Pck2 deletion completely abolished Pmk1 activation in the presence of KCl (Figure 2B). Despite showing very low MAPK levels, the growth sensitivity to caspofungin of S. japonicus pck2Δ cells was negligible as compared to that of pmk1Δ cells (Figure 1B). Notably, and in contrast to the S. pombe pck1Δ mutant, S. japonicus cells lacking Pck1 were growth resistant to this compound (Figure 1B). In S. pombe, Pck2 is a major factor responsible for Pmk1 activation during cell wall damage induced with caspofungin, while Pck1 likely plays a secondary role [7]. Instead, caspofungin-induced Pmk1 activation in S. japonicus was strictly Pck2-dependent, as Pck1 absence did not reduce MAPK activation and/or dynamics (Figure 2B). Together, the above findings suggest that respective positive and negative modulation of CIP activity by Pck2 and Pck1 has an opposite impact on S. japonicus cell wall integrity, and that MAPK activation in response to cell wall damage is transduced to the MAPK module exclusively through Pck2.
3.2. Fine-Tuning of Pmk1 Activity by Pck1 and Pck2 Modulates Antagonistically S. japonicus Hyphal Differentiation
As compared to wild-type cells, S. japonicus null mutants lacking the putative CIP MAP kinase module components Mkh1 (MAPKKK), Pek1 (MAPKK), and Pmk1 (MAPK), show defective mycelia formation when growing in a malt-extract-based solid medium (YEMA) [22], suggesting that the CIP plays a positive role to modulate yeast to hypha dimorphic transition. However, the possible role of the upstream activators of the MAP kinase module Pck1 and Pck2, and the functional relevance of this control, have not been addressed. We note that the average cell length after 6 h of incubation in rich medium supplemented with 0.2 μM CPT, which induces a quick yeast to hyphae transition, was significantly higher in S. japonicus pck1Δ cells than in wild-type cells (81.5 + 15.4 μm in pck1Δ cells versus 55.1 + 24.9 μm in wild-type cells; Figure 2C,D). On the contrary, CPT-induced differentiation was markedly impaired to a similar extent in both pck2Δ and pmk1Δ mutants (34.2 + 12.6 and 37.6 + 16.2 μm in pck2Δ and pmk1Δ cells, respectively; Figure 2C,D). Total and activated basal Pmk1 levels remained unchanged during the early stages of hyphal initiation (Figure 2E). Interestingly, the enhanced hyphal differentiation of pck1Δ cells was suppressed in the absence of Pmk1 function only partially (47.2 + 16.2 μm in pck1Δ pmk1Δ double mutant; Figure 2C,D). In line with the above findings, the pck1Δ mutant produced a higher ratio of filaments and/or hypha than the wild-type strain after 12 h of incubation with CPT, whereas this percentage became significantly reduced in pck2Δ and pmk1Δ mutants, but not in a pck1Δ pmk1Δ strain (Figure 2F).
The mycelial area of expansion of the pmk1Δ mutant, but not that of the pck2Δ strain, decreased significantly with respect to the wild type when incubated for six days in YEMA or RGE plates (Figure 2G). In contrary, mycelium production was enhanced in pck1Δ cells growing in both inducing media, and this phenotype was fully suppressed by simultaneous deletion of Pmk1 (Figure 2G). Taken together, the above findings suggest that Pmk1 plays a major positive role to control both the early and late phases of hyphal differentiation in S. japonicus, whereas the positive impact of Pck2 signaling seems restricted to the initial differentiation step. Conversely, Pck1 negatively modulates this process by a mechanism that fully depends on the reduction of Pmk1 activity during the late stages of mycelium production (plate assays), but not at the early stages of the dimorphic switch (CPT treatment).
3.3. Species-Specific N-lobe Domain Architecture Accounts for Differential Control of Pmk1 Function in S. pombe and S. japonicus during Growth, Stress, and Hyphal Differentiation
The unusual secondary structure of S. japonicus Pmk1 at its N-lobe lacking most parts of subdomains IV and V (Figure S2), together with the results obtained so far, prompted us to thoroughly explore the possible impact of MAPK conformation on the evolutionary conservation of Pmk1 functions in the two fission yeast species. To this end, we constructed S. pombe and S. japonicus pmk1Δ mutants expressing distinct genomic Pmk1–HA fused chimeras under the control of their respective wild-type promoters (named as Pmk1Sp and Pmk1Sj thereafter) (Figure 3A). In contrast to the wild-type PmkSp–HA fusion, expression in S. pombe pmk1Δ cells of a MAPK version lacking the conserved N-lobe 24 aa motif (Pmk1Sp (Δ82-105)–HA; Figure 3A), did not show detectable phosphorylation during unperturbed growth (Figure 3B), suggesting that this motif is essential for kinase activation and/or activity. Accordingly, Pmk1Sp (Δ82-105)–HA failed to suppress both the VIC and multiseptated phenotypes of S. pombe pmk1Δ cells (Figure 3C,D), and was not activated in response to saline stress (Figure 3E). Indeed, the relative basal activity of a S. japonicus Pmk1 fusion expressed in S. pombe (Pmk1Sj–HA; Figure 3A) was significantly higher than that of the endogenous MAPK (Figure 3B). Pmk1Sj–HA suppressed the VIC and multiseptated phenotypes of S. pombe pmk1Δ cells to a similar extent as Pmk1Sp–HA (Figure 3C,D), but displayed a slightly lower activation threshold in response to an osmotic-saline stress (Figure 3E). Moreover, insertion of the N-lobe 24 aa motif within S. japonicus Pmk1 (Pmk1Sj (ins82-105)–HA), resulted in a total lack of MAPK activity and functions (Figure 3B–E). Taking into account these findings, we hypothesized that the differences in biological activity among the different Pmk1 chimeras might result from changes in their binding affinity to the upstream activating MAPKK Pek1 [1,5]. Indeed, co-IP experiments revealed that the In Vivo association of Pmk1Sj–HA to a genomic Pek1–GFP fusion was enhanced as compared to that of Pmk1Sp–HA, and absent in cells expressing either Pmk1Sp(Δ82-105)–HA or Pmk1Sj-ins(82-105)–HA fusions (Figure 3F). Together, these observations indicate that Pmk1Sj is fully functional in S. pombe, and that absence of the subdomain IV–V motif within the ATP-binding pocket at the N-terminal lobe is required for MAPKK binding and functional activation of the MAPK. They also suggest that the enhanced MAPKK binding and altered activity and activation pattern of Pmk1Sj with regard to the S. pombe isoform (i.e. increased basal activity and reduced activation in response to stress), may rely upon structural constraints determined by its atypical N-terminal lobe.

Figure 3.
S japonicus Pmk1 is fully functional in S. pombe. (A) S. pombe and S. japonicus Pmk1–HA chimeric constructs. Pmk1Sp–HA: S. pombe wild-type Pmk1 construct showing the N-lobe subdomain IV–V conserved 24 aa motif (light green) is shown. Pmk1Sp (Δ82-105)–HA: S. pombe Pmk1 version lacking the conserved N-lobe conserved motif. Pmk1Sj–HA: S. japonicus wild-type Pmk1 construct that lacks the conserved N-lobe conserved motif. Pmk1Sj (ins82-105)–HA: S. japonicus Pmk1 version with an insertion of the S. pombe Pmk1 N-lobe 24 aa motif (light green); (B) S. pombe pmk1Δ cells expressing the Pmk1–HA chimeric constructs described in (A) were grown in YES medium to mid-log phase, and activated/total Pmk1 were detected with anti-phospho-p44/42 and anti-HA antibodies, respectively. Relative units as mean ± SD (biological triplicates) for Pmk1 phosphorylation (anti-phospho-p44/42 blot), were determined with respect to the anti-HA blot (total Pmk1). ***, p < 0.001; as calculated by unpaired Student’s t-test; (C) Decimal dilutions of S. pombe pmk1Δ cells (-) and those expressing the indicated Pmk1–HA chimeric constructs, were spotted onto YES plates supplemented with 0.5 µg/mL FK506 plus 0.2 M MgCl2 (VIC assay), and incubated for 3 days at 28 °C before being photographed. A representative experiment is shown; (D) S. pombe pmk1Δ cells (-) and those expressing the indicated Pmk1–HA chimeric constructs were grown for 24 h in YES medium plus 1M sorbitol, and the percentage of septated/multiseptated cells (represented as mean ± SD from biological triplicates; number of cells ≥ 200) was determined by fluorescence microscopy after calcofluor white staining; (E) S. pombe strains expressing the indicated Pmk1–HA chimeric constructs were grown in YES medium to mid-log phase and treated with 0.6 M KCl for the indicated times. Activated/total Pmk1 were detected with anti-phospho-p44/42 and anti-HA antibodies, respectively. Relative units as mean ± SD (biological duplicates) for Pmk1 phosphorylation (anti-phospho-p44/42 blot) were determined with respect to the anti-HA blot (total Pmk1). Either representative results or those obtained from biological triplicates are shown; (F) Co-immunoprecipitation of Pmk1–HA chimeric constructs and a Pek1–GFP (MAPKK) genomic fusion from yeast extracts obtained from vegetatively growing cultures from the indicated strains. Results from a representative experiment are shown. IP, immunoprecipitation; WB, Western blot; *, specific Pek1-GFP fusion.
Insertion of the S. pombe 24 aa motif within S. japonicus Pmk1 (Pmk1Sj (ins82-105)–HA), resulted in a clear reduction in basal MAPK activity as compared to the wild-type Pmk1Sj–HA fusion when expressed in a S. japonicus pmk1Δ background (Figure 4A). In contrary, MAPK activation during osmotic–saline stress remained unaffected (Figure 4B). However, Pmk1Sj(ins82-105)–HA did not abolish the VIC phenotype of S. japonicus pmk1Δ cells as the Pmk1Sj–HA version did (Figure 4C). Interestingly, a genomic Pmk1Sp–HA chimera expressed in S. japonicus showed a relative basal MAPK activity comparable to that of the endogenous Pmk1Sj–HA construct (Figure 4A), and became activated to a similar threshold in response to KCl stress (Figure 4D). Nonetheless, Pmk1Sp was not fully functional in S. japonicus, because it suppressed the VIC phenotype of the pmk1Δ mutant only partially (Figure 4B). Finally, deletion of the Pmk1Sp N-lobe motif (Pmk1Sp (Δ82-105)–HA) resulted in a biologically inactive and non-functional MAPK chimera (Figure 4A–C).
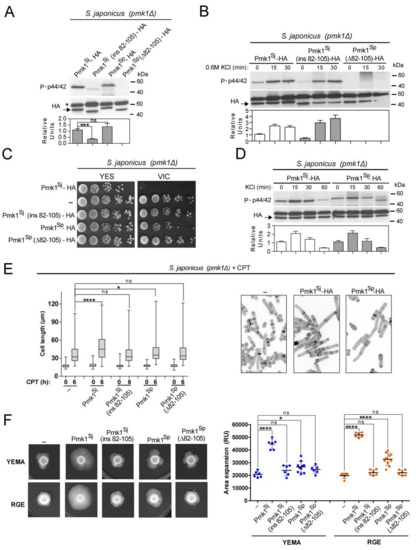
Figure 4.
S. pombe Pmk1 shows little ability to execute CIP functions and to promote mycelial development in S. japonicus. (A) S. japonicus pmk1Δ cells expressing the Pmk1–HA chimeric constructs described in Figure 3A were grown in YES medium to mid-log phase, and activated/total Pmk1 were detected with anti-phospho-p44/42 and anti-HA antibodies, respectively. Relative units as mean ± SD (biological triplicates) for Pmk1 phosphorylation (anti-phospho-p44/42 blot) were determined with respect to the anti-HA blot (total Pmk1). ***, p < 0.001; ns, not significant, as calculated by unpaired Student’s t-test; (B) S. japonicus pmk1Δ strains expressing the Pmk1–HA chimeric constructs were grown in YES medium to mid-log phase and treated with 0.6 M KCl for the indicated times. Activated/total Pmk1 were detected with anti-phospho-p44/42 and anti-HA antibodies, respectively. Relative units as mean ± SD (biological duplicates) for Pmk1 phosphorylation (anti-phospho-p44/42 blot) were determined with respect to the anti-HA blot (total Pmk1); (C) Decimal dilutions of S. japonicus pmk1Δ cells (-), and those expressing Pmk1–HA chimeric constructs, were spotted onto YES plates supplemented with 0.5 µg/mL FK506 plus 0.2 M MgCl2 (VIC), and incubated for 3 days at 30 °C before being photographed. A representative experiment is shown; (D) S. japonicus pmk1Δ strains expressing the Pmk1–HA chimeric constructs were grown in YES medium to mid-log phase and treated with 0.6 M KCl for the indicated times. Activated/total Pmk1 were detected with anti-phospho-p44/42 and anti-HA antibodies, respectively. Relative units as mean ± SD (biological duplicates) for Pmk1 phosphorylation (anti-phospho-p44/42 blot) were determined with respect to the anti-HA blot (total Pmk1); (E) Left: Exponentially growing S. japonicus pmk1Δ cells (-), and those expressing the indicated Pmk1–HA chimeric constructs were inoculated at an initial cell density of 106 cells/mL in YES medium with 6% glucose and incubated for 0 and 6 h with 0.2 μM CPT. Cell length at each time point is represented as box and whisker plots. Data obtained after quantification of one experiment performed per triplicate (n ≥ 400 cells/strain) are shown. *, p < 0.05; ****, p < 0.0001; ns, not significant, as calculated by one-way ANOVA. Right: representative images were obtained by fluorescence microscopy after calcofluor white staining; (F) Left: cells from log-phase cultures of S. japonicus pmk1Δ cells (-), and those expressing Pmk1–HA chimeric constructs growing in YES medium (~2.106 total cells), were spotted on YEMA and RGE plates, incubated at 30 °C for 7 days, and then photographed. Right: the total area of mycelial expansion (expressed as relative units) was measured for each strain genotype (n ≥ 6) and is represented as a scatter plot. *, p < 0.05; ****, p < 0.0001; ns, not significant, as calculated by one-way ANOVA.
As compared to endogenous Pmk1Sj–HA, expression of Pmk1Sp–HA showed a limited ability to promote filamentation in S. japonicus pmk1Δ cells during the initial steps of yeast to hypha differentiation induced with CPT (Figure 4E), and also at later stages, as confirmed by comparatively scoring the mycelial area of expansion of strains expressing both constructs growing on YEMA and RGE plates (Figure 4F). As might be expected, neither Pmk1Sj (ins82-105)–HA nor Pmk1Sp (Δ82-105)–HA fusions were able to alleviate the myceliation defect of the pmk1Δ mutant (Figure 4F). Therefore, these observations suggest that in S. japonicus, evolutionary divergence of MAPK Pmk1 secondary structure at the N-lobe has had a significant impact on its biological functions including the positive modulation of dimorphism.
4. Discussion
In this work, we have investigated the signaling architecture and biological functions of the cell integrity MAPK signaling pathway (CIP) in the dimorphic fission yeast S. japonicus. Comparative phenotypic assessment of S. japonicus and S. pombe pmk1Δ mutants revealed that control of ionic homeostasis and cell wall integrity is a conserved regulatory feature of the CIP in both fission yeast species. In contrast to S. pombe, Pmk1 did not localize to the septum in S. japonicus, and pmk1Δ cells only displayed cytokinetic defects in response to specific environmental cues (i.e. with KCl), suggesting that the CIP may not play a similar role during control of septation in both fission yeasts [1,34]. Pmk1-dependent regulation of cell separation in S. pombe operates in a manner mostly dependent on its localization at the division area; however, transcriptional control by nuclear Pmk1 has also been proposed to play some role during cytokinetic control [34]. Moreover, it has been recently shown that CIP/Pmk1 signaling participates in a checkpoint that relies on the SIN (septation initiation network), which ensures that S. pombe cytokinesis terminates successfully [35]. This regulatory mechanism could not have evolved in S. japonicus, since both fission species show marked differences in the timely coordination of mitosis and cytokinesis, with S. japonicus delaying the assembly of the CAR until late anaphase when chromosome segregation is complete [42]. Hence, it might be possible that Pmk1-dependent regulation of cytokinesis in S. japonicus is exerted at a transcriptional level.
S. japonicus Pmk1 becomes activated as with its S. pombe MAPK counterpart in response to environmental cues including saline and cell wall stress, and is negatively regulated by the SAPK pathway, as initially described in S. pombe [4]. However, unlike S. pombe Pmk1, which becomes strongly activated upon glucose withdrawal [3], Pmk1 activity undergoes a sudden decrease in S. japonicus cells after carbon source removal from the growth medium. Pmk1 activation in the absence of glucose is required in S. pombe for adaptive growth from fermentative to respiratory metabolism, and the use of alternative carbon sources such as glycerol [3]. Lack of Pmk1 activation in S. japonicus might thus be explained from an evolutionary perspective, since this fission yeast species is respiration defective, and utilizes glucose exclusively via fermentation [17].
The role of PKC orthologs Pck1 and Pck2 as key upstream regulators of the CIP pathway is maintained in S. japonicus as originally shown in S. pombe [1,6,7,39], although with several differences. As in S. pombe, S. japonicus Pck2 is the major activator of Pmk1 during unperturbed growth, whereas Pck1 deletion prompts increased Pmk1 phosphorylation. In S. pombe, this response has been shown to arise from a cell wall stress that is transduced to the CIP by Rho GTPases Rho1 and Rho2 [7], and whose existence in S. japonicus remains unknown. However, Pmk1 activation during cell wall damage, which in S. pombe relies on both Pck1 and Pck2, is in S. japonicus exclusively Pck2 dependent. These findings, together with the observation that pck1Δ cells are growth resistant to caspofungin, suggest that opposite control of CIP/Pmk1 activity by Pck1 and Pck2 has a meaningful impact on S. japonicus cell wall structure and integrity.
S. japonicus yeast cells readily differentiate to hyphae in response to different environmental cues [18,19,20,21]. This led us to thoroughly study the putative role of CIP signaling during regulation of the dimorphic transition in this organism. As proposed earlier [22], Pmk1 MAPK is an essential and positive modulator of both the early and late stages of S. japonicus yeast to hyphae differentiation. This role is similar to that played by cell integrity MAPKs during hyphal or pseudohyphal differentiation in other yeast species, including Candida albicans [43,44], Cryptococcus neoformans [45], and Saccharomyces cerevisiae [46]. However, our data also suggest that Pck1 and Pck2 may regulate CIP signaling during S. japonicus dimorphic switch in an intricate fashion. Deletion of Pck2, which strongly reduces Pmk1 basal activity, had a notoriously negative impact during the early steps of yeast to hyphae differentiation, and was very similar to that imposed by Pmk1 deletion. This suggests that the cellular requirement for basal CIP/Pmk1 signaling is maximal during the early stages of S. japonicus hyphal differentiation. On the contrary, Pck2 absence did not affect late mycelia expansion, indicating that only a minimal CIP activity threshold is necessary at the later stages of mycelia formation. On the other hand, Pck1 deletion, which increases basal Pmk1 activity, resulted both in increased hyphal elongation and myceliation during the whole differentiation process. Intriguingly, Pck1 negative regulation of S. japonicus yeast to hyphae switch is totally dependent on Pmk1 downregulation during late mycelial expansion, but appears to operate independently of the MAPK at the early stages of yeast differentiation. Altogether, these findings point to the existence of an antagonistic and biologically relevant control of S. japonicus hyphal differentiation by Pck1 and Pck2, which operates by fine-tuning of Pmk1 activity during the different stages of mycelia formation, but may also function independently of CIP signaling.
MAPKs adopt a typical kinase fold including N- and C-terminal lobes that mediate, respectively, ATP and effector/substrate binding, and are connected via a conserved hinge region at subdomain V [47]. Activation loop phosphorylation by the upstream MAPKKs results in MAPKs rotation of the N- and C-terminal lobes through this hinge region to facilitate phosphoryl transfer, and also triggers global changes in their conformational exchange dynamics [47]. Therefore, the atypical secondary structure of the N-lobe in S. japonicus Pmk1, which lacks a highly conserved motif corresponding to subdomain IV and part of the hinge region including the putative gatekeeper residue, raised the question of to what extent has evolutionary divergence modeled the biological functions of this MAPK? By performing kinase swapping experiments, we found that Pmk1Sj is totally functional when expressed in S. pombe, as evidenced by its ability to become fully activated during growth and stress, and to suppress the altered ionic homeostasis and defective cytokinesis of S. pombe pmk1Δ cells. On the contrary, the ability of Pmk1Sp to relieve the VIC phenotype and the hyphal differentiation defect of S. japonicus pmk1Δ cells was minimal. This behavior does not result from limited binding and activation of Pmk1Sp by the MAPKK, since its basal activity and activation pattern in response to stress were very similar to those shown by Pmk1Sj. Thus, with its atypical structure, Pmk1Sj seems more efficient for downstream substrate and/or effector docking than Pmk1Sp. Significantly, Pmk1Sj conformational change, elicited by insertion of the highly conserved Pmk1Sp N-lobe motif, resulted in a total lack of kinase function when expressed in S. pombe due to abrogated binding and activation by Pek1 MAPKK, but also in S. japonicus, despite becoming activated during growth and stress to a certain threshold. These results suggest that evolutionary divergence within the Schizosaccharomyces clade in the architecture of the Pmk1 N-terminal lobe has exerted a profound impact on MAPK binding/docking to both upstream (MAPKK) and downstream substrates/effectors. In support of this prediction, bioinformatics and structural studies with mammalian MAPKs have shown that co-evolution between the subdomains IV–V, the D-domain docking site, and the C-tail, play a major role for tight and allosteric regulation of MAPK activity [48].
In conclusion, while some of the functional roles of the CIP earlier described in S. pombe are conserved in S. japonicus, others have evolved separately in both fission yeast species. This includes the apparent inability of S. japonicus Pmk1 to become activated in response to glucose deprivation and the differential regulation of cytokinesis. Our findings also reveal an antagonistic role of PKC orthologs Pck1 and Pck2 in controlling MAPK basal activity during the regulation of yeast to hyphae dimorphic switch, and suggest that conformational changes in the N-lobe of S. japonicus Pmk1 have modeled CIP signaling to fulfill the specific developmental requirements of this fission yeast species.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/jof7060482/s1; Figure S1: Septation phenotypes of S. japonicus pmk1Δ cells; Figure S2: Conserved structural features of Pmk1 MAPKs in S. pombe and S. japonicus; Figure S3: Conserved sequences surrounding the N-lobe subdomains IV and V in several fungal CIP MAPKs; Figure S4: Conserved structural features of Pck1 orthologs in S. pombe and S. japonicus; Figure S5: Conserved structural features of Pck2 orthologs in S. pombe and S. japonicus; Table S1: S. japonicus and S. pombe strains used in this work; Table S2: Oligonucleotides used in this work.
Author Contributions
Conceptualization, T.S.; Data curation, E.G.-G., A.F., J.V.-S., and J.C.; Formal analysis, E.G.-G., A.F., F.P.-R., A.P.-D., M.M., T.S., and J.C.; Funding acquisition, B.V.-M., T.S., and J.C.; Investigation, E.G.-G., B.V.-M., F.P.-R., A.P.-D., J.V.-S., M.M., T.S., and J.C.; Methodology, E.G.-G., A.F., B.V.-M., A.P.-D., J.V.-S., M.M., and T.S.; Project administration, F.P.-R., J.V.-S., T.S., and J.C.; Resources, A.F. and J.C.; Supervision, T.S. and J.C.; Validation, E.G.-G., M.M., T.S., and J.C.; Visualization, E.G.-G.; Writing—original draft, J.C.; Writing—review and editing, E.G.-G., A.F., and T.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Agencia Estatal de Investigación and Ministerio de Ciencia, Innovación y Universidades, Spain, grant number BFU2017-82423-P, Fundación Séneca de la Región de Murcia, Spain, grant number 20856/PI/18, and European Regional Development Fund (ERDF) co-funding from the European Union.
Data Availability Statement
The data presented in this study are available on request from the corresponding authors.
Acknowledgments
We thank Snezhana Oliferenko and Hironori Niki for S. japonicus yeast strains and plasmids. Elisa Gómez-Gil and Francisco Prieto-Ruiz are, respectively, Formación de Personal Investigador (FPI) and Formación de Profesorado Universitario (FPU) PhD fellows from the Universidad de Murcia and Ministerio de Educación y Formación Profesional, Spain.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Perez, P.; Cansado, J. Cell Integrity Signaling and Response to Stress in Fission Yeast. Curr. Protein Pept. Sci. 2010, 11, 680–692. [Google Scholar] [CrossRef] [PubMed]
- Kabeche, R.; Madrid, M.; Cansado, J.; Moseley, J.B. Eisosomes Regulate Phosphatidylinositol 4,5-Bisphosphate (PI(4,5)P2) Cortical Clusters and Mitogen-activated Protein (MAP) Kinase Signaling upon Osmotic Stress. J. Biol. Chem. 2015, 290, 25960–25973. [Google Scholar] [CrossRef] [PubMed]
- Madrid, M.; Fernandez-Zapata, J.; Sanchez-Mir, L.; Soto, T.; Franco, A.; Vicente-Soler, J.; Gacto, M.; Cansado, J. Role of the fission yeast cell integrity MAPK pathway in response to glucose limitation. BMC Microbiol. 2013, 13. [Google Scholar] [CrossRef] [PubMed]
- Madrid, M.; Nunez, A.; Soto, T.; Vicente-Soler, J.; Gacto, M.; Cansado, J. Stress-activated protein kinase-mediated down-regulation of the cell integrity pathway mitogen-activated protein kinase Pmk1p by protein phosphatases. Mol. Biol. Cell 2007, 18, 4405–4419. [Google Scholar] [CrossRef]
- Madrid, M.; Soto, T.; Khong, H.; Franco, A.; Vicente, J.; Perez, P.; Gacto, M.; Cansado, J. Stress-induced response, localization, and regulation of the Pmk1 cell integrity pathway in Schizosaccharomyces pombe. J. Biol. Chem. 2006, 281, 2033–2043. [Google Scholar] [CrossRef]
- Barba, G.; Soto, T.; Madrid, M.; Nunez, A.; Vicente, J.; Gacto, M.; Cansado, J.; Grp, Y.P.; Grp, Y.P. Activation of the cell integrity pathway is channelled through diverse signalling elements in fission yeast. Cell. Signal. 2008, 20, 748–757. [Google Scholar] [CrossRef]
- Sanchez-Mir, L.; Soto, T.; Franco, A.; Madrid, M.; Viana, R.; Vicente, J.; Gacto, M.; Perez, P.; Cansado, J. Rho1 GTPase and PKC Ortholog Pck1 Are Upstream Activators of the Cell Integrity MAPK Pathway in Fission Yeast. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Nunez, A.; Franco, A.; Madrid, M.; Soto, T.; Vicente, J.; Gacto, M.; Cansado, J. Role for RACK1 Orthologue Cpc2 in the Modulation of Stress Response in Fission Yeast. Mol. Biol. Cell 2009, 20, 3996–4009. [Google Scholar] [CrossRef]
- Soto, T.; Villar-Tajadura, M.; Madrid, M.; Vicente, J.; Gacto, M.; Perez, P.; Cansado, J. Rga4 Modulates the Activity of the Fission Yeast Cell Integrity MAPK Pathway by Acting as a Rho2 GTPase-activating Protein. J. Biol. Chem. 2010, 285, 11516–11525. [Google Scholar] [CrossRef]
- Villar-Tajadura, M.; Coll, P.; Madrid, M.; Cansado, J.; Santos, B.; Perez, P. Rga2 is a Rho2 GAP that regulates morphogenesis and cell integrity in S-pombe. Mol. Microbiol. 2008, 70, 867–881. [Google Scholar] [CrossRef]
- Sanchez-Mir, L.; Franco, A.; Martin-Garcia, R.; Madrid, M.; Vicente-Soler, J.; Soto, T.; Gacto, M.; Perez, P.; Cansado, J. Rho2 palmitoylation is required for plasma membrane localization and proper signaling to the fission yeast cell integrity MAPK pathway. Mol. Cell. Biol. 2014. [Google Scholar] [CrossRef]
- Garcia, P.; Tajadura, V.; Sanchez, Y. The Rho1p exchange factor Rgf1p signals upstream from the Pmk1 mitogen-activated protein kinase pathway in fission yeast. Mol. Biol. Cell 2009, 20, 721–731. [Google Scholar] [CrossRef][Green Version]
- Madrid, M.; Vazquez-Marin, B.; Franco, A.; Soto, T.; Vicente-Soler, J.; Gacto, M.; Cansado, J. Multiple crosstalk between TOR and the cell integrity MAPK signaling pathway in fission yeast. Sci. Rep. 2016, 6, 37515. [Google Scholar] [CrossRef]
- Prieto-Ruiz, F.; Vicente-Soler, J.; Franco, A.; Gomez-Gil, E.; Sanchez-Marinas, M.; Vazquez-Marin, B.; Aligue, R.; Madrid, M.; Moreno, S.; Soto, T.; et al. RNA-Binding Protein Rnc1 Regulates Cell Length at Division and Acute Stress Response in Fission Yeast through Negative Feedback Modulation of the Stress-Activated Mitogen-Activated Protein Kinase Pathway. mBio 2020, 11. [Google Scholar] [CrossRef]
- Makarova, M.; Peter, M.; Balogh, G.; Glatz, A.; MacRae, J.I.; Lopez Mora, N.; Booth, P.; Makeyev, E.; Vigh, L.; Oliferenko, S. Delineating the Rules for Structural Adaptation of Membrane-Associated Proteins to Evolutionary Changes in Membrane Lipidome. Curr. Biol. 2020, 30, 367–380.e368. [Google Scholar] [CrossRef]
- Gu, Y.; Yam, C.; Oliferenko, S. Divergence of mitotic strategies in fission yeasts. Nucleus 2012, 3, 220–225. [Google Scholar] [CrossRef]
- Kaino, T.; Tonoko, K.; Mochizuki, S.; Takashima, Y.; Kawamukai, M. Schizosaccharomyces japonicus has low levels of CoQ10 synthesis, respiration deficiency, and efficient ethanol production. Biosci. Biotechnol. Biochem. 2018, 82, 1031–1042. [Google Scholar] [CrossRef]
- Sipiczki, M.; Takeo, K.; Yamaguchi, M.; Yoshida, S.; Miklos, I. Environmentally controlled dimorphic cycle in a fission yeast. Microbiology 1998, 144 Pt 5, 1319–1330. [Google Scholar] [CrossRef]
- Papp, L.; Sipiczki, M.; Holb, I.J.; Miklos, I. Optimal conditions for mycelial growth of Schizosaccharomyces japonicus cells in liquid medium: It enables the molecular investigation of dimorphism. Yeast 2014, 31, 475–482. [Google Scholar] [CrossRef]
- Aoki, K.; Furuya, K.; Niki, H. Schizosaccharomyces japonicus: A Distinct Dimorphic Yeast among the Fission Yeasts. Cold Spring Harb. Protoc. 2017, 2017, 082651. [Google Scholar] [CrossRef]
- Niki, H. Induction of Hyphal Growth in Schizosaccharomyces japonicus. Cold Spring Harb. Protoc. 2017, 2017, 091868. [Google Scholar] [CrossRef]
- Nozaki, S.; Furuya, K.; Niki, H. The Ras1-Cdc42 pathway is involved in hyphal development of Schizosaccharomyces japonicus. FEMS Yeast Res. 2018, 18. [Google Scholar] [CrossRef]
- Gomez-Gil, E.; Franco, A.; Madrid, M.; Vazquez-Marin, B.; Gacto, M.; Fernandez-Breis, J.; Vicente-Soler, J.; Soto, T.; Cansado, J. Quorum sensing and stress-activated MAPK signaling repress yeast to hypha transition in the fission yeast Schizosaccharomyces japonicus. PLoS Genet. 2019, 15, e1008192. [Google Scholar] [CrossRef]
- Moreno, S.; Klar, A.; Nurse, P. Molecular Genetic-Analysis Of Fission Yeast Schizosaccharomyces pombe. Methods Enzymol. 1991, 194, 795–823. [Google Scholar]
- Petersen, J.; Russell, P. Growth and the Environment of Schizosaccharomyces pombe. Cold Spring Harb. Protoc. 2016, 2016, 079764. [Google Scholar] [CrossRef]
- Kinnaer, C.; Dudin, O.; Martin, S.G. Yeast-to-hypha transition of Schizosaccharomyces japonicus in response to environmental stimuli. Mol. Biol. Cell 2019, 30, 975–991. [Google Scholar] [CrossRef]
- Aoki, K.; Niki, H. Transformation of Schizosaccharomyces japonicus. Cold Spring Harb. Protoc. 2017, 2017, 091850. [Google Scholar] [CrossRef]
- Furuya, K.; Niki, H. Isolation of heterothallic haploid and auxotrophic mutants of Schizosaccharomyces japonicus. Yeast 2009, 26, 221–233. [Google Scholar] [CrossRef]
- Hentges, P.; Van Driessche, B.; Tafforeau, L.; Vandenhaute, J.; Carr, A.M. Three novel antibiotic marker cassettes for gene disruption and marker switching in Schizosaccharomyces pombe. Yeast 2005, 22, 1013–1019. [Google Scholar] [CrossRef]
- Krawchuk, M.D.; Wahls, W.P. High-efficiency gene targeting in Schizosaccharomyces pombe using a modular, PCR-based approach with long tracts of flanking homology. Yeast 1999, 15, 1419–1427. [Google Scholar] [CrossRef]
- Madrid, M.; Jimenez, R.; Sanchez-Mir, L.; Soto, T.; Franco, A.; Vicente-Soler, J.; Gacto, M.; Perez, P.; Cansado, J. Multiple layers of regulation influence cell integrity control by the PKC ortholog Pck2 in fission yeast. J. Cell. Sci. 2015, 128, 266–280. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.; Rasband, W.; Eliceiri, K. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Hagan, I.M. Immunofluorescence Microscopy of Schizosaccharomyces pombe Using Chemical Fixation. Cold Spring Harb. Protoc. 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Mir, L.; Franco, A.; Madrid, M.; Vicente-Soler, J.; Antonia Villar-Tajadura, M.; Soto, T.; Perez, P.; Gacto, M.; Cansado, J. Biological Significance of Nuclear Localization of Mitogen-activated Protein Kinase Pmk1 in Fission Yeast. J. Biol. Chem. 2012, 287, 26038–26051. [Google Scholar] [CrossRef]
- Edreira, T.; Celador, R.; Manjón, E.; Sánchez, Y. A novel checkpoint pathway controls actomyosin ring constriction trigger in fission yeast. eLife 2020, 9. [Google Scholar] [CrossRef]
- Zaitsevskaya-Carter, T.; Cooper, J.A. Spm1, a stress-activated MAP kinase that regulates morphogenesis in S.pombe. EMBO J. 1997, 16, 1318–1331. [Google Scholar] [CrossRef]
- Sugiura, R.; Toda, T.; Dhut, S.; Shuntoh, H.; Kuno, T. The MAPK kinase Pek1 acts as a phosphorylation-dependent molecular switch. Nature 1999, 399, 479–483. [Google Scholar] [CrossRef]
- Toda, T.; Dhut, S.; SupertiFurga, G.; Gotoh, Y.; Nishida, E.; Sugiura, R.; Kuno, T. The fission yeast pmk1(+) gene encodes a novel mitogen-activated protein kinase homolog which regulates cell integrity and functions coordinately with the protein kinase C pathway. Mol. Cell. Biol. 1996, 16, 6752–6764. [Google Scholar] [CrossRef]
- Madrid, M.; Vazquez-Marin, B.; Soto, T.; Franco, A.; Gomez-Gil, E.; Vicente-Soler, J.; Gacto, M.; Perez, P.; Cansado, J. Differential functional regulation of PKC orthologs in fission yeast. J. Biol. Chem. 2017. [Google Scholar] [CrossRef]
- Arellano, M.; Valdivieso, M.; Calonge, T.; Coll, P.; Duran, A.; Perez, P. Schizosaccharomyces pombe protein kinase C homologues, pck1p and pck2p, are targets of rho1p and rho2p and differentially regulate cell integrity. J. Cell Sci. 1999, 112, 3569–3578. [Google Scholar] [CrossRef]
- Calonge, T.; Nakano, K.; Arellano, M.; Arai, R.; Katayama, S.; Toda, T.; Mabuchi, I.; Perez, P. Schizosaccharomyces pombe Rho2p GTPase regulates cell wall alpha-glucan biosynthesis through the protein kinase Pck2p. Mol. Biol. Cell 2000, 11, 4393–4401. [Google Scholar] [CrossRef]
- Gu, Y.; Oliferenko, S. Comparative biology of cell division in the fission yeast clade. Curr. Opin. Microbiol. 2015, 28, 18–25. [Google Scholar] [CrossRef]
- Kumamoto, C.A. A contact-activated kinase signals Candida albicans invasive growth and biofilm development. Proc. Natl. Acad. Sci. USA 2005, 102, 5576–5581. [Google Scholar] [CrossRef]
- Monge, R.A.; Román, E.; Nombela, C.; Pla, J. The MAP kinase signal transduction network in Candida albicans. Microbiology 2006, 152, 905–912. [Google Scholar] [CrossRef]
- Kraus, P.R.; Fox, D.S.; Cox, G.M.; Heitman, J. The Cryptococcus neoformans MAP kinase Mpk1 regulates cell integrity in response to antifungal drugs and loss of calcineurin function. Mol. Microbiol. 2003, 48, 1377–1387. [Google Scholar] [CrossRef]
- Birkaya, B.; Maddi, A.; Joshi, J.; Free, S.J.; Cullen, P.J. Role of the cell wall integrity and filamentous growth mitogen-activated protein kinase pathways in cell wall remodeling during filamentous growth. Eukaryot. Cell 2009, 8, 1118–1133. [Google Scholar] [CrossRef]
- Kumar, G.S.; Clarkson, M.W.; Kunze, M.B.A.; Granata, D.; Wand, A.J.; Lindorff-Larsen, K.; Page, R.; Peti, W. Dynamic activation and regulation of the mitogen-activated protein kinase p38. Proc. Natl. Acad. Sci. USA 2018, 115, 4655–4660. [Google Scholar] [CrossRef]
- Nguyen, T.; Ruan, Z.; Oruganty, K.; Kannan, N. Co-conserved MAPK features couple D-domain docking groove to distal allosteric sites via the C-terminal flanking tail. PLoS ONE 2015, 10, e0119636. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).